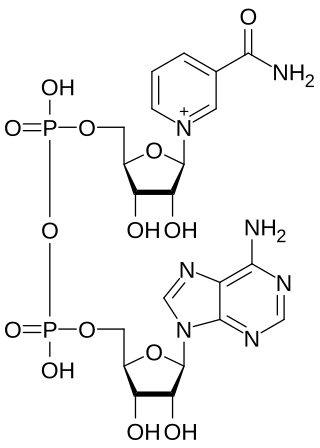
Nicotinamide adenine dinucleotide (NAD) is a coenzyme central to metabolism. Found in all living cells, NAD is called a dinucleotide because it consists of two nucleotides joined through their phosphate groups. One nucleotide contains an adenine nucleobase and the other, nicotinamide. NAD exists in two forms: an oxidized and reduced form, abbreviated as NAD+ and NADH (H for hydrogen), respectively.
Histone deacetylases (EC 3.5.1.98, HDAC) are a class of enzymes that remove acetyl groups (O=C-CH3) from an ε-N-acetyl lysine amino acid on both histone and non-histone proteins. HDACs allow histones to wrap the DNA more tightly. This is important because DNA is wrapped around histones, and DNA expression is regulated by acetylation and de-acetylation. HDAC's action is opposite to that of histone acetyltransferase. HDAC proteins are now also called lysine deacetylases (KDAC), to describe their function rather than their target, which also includes non-histone proteins. In general, they suppress gene expression.
Leonard Pershing Guarente is an American biologist best known for his research on life span extension in the budding yeast Saccharomyces cerevisiae, roundworms, and mice. He is a Novartis Professor of Biology at the Massachusetts Institute of Technology.
Histone deacetylase inhibitors are chemical compounds that inhibit histone deacetylases. Since deacetylation of histones produces transcriptionally silenced heterochromatin, HDIs can render chromatin more transcriptionally active and induce epigenomic changes.
Chromatin remodeling is the dynamic modification of chromatin architecture to allow access of condensed genomic DNA to the regulatory transcription machinery proteins, and thereby control gene expression. Such remodeling is principally carried out by 1) covalent histone modifications by specific enzymes, e.g., histone acetyltransferases (HATs), deacetylases, methyltransferases, and kinases, and 2) ATP-dependent chromatin remodeling complexes which either move, eject or restructure nucleosomes. Besides actively regulating gene expression, dynamic remodeling of chromatin imparts an epigenetic regulatory role in several key biological processes, egg cells DNA replication and repair; apoptosis; chromosome segregation as well as development and pluripotency. Aberrations in chromatin remodeling proteins are found to be associated with human diseases, including cancer. Targeting chromatin remodeling pathways is currently evolving as a major therapeutic strategy in the treatment of several cancers.

ADP-ribosylation is the addition of one or more ADP-ribose moieties to a protein. It is a reversible post-translational modification that is involved in many cellular processes, including cell signaling, DNA repair, gene regulation and apoptosis. Improper ADP-ribosylation has been implicated in some forms of cancer. It is also the basis for the toxicity of bacterial compounds such as cholera toxin, diphtheria toxin, and others.

Nicotinamide phosphoribosyltransferase, formerly known as pre-B-cell colony-enhancing factor 1 (PBEF1) or visfatin for its extracellular form (eNAMPT), is an enzyme that in humans is encoded by the NAMPT gene. The intracellular form of this protein (iNAMPT) is the rate-limiting enzyme in the nicotinamide adenine dinucleotide (NAD+) salvage pathway that converts nicotinamide to nicotinamide mononucleotide (NMN) which is responsible for most of the NAD+ formation in mammals. iNAMPT can also catalyze the synthesis of NMN from phosphoribosyl pyrophosphate (PRPP) when ATP is present. eNAMPT has been reported to be a cytokine (PBEF) that activates TLR4, that promotes B cell maturation, and that inhibits neutrophil apoptosis.
Sirtuin 1, also known as NAD-dependent deacetylase sirtuin-1, is a protein that in humans is encoded by the SIRT1 gene.

NAD-dependent deacetylase sirtuin 2 is an enzyme that in humans is encoded by the SIRT2 gene. SIRT2 is an NAD+ -dependent deacetylase. Studies of this protein have often been divergent, highlighting the dependence of pleiotropic effects of SIRT2 on cellular context. The natural polyphenol resveratrol is known to exert opposite actions on neural cells according to their normal or cancerous status. Similar to other sirtuin family members, SIRT2 displays a ubiquitous distribution. SIRT2 is expressed in a wide range of tissues and organs and has been detected particularly in metabolically relevant tissues, including the brain, muscle, liver, testes, pancreas, kidney, and adipose tissue of mice. Of note, SIRT2 expression is much higher in the brain than all other organs studied, particularly in the cortex, striatum, hippocampus, and spinal cord.

Nicotinamide N-methyltransferase (NNMT) is an enzyme that in humans is encoded by the NNMT gene. NNMT catalyzes the methylation of nicotinamide and similar compounds using the methyl donor S-adenosyl methionine (SAM-e) to produce S-adenosyl-L-homocysteine (SAH) and 1-methylnicotinamide.

NAD-dependent deacetylase sirtuin-3, mitochondrial also known as SIRT3 is a protein that in humans is encoded by the SIRT3 gene [sirtuin 3 ]. SIRT3 is member of the mammalian sirtuin family of proteins, which are homologs to the yeast Sir2 protein. SIRT3 exhibits NAD+-dependent deacetylase activity.

Sirtuin 5 , also known as SIRT5 is a protein which in humans in encoded by the SIRT5 gene and in other species by the orthologous Sirt5 gene.
NAD-dependent deacetylase sirtuin 7 is an enzyme that in humans is encoded by the SIRT7 gene. SIRT7 is member of the mammalian sirtuin family of proteins, which are homologs to the yeast Sir2 protein.
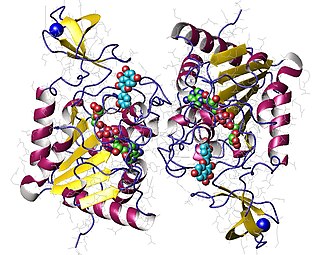
Sirtuin 6 is a stress responsive protein deacetylase and mono-ADP ribosyltransferase enzyme encoded by the SIRT6 gene. In laboratory research, SIRT6 appears to function in multiple molecular pathways related to aging, including DNA repair, telomere maintenance, glycolysis and inflammation. SIRT6 is member of the mammalian sirtuin family of proteins, which are homologs to the yeast Sir2 protein.

Sirtuin 4, also known as SIRT4, is a mitochondrial protein which in humans is encoded by the SIRT4 gene. SIRT4 is member of the mammalian sirtuin family of proteins, which are homologs to the yeast Sir2 protein. SIRT4 exhibits NAD+-dependent deacetylase activity.
Sirtuin-activating compounds (STAC) are chemical compounds having an effect on sirtuins, a group of enzymes that use NAD+ to remove acetyl groups from proteins. They are caloric restriction mimetic compounds that may be helpful in treating various aging-related diseases.

CobB is a bacterial protein that belongs to the sirtuin family, a broadly conserved family of NAD+-dependent protein deacetylases.

Genetics of aging is generally concerned with life extension associated with genetic alterations, rather than with accelerated aging diseases leading to reduction in lifespan.
SilentInformationRegulator (SIR) proteins are involved in regulating gene expression. SIR proteins organize heterochromatin near telomeres, ribosomal DNA (rDNA), and at silent loci including hidden mating type loci in yeast. The SIR family of genes encodes catalytic and non-catalytic proteins that are involved in de-acetylation of histone tails and the subsequent condensation of chromatin around a SIR protein scaffold. Some SIR family members are conserved from yeast to humans.

MicroRNA 34a (miR-34a) is a microRNA that in humans is encoded by the MIR34A gene.
















