A DNA polymerase is a member of a family of enzymes that catalyze the synthesis of DNA molecules from nucleoside triphosphates, the molecular precursors of DNA. These enzymes are essential for DNA replication and usually work in groups to create two identical DNA duplexes from a single original DNA duplex. During this process, DNA polymerase "reads" the existing DNA strands to create two new strands that match the existing ones. These enzymes catalyze the chemical reaction
DNA primase is an enzyme involved in the replication of DNA and is a type of RNA polymerase. Primase catalyzes the synthesis of a short RNA segment called a primer complementary to a ssDNA template. After this elongation, the RNA piece is removed by a 5' to 3' exonuclease and refilled with DNA.

DNA repair is a collection of processes by which a cell identifies and corrects damage to the DNA molecules that encode its genome. In human cells, both normal metabolic activities and environmental factors such as radiation can cause DNA damage, resulting in tens of thousands of individual molecular lesions per cell per day. Many of these lesions cause structural damage to the DNA molecule and can alter or eliminate the cell's ability to transcribe the gene that the affected DNA encodes. Other lesions induce potentially harmful mutations in the cell's genome, which affect the survival of its daughter cells after it undergoes mitosis. As a consequence, the DNA repair process is constantly active as it responds to damage in the DNA structure. When normal repair processes fail, and when cellular apoptosis does not occur, irreparable DNA damage may occur. This can eventually lead to malignant tumors, or cancer as per the two-hit hypothesis.

DNA polymerase II is a prokaryotic DNA-dependent DNA polymerase encoded by the PolB gene.

Poland syndrome is a birth defect characterized by an underdeveloped chest muscle and short webbed fingers on one side of the body. There may also be short ribs, less fat, and breast and nipple abnormalities on the same side of the body. Typically, the right side is involved. Those affected generally have normal movement and health.

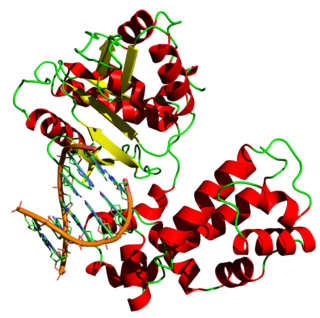
Proliferating cell nuclear antigen (PCNA) is a DNA clamp that acts as a processivity factor for DNA polymerase δ in eukaryotic cells and is essential for replication. PCNA is a homotrimer and achieves its processivity by encircling the DNA, where it acts as a scaffold to recruit proteins involved in DNA replication, DNA repair, chromatin remodeling and epigenetics.

Mitotic spindle assembly checkpoint protein MAD1 is a protein that in humans is encoded by the MAD1L1 gene.

DNA polymerase iota is an enzyme that in humans is encoded by the POLI gene. It is found in higher eukaryotes, and is believed to have arisen from a gene duplication from Pol η. Pol ι, is a Y family polymerase that is involved in translesion synthesis. It can bypass 6-4 pyrimidine adducts and abasic sites and has a high frequency of wrong base incorporation. Like many other Y family polymerases Pol ι, has low processivity, a large DNA binding pocket and doesn't undergo conformational changes when DNA binds. These attributes are what allow Pol ι to carry out its task as a translesion polymerase. Pol ι only uses Hoogsteen base pairing, during DNA synthesis, it will add adenine opposite to thymine in the syn conformation and can add both cytosine and thymine in the anti conformation across guanine, which it flips to the syn conformation.

DNA polymerase kappa is a DNA polymerase that in humans is encoded by the POLK gene. It is involved in translesion synthesis.

Mitotic spindle assembly checkpoint protein MAD2A is a protein that in humans is encoded by the MAD2L1 gene.

DNA polymerase delta catalytic subunit(DPOD1) is an enzyme that is encoded in the human by the POLD1 gene, in the DNA polymerase delta complex. DPOD1 is responsible for synthesizing the lagging strand of DNA, and has also been implicated in some activities at the leading strand. The DPOD1 subunit encodes both DNA polymerizing and exonuclease domains, which provide the protein an important second function in proofreading to ensure replication accuracy during DNA synthesis, and in a number of types of replication-linked DNA repair following DNA damage.

Mitotic spindle assembly checkpoint protein MAD2B is a protein that in humans is encoded by the MAD2L2 gene.

DNA repair protein REV1 is a protein that in humans is encoded by the REV1 gene.

DNA polymerase theta is an enzyme that in humans is encoded by the POLQ gene. This polymerase plays a key role in one of the three major double strand break repair pathways: theta-mediated end joining (TMEJ). Most double-strand breaks are repaired by non-homologous end joining (NHEJ) or homology directed repair (HDR). However, in some contexts, NHEJ and HR are insufficient and TMEJ is the only solution to repair the break. TMEJ is often described as alternative NHEJ, but differs in that it lacks a requirement for the Ku heterodimer, and it can only act on resected DNA ends. Following annealing of short regions on the DNA overhangs, DNA polymerase theta catalyzes template-dependent DNA synthesis across the broken ends, stabilizing the paired structure.

Tumor suppressor candidate 3 is a protein that in humans is encoded by the TUSC3 gene.

DNA polymerase eta, is a protein that in humans is encoded by the POLH gene.

Ribonuclease H2, subunit B is a protein in humans is encoded by the RNASEH2B gene. RNase H2 is composed of a single catalytic subunit (A) and two non-catalytic subunits, and degrades the RNA of RNA:DNA hybrids. The non-catalytic B subunit of RNase H2 is thought to play a role in DNA replication.

In molecular biology, kataegis describes a pattern of localized hypermutations identified in some cancer genomes, in which a large number of highly patterned basepair mutations occur in a small region of DNA. The mutational clusters are usually several hundred basepairs long, alternating between a long range of C→T substitutional pattern and a long range of G→A substitutional pattern. This suggests that kataegis is carried out on only one of the two template strands of DNA during replication. Compared to other cancer-related mutations, such as chromothripsis, kataegis is more commonly seen; it is not an accumulative process but likely happens during one cycle of replication.
DNA Polymerase V is a polymerase enzyme involved in DNA repair mechanisms in bacteria, such as Escherichia coli. It is composed of a UmuD' homodimer and a UmuC monomer, forming the UmuD'2C protein complex. It is part of the Y-family of DNA Polymerases, which are capable of performing DNA translesion synthesis (TLS). Translesion polymerases bypass DNA damage lesions during DNA replication - if a lesion is not repaired or bypassed the replication fork can stall and lead to cell death. However, Y polymerases have low sequence fidelity during replication. When the UmuC and UmuD' proteins were initially discovered in E. coli, they were thought to be agents that inhibit faithful DNA replication and caused DNA synthesis to have high mutation rates after exposure to UV-light. The polymerase function of Pol V was not discovered until the late 1990s when UmuC was successfully extracted, consequent experiments unequivocally proved UmuD'2C is a polymerase. This finding lead to the detection of many Pol V orthologs and the discovery of the Y-family of polymerases.

PrimPol is a protein encoded by the PRIMPOL gene in humans. PrimPol is a eukaryotic protein with both DNA polymerase and DNA Primase activities involved in translesion DNA synthesis. It is the first eukaryotic protein to be identified with priming activity using deoxyribonucleotides. It is also the first protein identified in the mitochondria to have translesion DNA synthesis activities.























