
In biology, histones are highly basic proteins abundant in lysine and arginine residues that are found in eukaryotic cell nuclei. They act as spools around which DNA winds to create structural units called nucleosomes. Nucleosomes in turn are wrapped into 30-nanometer fibers that form tightly packed chromatin. Histones prevent DNA from becoming tangled and protect it from DNA damage. In addition, histones play important roles in gene regulation and DNA replication. Without histones, unwound DNA in chromosomes would be very long. For example, each human cell has about 1.8 meters of DNA if completely stretched out, however when wound about histones, this length is reduced to about 90 micrometers (0.09 mm) of 30 nm diameter chromatin fibers.
Heterochromatin is a tightly packed form of DNA or condensed DNA, which comes in multiple varieties. These varieties lie on a continuum between the two extremes of constitutive heterochromatin and facultative heterochromatin. Both play a role in the expression of genes. Because it is tightly packed, it was thought to be inaccessible to polymerases and therefore not transcribed, however according to Volpe et al. (2002), and many other papers since, much of this DNA is in fact transcribed, but it is continuously turned over via RNA-induced transcriptional silencing (RITS). Recent studies with electron microscopy and OsO4 staining reveal that the dense packing is not due to the chromatin.
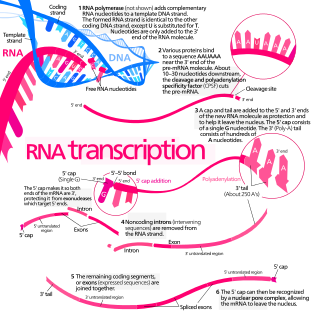
Transcription is the process of copying a segment of DNA into RNA. The segments of DNA transcribed into RNA molecules that can encode proteins are said to produce messenger RNA (mRNA). Other segments of DNA are copied into RNA molecules called non-coding RNAs (ncRNAs). Averaged over multiple cell types in a given tissue, the quantity of mRNA is more than 10 times the quantity of ncRNA. The general preponderance of mRNA in cells is valid even though less than 2% of the human genome can be transcribed into mRNA, while at least 80% of mammalian genomic DNA can be actively transcribed, with the majority of this 80% considered to be ncRNA.
In molecular biology, RNA polymerase, is an enzyme that synthesizes RNA from a DNA template.
In molecular biology and genetics, transcriptional regulation is the means by which a cell regulates the conversion of DNA to RNA (transcription), thereby orchestrating gene activity. A single gene can be regulated in a range of ways, from altering the number of copies of RNA that are transcribed, to the temporal control of when the gene is transcribed. This control allows the cell or organism to respond to a variety of intra- and extracellular signals and thus mount a response. Some examples of this include producing the mRNA that encode enzymes to adapt to a change in a food source, producing the gene products involved in cell cycle specific activities, and producing the gene products responsible for cellular differentiation in multicellular eukaryotes, as studied in evolutionary developmental biology.

The preinitiation complex is a complex of approximately 100 proteins that is necessary for the transcription of protein-coding genes in eukaryotes and archaea. The preinitiation complex positions RNA polymerase II at gene transcription start sites, denatures the DNA, and positions the DNA in the RNA polymerase II active site for transcription.

In molecular genetics, a repressor is a DNA- or RNA-binding protein that inhibits the expression of one or more genes by binding to the operator or associated silencers. A DNA-binding repressor blocks the attachment of RNA polymerase to the promoter, thus preventing transcription of the genes into messenger RNA. An RNA-binding repressor binds to the mRNA and prevents translation of the mRNA into protein. This blocking or reducing of expression is called repression.

RNA polymerase II is a multiprotein complex that transcribes DNA into precursors of messenger RNA (mRNA) and most small nuclear RNA (snRNA) and microRNA. It is one of the three RNAP enzymes found in the nucleus of eukaryotic cells. A 550 kDa complex of 12 subunits, RNAP II is the most studied type of RNA polymerase. A wide range of transcription factors are required for it to bind to upstream gene promoters and begin transcription.
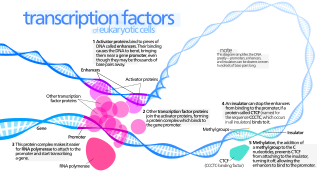
General transcription factors (GTFs), also known as basal transcriptional factors, are a class of protein transcription factors that bind to specific sites (promoter) on DNA to activate transcription of genetic information from DNA to messenger RNA. GTFs, RNA polymerase, and the mediator constitute the basic transcriptional apparatus that first bind to the promoter, then start transcription. GTFs are also intimately involved in the process of gene regulation, and most are required for life.
The family of heterochromatin protein 1 (HP1) consists of highly conserved proteins, which have important functions in the cell nucleus. These functions include gene repression by heterochromatin formation, transcriptional activation, regulation of binding of cohesion complexes to centromeres, sequestration of genes to nuclear periphery, transcriptional arrest, maintenance of heterochromatin integrity, gene repression at the single nucleosome level, gene repression by heterochromatization of euchromatin and DNA repair. HP1 proteins are fundamental units of heterochromatin packaging that are enriched at the centromeres and telomeres of nearly all Eukaryotic chromosomes with the notable exception of budding yeast, in which a yeast-specific silencing complex of SIR proteins serve a similar function. Members of the HP1 family are characterized by an N-terminal chromodomain and a C-terminal chromoshadow domain, separated by a Hinge region. HP1 is also found at euchromatic sites, where its binding correlates with gene repression. HP1 was originally discovered by Tharappel C James and Sarah Elgin in 1986 as a factor in the phenomenon known as position effect variegation in Drosophila melanogaster.
RNA-induced transcriptional silencing (RITS) is a form of RNA interference by which short RNA molecules – such as small interfering RNA (siRNA) – trigger the downregulation of transcription of a particular gene or genomic region. This is usually accomplished by posttranslational modification of histone tails which target the genomic region for heterochromatin formation. The protein complex that binds to siRNAs and interacts with the methylated lysine 9 residue of histones H3 is the RITS complex.
RNA silencing or RNA interference refers to a family of gene silencing effects by which gene expression is negatively regulated by non-coding RNAs such as microRNAs. RNA silencing may also be defined as sequence-specific regulation of gene expression triggered by double-stranded RNA (dsRNA). RNA silencing mechanisms are highly conserved in most eukaryotes. The most common and well-studied example is RNA interference (RNAi), in which endogenously expressed microRNA (miRNA) or exogenously derived small interfering RNA (siRNA) induces the degradation of complementary messenger RNA. Other classes of small RNA have been identified, including piwi-interacting RNA (piRNA) and its subspecies repeat associated small interfering RNA (rasiRNA).
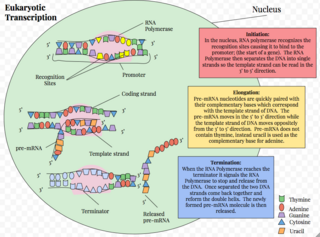
Eukaryotic transcription is the elaborate process that eukaryotic cells use to copy genetic information stored in DNA into units of transportable complementary RNA replica. Gene transcription occurs in both eukaryotic and prokaryotic cells. Unlike prokaryotic RNA polymerase that initiates the transcription of all different types of RNA, RNA polymerase in eukaryotes comes in three variations, each translating a different type of gene. A eukaryotic cell has a nucleus that separates the processes of transcription and translation. Eukaryotic transcription occurs within the nucleus where DNA is packaged into nucleosomes and higher order chromatin structures. The complexity of the eukaryotic genome necessitates a great variety and complexity of gene expression control.
FACT is a heterodimeric protein complex that affects eukaryotic RNA polymerase II transcription elongation both in vitro and in vivo. It was discovered in 1998 as a factor purified from human cells that was essential for productive, in vitro Pol II transcription on a chromatinized DNA template.
RNA polymerase II holoenzyme is a form of eukaryotic RNA polymerase II that is recruited to the promoters of protein-coding genes in living cells. It consists of RNA polymerase II, a subset of general transcription factors, and regulatory proteins known as SRB proteins.
Transposable elements are short strands of repetitive DNA that can self-replicate and translocate within the eukaryotic genome, and are generally perceived as parasitic in nature. Their transcription can lead to the production of dsRNAs, which resemble retroviruses transcripts. While most host cellular RNA has a singular, unpaired sense strand, dsRNA possesses sense and anti-sense transcripts paired together, and this difference in structure allows an host organism to detect dsRNA production, and thereby the presence of transposons. Plants lack distinct divisions between somatic cells and reproductive cells, and also have, generally, larger genomes than animals, making them an intriguing case-study kingdom to be used in attempting to better understand the epigenetics function of transposable elements.
RNA polymerase V is a multisubunit plant specific RNA polymerase found in nucleus. Together with RNA polymerase IV required for normal function and biogenesis of small interfering RNA (siRNA). Pol V is involved in siRNA-directed DNA methylation pathway which leads to heterochromatic silencing.
DDM1, Decreased DNA Methylation I, is a plant gene that encodes a nucleosome remodeler which facilitates DNA methylation. The DDM1 gene has been described extensively in Arabidopsis thaliana and also in maize. The protein has been described to be similar to the SWI2/SNF2 chromatin remodeling proteins.

Julie Law is an American molecular and cellular biologist. Law's pioneering work on DNA methylation patterns led to the discovery of the role of the CLASSY protein family in DNA methylation. Law is currently an associate professor at the Salk Institute for Biological Studies.

RNA-directed DNA methylation (RdDM) is a biological process in which non-coding RNA molecules direct the addition of DNA methylation to specific DNA sequences. The RdDM pathway is unique to plants, although other mechanisms of RNA-directed chromatin modification have also been described in fungi and animals. To date, the RdDM pathway is best characterized within angiosperms, and particularly within the model plant Arabidopsis thaliana. However, conserved RdDM pathway components and associated small RNAs (sRNAs) have also been found in other groups of plants, such as gymnosperms and ferns. The RdDM pathway closely resembles other sRNA pathways, particularly the highly conserved RNAi pathway found in fungi, plants, and animals. Both the RdDM and RNAi pathways produce sRNAs and involve conserved Argonaute, Dicer and RNA-dependent RNA polymerase proteins.











