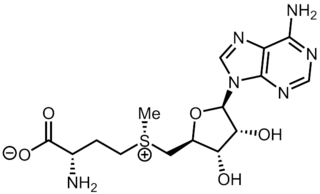
S-Adenosyl methionine (SAM), also known under the commercial names of SAMe, SAM-e, or AdoMet, is a common cosubstrate involved in methyl group transfers, transsulfuration, and aminopropylation. Although these anabolic reactions occur throughout the body, most SAM is produced and consumed in the liver. More than 40 methyl transfers from SAM are known, to various substrates such as nucleic acids, proteins, lipids and secondary metabolites. It is made from adenosine triphosphate (ATP) and methionine by methionine adenosyltransferase. SAM was first discovered by Giulio Cantoni in 1952.

Adenosine deaminase deficiency is a metabolic disorder that causes immunodeficiency. It is caused by mutations in the ADA gene. It accounts for about 10–20% of all cases of autosomal recessive forms of severe combined immunodeficiency (SCID) after excluding disorders related to inbreeding.
S-Adenosyl-L-homocysteine (SAH) is the biosynthetic precursor to homocysteine. SAH is formed by the demethylation of S-adenosyl-L-methionine. Adenosylhomocysteinase converts SAH into homocysteine and adenosine.
In enzymology, a carnosine N-methyltransferase is an enzyme that catalyzes the chemical reaction

Guanidinoacetate N-methyltransferase is an enzyme that catalyzes the chemical reaction and is encoded by gene GAMT located on chromosome 19p13.3.
In enzymology, a magnesium protoporphyrin IX methyltransferase is an enzyme that catalyzes the chemical reaction
In enzymology, a precorrin-4 C11-methyltransferase is an enzyme that catalyzes the chemical reaction
In enzymology, a protein-glutamate O-methyltransferase is an enzyme that catalyzes the chemical reaction
The isoprenylcysteine o-methyltransferase carries out carboxyl methylation of cleaved eukaryotic proteins that terminate in a CaaX motif. In Saccharomyces cerevisiae this methylation is carried out by Ste14p, an integral endoplasmic reticulum membrane protein. Ste14p is the founding member of the isoprenylcysteine carboxyl methyltransferase (ICMT) family, whose members share significant sequence homology.
In enzymology, a tRNA guanosine-2'-O-methyltransferase is an enzyme that catalyzes the chemical reaction

Nucleoside diphosphate kinase A is an enzyme that in humans is encoded by the NME1 gene. It is thought to be a metastasis suppressor.
The enzyme S-ribosylhomocysteine lyase catalyzes the reaction
In enzymology, an adenosylhomocysteine nucleosidase (EC 3.2.2.9) is an enzyme that catalyzes the chemical reaction
In enzymology, a S-adenosylhomocysteine deaminase (EC 3.5.4.28) is an enzyme that catalyzes the chemical reaction
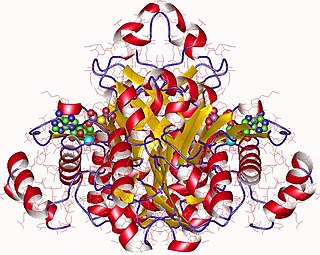
Adenosine kinase is an enzyme that catalyzes the transfer of gamma-phosphate from Adenosine triphosphate (ATP) to adenosine (Ado) leading to formation of Adenosine monophosphate (AMP). In addition to its well-studied role in controlling the cellular concentration of Ado, AdK also plays an important role in the maintenance of methylation reactions. All S-adenosylmethionine-dependent transmethylation reactions in cells lead to production of S-adenosylhomocysteine (SAH), which is cleaved by SAH hydrolase into Ado and homocysteine. The failure to efficiently remove these end products can result in buildup of SAH, which is a potent inhibitor of all transmethylation reactions. The disruption of AdK gene (-/-) in mice causes neonatal hepatic steatosis, a fatal condition characterized by rapid microvesicular fat infiltration, leading to early postnatal death. The liver was the main organ affected in these animals and in it the levels of adenine nucleotides were decreased, while those of SAH were elevated. Recently, missense mutations in the AdK gene in humans which result in AdK deficiency have also been shown to cause hypermethioninemia, encephalopathy and abnormal liver function.

Dihydropyrimidinase-related protein 3 is an enzyme that in humans is encoded by the DPYSL3 gene. A recent bioinformatics study suggested that the DPYSL3 gene might have a prognostic role in neuroblastoma.

Myosin-Ie (Myo1e) is a protein that in humans is encoded by the MYO1E gene.
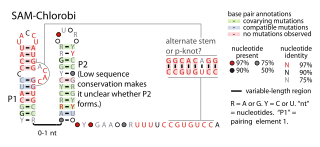
The SAM-Chlorobi RNA motif is a conserved RNA structure that was identified by bioinformatics. The RNAs are found only in bacteria classified as within the phylum Chlorobiota. These RNAs are always in the 5' untranslated regions of operons that contain metK and ahcY genes. metK genes encode methionine adenosyltransferase, which synthesizes S-adenosyl methionine (SAM), and ahcY genes encode S-adenosylhomocysteine hydrolase, which degrade the related metabolite S-Adenosyl-L-homocysteine (SAH). In fact all predicted metK and ahcY genes within Chlorobiota bacteria as of 2010 are preceded by predicted SAM-Chlorobi RNAs. Predicted promoter sequences are consistently found upstream of SAM-Chlorobi RNAs, and these promoter sequences imply that SAM-Chlorobi RNAs are indeed transcribed as RNAs. The promoter sequences are commonly associated with strong transcription in the phyla Chlorobiota and Bacteroidota, but are not used by most lineages of bacteria. The placement of SAM-Chlorobi RNAs suggests that they are involved in the regulation of the metK/ahcY operon through an unknown mechanism.
Erythromycin 3''-O-methyltransferase is an enzyme with systematic name S-adenosyl-L-methionine:erythromycin C 3''-O-methyltransferase. This enzyme catalyses the following chemical reaction

Deficiency of Adenosine deaminase 2 (DADA2) is a monogenic disease associated with systemic inflammation and vasculopathy that affects a wide variety of organs in different patients. As a result, it is hard to characterize a patient with this disorder. Manifestations of the disease include but are not limited to recurrent fever, livedoid rash, various cytopenias, stroke, immunodeficiency, and bone marrow failure. Symptoms often onset during early childhood, but some cases have been discovered as late as 65 years old.













