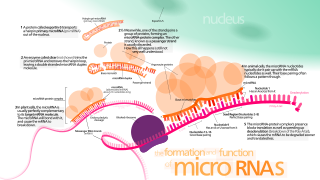
MicroRNA (miRNA) are small, single-stranded, non-coding RNA molecules containing 21 to 23 nucleotides. Found in plants, animals and some viruses, miRNAs are involved in RNA silencing and post-transcriptional regulation of gene expression. miRNAs base-pair to complementary sequences in mRNA molecules, then silence said mRNA molecules by one or more of the following processes:
- Cleavage of the mRNA strand into two pieces,
- Destabilization of the mRNA by shortening its poly(A) tail, or
- Reducing translation of the mRNA into proteins.
Gene silencing is the regulation of gene expression in a cell to prevent the expression of a certain gene. Gene silencing can occur during either transcription or translation and is often used in research. In particular, methods used to silence genes are being increasingly used to produce therapeutics to combat cancer and other diseases, such as infectious diseases and neurodegenerative disorders.
Gene knockdown is an experimental technique by which the expression of one or more of an organism's genes is reduced. The reduction can occur either through genetic modification or by treatment with a reagent such as a short DNA or RNA oligonucleotide that has a sequence complementary to either gene or an mRNA transcript.
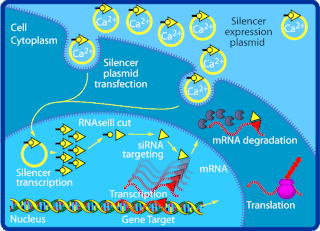
Small interfering RNA (siRNA), sometimes known as short interfering RNA or silencing RNA, is a class of double-stranded RNA at first non-coding RNA molecules, typically 20–24 base pairs in length, similar to miRNA, and operating within the RNA interference (RNAi) pathway. It interferes with the expression of specific genes with complementary nucleotide sequences by degrading mRNA after transcription, preventing translation. It was discovered in 1998, by Andrew Fire at Carnegie Institution for Science in Washington DC and Craig Mello at University of Massachusetts in Worcester.
The RNA-induced silencing complex, or RISC, is a multiprotein complex, specifically a ribonucleoprotein, which functions in gene silencing via a variety of pathways at the transcriptional and translational levels. Using single-stranded RNA (ssRNA) fragments, such as microRNA (miRNA), or double-stranded small interfering RNA (siRNA), the complex functions as a key tool in gene regulation. The single strand of RNA acts as a template for RISC to recognize complementary messenger RNA (mRNA) transcript. Once found, one of the proteins in RISC, Argonaute, activates and cleaves the mRNA. This process is called RNA interference (RNAi) and it is found in many eukaryotes; it is a key process in defense against viral infections, as it is triggered by the presence of double-stranded RNA (dsRNA).
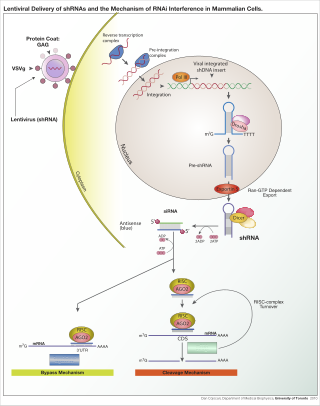
A short hairpin RNA or small hairpin RNA is an artificial RNA molecule with a tight hairpin turn that can be used to silence target gene expression via RNA interference (RNAi). Expression of shRNA in cells is typically accomplished by delivery of plasmids or through viral or bacterial vectors. shRNA is an advantageous mediator of RNAi in that it has a relatively low rate of degradation and turnover. However, it requires use of an expression vector, which has the potential to cause side effects in medicinal applications.
Ribonuclease III (RNase III or RNase C)(BRENDA 3.1.26.3) is a type of ribonuclease that recognizes dsRNA and cleaves it at specific targeted locations to transform them into mature RNAs. These enzymes are a group of endoribonucleases that are characterized by their ribonuclease domain, which is labelled the RNase III domain. They are ubiquitous compounds in the cell and play a major role in pathways such as RNA precursor synthesis, RNA Silencing, and the pnp autoregulatory mechanism.
The Argonaute protein family, first discovered for its evolutionarily conserved stem cell function, plays a central role in RNA silencing processes as essential components of the RNA-induced silencing complex (RISC). RISC is responsible for the gene silencing phenomenon known as RNA interference (RNAi). Argonaute proteins bind different classes of small non-coding RNAs, including microRNAs (miRNAs), small interfering RNAs (siRNAs) and Piwi-interacting RNAs (piRNAs). Small RNAs guide Argonaute proteins to their specific targets through sequence complementarity, which then leads to mRNA cleavage, translation inhibition, and/or the initiation of mRNA decay.
Drosha is a Class 2 ribonuclease III enzyme that in humans is encoded by the DROSHA gene. It is the primary nuclease that executes the initiation step of miRNA processing in the nucleus. It works closely with DGCR8 and in correlation with Dicer. It has been found significant in clinical knowledge for cancer prognosis. and HIV-1 replication.
Piwi genes were identified as regulatory proteins responsible for stem cell and germ cell differentiation. Piwi is an abbreviation of P-elementInduced WImpy testis in Drosophila. Piwi proteins are highly conserved RNA-binding proteins and are present in both plants and animals. Piwi proteins belong to the Argonaute/Piwi family and have been classified as nuclear proteins. Studies on Drosophila have also indicated that Piwi proteins have no slicer activity conferred by the presence of the Piwi domain. In addition, Piwi associates with heterochromatin protein 1, an epigenetic modifier, and piRNA-complementary sequences. These are indications of the role Piwi plays in epigenetic regulation. Piwi proteins are also thought to control the biogenesis of piRNA as many Piwi-like proteins contain slicer activity which would allow Piwi proteins to process precursor piRNA into mature piRNA.
RNA silencing or RNA interference refers to a family of gene silencing effects by which gene expression is negatively regulated by non-coding RNAs such as microRNAs. RNA silencing may also be defined as sequence-specific regulation of gene expression triggered by double-stranded RNA (dsRNA). RNA silencing mechanisms are conserved among most eukaryotes. The most common and well-studied example is RNA interference (RNAi), in which endogenously expressed microRNA (miRNA) or exogenously derived small interfering RNA (siRNA) induces the degradation of complementary messenger RNA. Other classes of small RNA have been identified, including piwi-interacting RNA (piRNA) and its subspecies repeat associated small interfering RNA (rasiRNA).

Protein argonaute-2 is a protein that in humans is encoded by the EIF2C2 gene.

Protein argonaute-1 is a protein that in humans is encoded by the EIF2C1 gene.
Trans-acting siRNA are a class of small interfering RNA (siRNA) that repress gene expression through post-transcriptional gene silencing in land plants. Precursor transcripts from TAS loci are polyadenylated and converted to double-stranded RNA, and are then processed into 21-nucleotide-long RNA duplexes with overhangs. These segments are incorporated into an RNA-induced silencing complex (RISC) and direct the sequence-specific cleavage of target mRNA. Ta-siRNAs are classified as siRNA because they arise from double-stranded RNA (dsRNA).
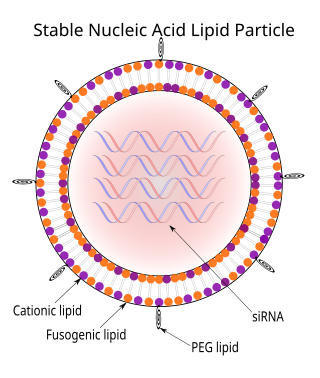
Stable nucleic acid lipid particles (SNALPs) are microscopic particles approximately 120 nanometers in diameter, smaller than the wavelengths of visible light. They have been used to deliver siRNAs therapeutically to mammals in vivo. In SNALPs, the siRNA is surrounded by a lipid bilayer containing a mixture of cationic and fusogenic lipids, coated with diffusible polyethylene glycol.

RNA interference (RNAi) is a biological process in which RNA molecules are involved in sequence-specific suppression of gene expression by double-stranded RNA, through translational or transcriptional repression. Historically, RNAi was known by other names, including co-suppression, post-transcriptional gene silencing (PTGS), and quelling. The detailed study of each of these seemingly different processes elucidated that the identity of these phenomena were all actually RNAi. Andrew Fire and Craig C. Mello shared the 2006 Nobel Prize in Physiology or Medicine for their work on RNAi in the nematode worm Caenorhabditis elegans, which they published in 1998. Since the discovery of RNAi and its regulatory potentials, it has become evident that RNAi has immense potential in suppression of desired genes. RNAi is now known as precise, efficient, stable and better than antisense therapy for gene suppression. Antisense RNA produced intracellularly by an expression vector may be developed and find utility as novel therapeutic agents.

Cas9 is a 160 kilodalton protein which plays a vital role in the immunological defense of certain bacteria against DNA viruses and plasmids, and is heavily utilized in genetic engineering applications. Its main function is to cut DNA and thereby alter a cell's genome. The CRISPR-Cas9 genome editing technique was a significant contributor to the Nobel Prize in Chemistry in 2020 being awarded to Emmanuelle Charpentier and Jennifer Doudna.
RDE-1 (RNAi-DEfective 1) is a primary Argonaute protein required for RNA-mediated interference (RNAi) in Caenorhabditis elegans. The rde-1 gene locus was first characterized in C. elegans mutants resistant to RNAi, and is a member of a highly conserved Piwi gene family that includes plant, Drosophila, and vertebrate homologs.

DCL1 is a gene in plants that codes for the DCL1 protein, a ribonuclease III enzyme involved in processing double-stranded RNA (dsRNA) and microRNA (miRNA). Although DCL1, also called Endoribonuclease Dicer homolog 1, is named for its homology with the metazoan protein Dicer, its role in miRNA biogenesis is somewhat different, due to substantial differences in miRNA maturation processes between plants and animals, as well due to additional downstream plant-specific pathways, where DCL1 paralogs like DCL4 participate, such Trans-acting siRNA biogenesis.

DCL2 is a gene in plants that codes for the DCL2 protein, a ribonuclease III enzyme involved in processing exogenous double-stranded RNA (dsRNA) into 22 nucleotide small interference RNAs (siRNAs).




















