
Ribozymes are RNA molecules that have the ability to catalyze specific biochemical reactions, including RNA splicing in gene expression, similar to the action of protein enzymes. The 1982 discovery of ribozymes demonstrated that RNA can be both genetic material and a biological catalyst, and contributed to the RNA world hypothesis, which suggests that RNA may have been important in the evolution of prebiotic self-replicating systems. The most common activities of natural or in vitro-evolved ribozymes are the cleavage or ligation of RNA and DNA and peptide bond formation. Within the ribosome, ribozymes function as part of the large subunit ribosomal RNA to link amino acids during protein synthesis. They also participate in a variety of RNA processing reactions, including RNA splicing, viral replication, and transfer RNA biosynthesis. Examples of ribozymes include the hammerhead ribozyme, the VS ribozyme, Leadzyme and the hairpin ribozyme.
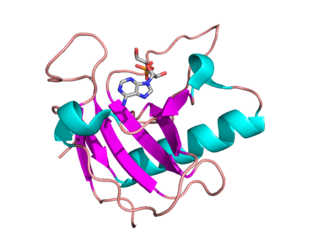
Ribonuclease is a type of nuclease that catalyzes the degradation of RNA into smaller components. Ribonucleases can be divided into endoribonucleases and exoribonucleases, and comprise several sub-classes within the EC 2.7 and 3.1 classes of enzymes.

A deoxyribonuclease is an enzyme that catalyzes the hydrolytic cleavage of phosphodiester linkages in the DNA backbone, thus degrading DNA. Deoxyribonucleases are one type of nuclease, a generic term for enzymes capable of hydrolyzing phosphodiester bonds that link nucleotides. A wide variety of deoxyribonucleases are known, which differ in their substrate specificities, chemical mechanisms, and biological functions.

A nuclease is an enzyme capable of cleaving the phosphodiester bonds between nucleotides of nucleic acids. Nucleases variously affect single and double stranded breaks in their target molecules. In living organisms, they are essential machinery for many aspects of DNA repair. Defects in certain nucleases can cause genetic instability or immunodeficiency. Nucleases are also extensively used in molecular cloning.
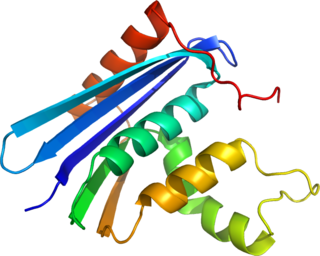
Ribonuclease H is a family of non-sequence-specific endonuclease enzymes that catalyze the cleavage of RNA in an RNA/DNA substrate via a hydrolytic mechanism. Members of the RNase H family can be found in nearly all organisms, from bacteria to archaea to eukaryotes.
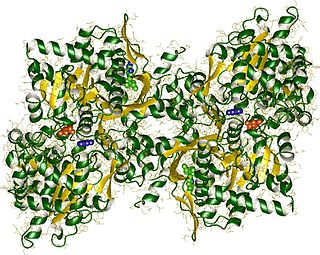
In biochemistry, phosphorylases are enzymes that catalyze the addition of a phosphate group from an inorganic phosphate (phosphate+hydrogen) to an acceptor.
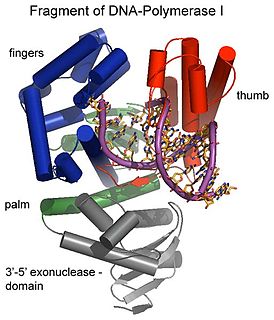
Exonucleases are enzymes that work by cleaving nucleotides one at a time from the end (exo) of a polynucleotide chain. A hydrolyzing reaction that breaks phosphodiester bonds at either the 3′ or the 5′ end occurs. Its close relative is the endonuclease, which cleaves phosphodiester bonds in the middle (endo) of a polynucleotide chain. Eukaryotes and prokaryotes have three types of exonucleases involved in the normal turnover of mRNA: 5′ to 3′ exonuclease (Xrn1), which is a dependent decapping protein; 3′ to 5′ exonuclease, an independent protein; and poly(A)-specific 3′ to 5′ exonuclease.
An esterase is a hydrolase enzyme that splits esters into an acid and an alcohol in a chemical reaction with water called hydrolysis.
Mung bean nuclease is a nuclease derived from sprouts of the mung bean that removes nucleotides in a step-wise manner from single-stranded DNA molecules (ssDNA) and is used in biotechnological applications to remove such ssDNA from a mixture also containing double-stranded DNA (dsDNA). This enzyme is useful for transcript mapping, removal of single-stranded regions in DNA hybrids or single-stranded overhangs produced by restriction enzymes, etc. It has an activity similar to Nuclease S1, but it has higher specificity for single-stranded molecules.
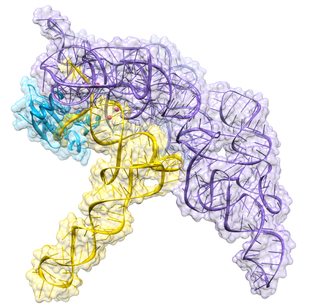
Ribonuclease P is a type of ribonuclease which cleaves RNA. RNase P is unique from other RNases in that it is a ribozyme – a ribonucleic acid that acts as a catalyst in the same way that a protein-based enzyme would. Its function is to cleave off an extra, or precursor, sequence of RNA on tRNA molecules. Further, RNase P is one of two known multiple turnover ribozymes in nature, the discovery of which earned Sidney Altman and Thomas Cech the Nobel Prize in Chemistry in 1989: in the 1970s, Altman discovered the existence of precursor tRNA with flanking sequences and was the first to characterize RNase P and its activity in processing of the 5' leader sequence of precursor tRNA. Recent findings also reveal that RNase P has a new function. It has been shown that human nuclear RNase P is required for the normal and efficient transcription of various small noncoding RNAs, such as tRNA, 5S rRNA, SRP RNA and U6 snRNA genes, which are transcribed by RNA polymerase III, one of three major nuclear RNA polymerases in human cells.
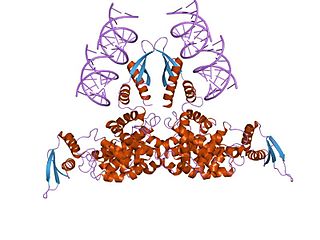
Ribonuclease III (BRENDA 3.1.26.3) is a type of ribonuclease that recognizes dsRNA and cleaves it at specific targeted locations to transform them into mature RNAs. These enzymes are a group of endoribonucleases that are characterized by their ribonuclease domain, which is labelled the RNase III domain. They are ubiquitous compounds in the cell and play a major role in pathways such as RNA precursor synthesis, RNA Silencing, and the pnp autoregulatory mechanism.
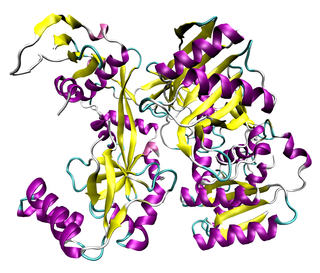
The Argonaute protein family plays a central role in RNA silencing processes, as essential components of the RNA-induced silencing complex (RISC). RISC is responsible for the gene silencing phenomenon known as RNA interference (RNAi). Argonaute proteins bind different classes of small non-coding RNAs, including microRNAs (miRNAs), small interfering RNAs (siRNAs) and Piwi-interacting RNAs (piRNAs). Small RNAs guide Argonaute proteins to their specific targets through sequence complementarity, which then leads to mRNA cleavage or translation inhibition.
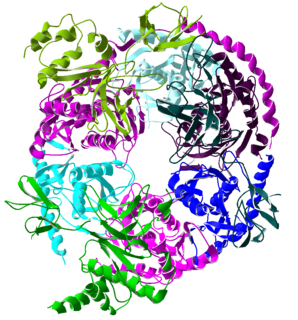
The exosome complex is a multi-protein intracellular complex capable of degrading various types of RNA molecules. Exosome complexes are found in both eukaryotic cells and archaea, while in bacteria a simpler complex called the degradosome carries out similar functions.
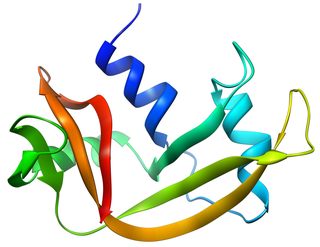
Pancreatic ribonuclease family is a superfamily of pyrimidine-specific endonucleases found in high quantity in the pancreas of certain mammals and of some reptiles.
Exoribonuclease II is an enzyme. This enzyme catalyses the following chemical reaction
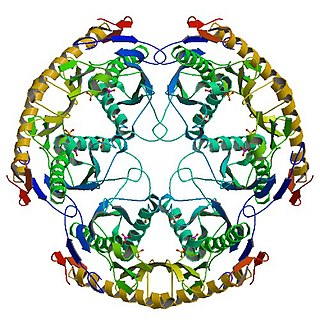
RNase PH is a tRNA nucleotidyltransferase, present in archaea and bacteria, that is involved in tRNA processing. Contrary to hydrolytic enzymes, it is a phosphorolytic enzyme, meaning that it uses inorganic phosphate as a reactant to cleave nucleotide-nucleotide bonds, releasing diphosphate nucleotides. The active structure of the proteins is a homohexameric complex, consisting of three ribonuclease (RNase) PH dimers. RNase PH has homologues in many other organisms, which are referred to as RNase PH-like proteins. The part of another larger protein with a domain that is very similar to RNase PH is called an RNase PH domain (RPD).
The degradosome is a multiprotein complex present in most bacteria that is involved in the processing of ribosomal RNA and the degradation of messenger RNA and is regulated by Non-coding RNA. It contains the proteins RNA helicase B, RNase E and Polynucleotide phosphorylase.
Ribonuclease E is a bacterial ribonuclease that participates in the processing of ribosomal RNA and the chemical degradation of bulk cellular RNA.
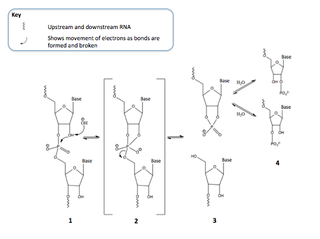
RNA hydrolysis is a reaction in which a phosphodiester bond in the sugar-phosphate backbone of RNA is broken, cleaving the RNA molecule. RNA is susceptible to this base-catalyzed hydrolysis because the ribose sugar in RNA has a hydroxyl group at the 2’ position. This feature makes RNA chemically unstable compared to DNA, which does not have this 2’ -OH group and thus is not susceptible to base-catalyzed hydrolysis.
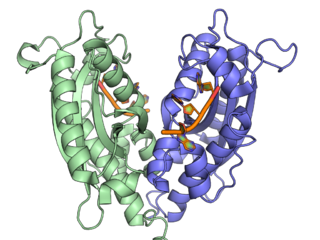
Ribonuclease T is a ribonuclease enzyme involved in the maturation of transfer RNA and ribosomal RNA in bacteria, as well as in DNA repair pathways. It is a member of the DnaQ family of exonucleases and non-processively acts on the 3' end of single-stranded nucleic acids. RNase T is capable of cleaving both DNA and RNA, with extreme sequence specificity discriminating against cytosine at the 3' end of the substrate.















