Volume 26, Number 12—December 2020
Research
Human-Pathogenic Kasokero Virus in Field-Collected Ticks
Abstract
Kasokero virus (KASV; genus Orthonairovirus) was first isolated in 1977 at Uganda Virus Research Institute from serum collected from Rousettus aegyptiacus bats captured at Kasokero Cave, Uganda. During virus characterization studies at the institute, 4 laboratory-associated infections resulted in mild to severe disease. Although orthonairoviruses are typically associated with vertebrate and tick hosts, a tick vector of KASV never has been reported. We tested 786 Ornithodoros (Reticulinasus) faini tick pools (3,930 ticks) for KASV. The ticks were collected from a large R. aegyptiacus bat roosting site in western Uganda. We detected KASV RNA in 43 tick pools and recovered 2 infectious isolates, 1 of which was derived from host blood–depleted ticks. Our findings suggest that KASV is maintained in an enzootic transmission cycle involving O. (R.) faini ticks and R. aegyptiacus bats and has the potential for incidental virus spillover to humans.
The genus Orthonairovirus (family Nairoviridae) comprises »40 viruses (1), including human pathogens such as Crimean-Congo hemorrhagic fever virus. The orthonairoviruses are assigned to 14 species (1), most of which have been associated with a single vertebrate subphylum (Vertebrata: bats, birds, rodents, shrews, or ungulates) and tick order (Ixodida: argasids or ixodids) (2). The species Kasokero orthonairovirus comprises 3 viruses isolated from bats belonging to the suborder Yinpterochiroptera (3,4) (Pteropodiformes) (5) in Africa, namely Yogue virus (YOGV; Rousettus aegyptiacus, Senegal), Leopards Hill virus (LPHV; Hipposideros gigas, Zambia), and Kasokero virus (KASV; Rousettus aegyptiacus, Uganda) (2,6,7).
KASV was first isolated in 1977 by scientists at Uganda Virus Research Institute (UVRI) from 2.7% (2/74) of serum samples collected from R. aegyptiacus bats captured at Kasokero Cave in Uganda (6). Two months after the KASV bat isolates were introduced to the UVRI laboratory and 3 weeks after the isolates were used in virus characterization assays, a laboratory staff member became ill. Shortly thereafter, 2 additional laboratory staff members became ill. Two of these laboratory staff members had participated in KASV characterization studies that involved virus antigen extraction and serologic testing; the third laboratory staff member had prepared KASV mouse brain suspensions for inoculation and examined virus-infected mice. Sixteen days after symptom onset in the index patient, a UVRI driver who reported no direct contact with the laboratory rooms used to handle the KASV isolates became ill. Manifestations ranged in severity from mild febrile illness to prolonged systemic disease characterized by fever, headache, myalgia, arthralgia, abdominal pain, nausea, diarrhea, chest pain, coughing, and hyperactive reflexes. Intracerebral inoculation of suckling mice with acute phase blood specimens collected from each of the 4 humans yielded a KASV isolate. KASV-specific antibodies were detected in serum from the 4 patients at various times after illness, as well as in 9.5% (10/105) of serum samples collected from other UVRI laboratory staff members and 67.6% (50/74) of the original R. aegyptiacus serum. In susceptibility studies, KASV killed suckling and adult mice by intracerebral and intraperitoneal inoculation within 8 days (6). In addition, nearly all naive adult mice that nursed KASV-inoculated suckling mice died of KASV infection, indicating horizontal transmission of virus infection.
The home range of R. aegyptiacus bats extends throughout sub-Saharan Africa; the bats prefer subterranean environments, such as caves or mines. At multiple locations, Ornithodoros (Reticulinasus) faini ticks (family Argasidae) (8) have been observed living within rock crevices and feeding on R. aegyptiacus bats (9–13). Although chiropteran ticks typically exhibit high host-specificity (9,14), miners, researchers, and other persons entering R. aegyptiacus bat roosts have reported being bitten by O. (R.) faini ticks (13). Because most orthonairoviruses have been associated with a tick host, O. (R.) faini ticks are likely to be involved in the enzootic transmission and maintenance of KASV and have the potential to be vectors for virus spillover into humans. In 1994 and 1995, KASV was isolated by 1 author (R.S.) from O. (R.) faini ticks collected in Lanner Gorge Cave (22.450°S, 31.150°E) in South Africa, where R. aegyptiacus bats roosted. The isolations were made by intracerebral inoculation of suckling mice and identified in cross-neutralization tests in mice using homologous and reference mouse antiserum and the prototype KASV UG Z-52969 isolate obtained from Yale Arbovirus Research Unit (New Haven, CT, USA) and methods described by Shope and Sather (15). At the time, KASV was considered a possible bunyavirus, and the isolations remained unpublished. No molecular studies were attempted, and the isolations are no longer available for sequencing. Members of the team that entered Lannar Gorge Cave were bitten by ticks, and in 2 team members, a moderately severe, transient febrile illness developed with headache, malaise, and myalgia a few days later; they refused to seek medical attention or to donate blood samples for virologic examination.
In this study, we tested 786 tick pools (3,930 total O. [R.] faini ticks) for KASV. We collected the ticks from a large R. aegyptiacus bat roosting site in western Uganda in 2013 and 2017.
Tick Collection and Processing
After obtaining approval from the Uganda Wildlife Authority, we collected adult and nymph O. (R.) faini ticks with forceps from rock crevices in Python Cave, Queen Elizabeth National Park, Uganda, over 4 days in April 2013 (12) and 1 day in September 2017. A chiropteran population consisting solely of »40,000 R. aegyptiacus bats inhabit the cave (11). Ticks collected in 2013 were pooled in groups of 5, placed directly into grinding vials (OPS Diagnostics, https://opsdiagnostics.com) containing 250 μL of a 1:1 ratio of MagMax Lysis Binding Solution Concentrate (Thermo Fisher Scientific, https://www.thermofisher.com) to 100% isopropanol (MagMax Lysis Binding Buffer) and then homogenized using the GenoGrinder 2000 (OPS Diagnostics). After we added 550 μL of MagMax Lysis Binding Buffer, we transferred the tick pool lysates to cryovials and stored them under liquid nitrogen (12).
Ticks collected in 2017, also pooled in groups of 5, were placed directly into cryovials containing Dulbecco’s Modified Eagle Medium supplemented with 20% heat-inactivated fetal bovine serum (FBS) and antimicrobial drugs and then stored under liquid nitrogen. After thawing the tick pools, we transferred the contents to grinding vials, homogenized them using the GenoGrinder 2000, and then transferred them to a cryovial containing 250 μL of Dulbecco’s Modified Eagle Medium supplemented with 2% heat-inactivated FBS and antimicrobial drugs. We transferred a portion of each tick pool homogenate (100-μL) into a 400-μL aliquot of MagMax Lysis Binding Buffer.
RNA Extraction and Quantitative Reverse Transcriptase PCR
We extracted RNA (90 μL) from the 2013 (800 μL) and 2017 (500 μL) tick pool lysates using the MagMax Pathogen RNA/DNA Kit on the MagMax Express-96 Deep Well Magnetic Particle Processor (Thermo Fisher Scientific). KASV has an 18.3-kb single-stranded, negative-sense, trisegmented RNA genome comprising large segment that encodes for the viral RNA-dependent RNA polymerase (RdRp), medium segment that encodes for the glycoprotein precursor (GP), and small segment that encodes for the nucleoprotein (N) (2). We analyzed RNA by quantitative reverse transcription PCR (qRT-PCR) using the SuperScript III Platinum One-Step qRT-PCR Kit (Thermo Fisher Scientific) with primers and probes (Appendix Table 1) targeting the KASV N gene, tick mitochondrial 16S ribosomal RNA (rRNA) gene (16), and eukaryotic 18S rRNA gene (Thermo Fisher Scientific; 2017 tick pools only). Relative KASV RNA copies/tick pool were interpolated from a standard curve generated from a serial dilution of a known concentration of a synthetic KASV RNA oligo.
KASV Infection Prevalence Calculations
We calculated maximum-likelihood estimates of KASV infection prevalence in individual ticks with exact 95% CIs using an online pooled prevalence calculator (https://epitools.ausvet.com.au). The calculator implemented a frequentist approach and assumed a fixed tick pool size (n = 5) and 100% KASV qRT-PCR sensitivity and specificity (17,18).
Virus Isolation and Immunofluorescence Assay
We attempted virus isolation on the four 2017 KASV RNA-positive tick pools. After clarifying the tick pool homogenates (650 μL) by centrifugation, we transferred 200 μL supernatant to a vial containing antimicrobials and incubated it at room temperature for 1 h. Monolayers of Vero E6 cells in 12-well plates were inoculated with 210 μL of antimicrobial-treated supernatant and incubated for 1 h at 37°C under 5% CO2. After the addition of 1.3 mL maintenance media, cultures were incubated at 37°C under 5% CO2 and monitored daily for cytopathic effect. After 7 d, we transferred 1 mL culture media to a cryovial and replaced with an equal volume of fresh maintenance media. We transferred a portion of the day 7 media (100 μL) into MagMax Lysis Binding Buffer (400 μL) for RNA extraction and qRT-PCR. After 9–10 d, tissue cultures monolayers that were KASV RNA positive at day 7 were scraped to release virus-infected cells. Part of each cellular medium (1 mL) was suspended in 5 mL of borate saline, and 100 μL was placed into MagMax Lysis Binding Buffer (400 μL) for RNA extraction and qRT-PCR. After the cell suspensions were pelleted by centrifugation, the borate saline was decanted, the cells were resuspended in 500 μL borate saline, and 12-well spot slides were spotted with 25 μL of the cellular suspensions. The slides were fixed in acetone before receiving 2 megarads of γ-irradiation.
Six spots on each slide were incubated with 25 μL of a 1:100 dilution of KASV mouse immune ascitic fluid (World Reference Center for Emerging Viruses and Arboviruses, https://www.utmb.edu/gnl/research/wrceva), and the other 6 spots were incubated with normal mouse ascitic fluid for 30 min at 37°C. After the incubation, the spot slides were rinsed 2 times with phosphate buffered saline (PBS), incubated with 24 μL of a 1:40 dilution of goat anti-mouse fluorescein isothiocyanate (MP Biomedicals, https://www.mpbio.com) for 30 min at 37°C, rinsed with PBS, stained with Eriochrome Black T, rinsed with PBS, and then observed under a fluorescence microscope.
KASV Genome Sequencing
First-strand cDNA was synthesized directly from RNA extracted from 9 of the 2013 KASV RNA-positive tick pools using the qScript XLT cDNA SuperMix Kit (Quantabio, https://www.quantabio.com). KASV amplicons were generated from first-strand cDNA using the Q5 High-Fidelity 2X Master Mix (New England BioLabs, https://www.neb.com) and 6 multiplex pools of KASV-specific tiling primers (Appendix Table 2) that were designed using the Primal Scheme software (http://primal.zibraproject.org) (19). We prepared purified KASV amplicons for sequencing using the Accel-NGS 2S DNA Library Kit (Swift Biosciences, https://swiftbiosci.com). Indexed DNA libraries were pooled and then pair-end sequenced using a 500-cycle MiSeq Reagent Kit v2 on the MiSeq System (Illumina, https://www.illumina.com).
After thawing media collected from the two 2017 KASV isolates, we clarified the media by centrifugation and transferred 100 μL supernatant into 400 μL TriPure Isolation Reagent (MilliporeSigma, https://www.emdmillipore.com). We extracted RNA from the KASV isolate lysates using the 5PRIME Phase Lock Gel (Quantabio) system and then purified it using the Monarch Total RNA Miniprep Kit (New England Biolabs). We prepared purified RNA for sequencing using the NEB rRNA Depletion and NEBNext Ultra II RNA Library Kits for Illumina (New England Biolabs). Indexed DNA libraries were pooled and then pair-end sequenced using a 300-cycle MiSeq Reagent Kit v2 on the MiSeq System (Illumina).
Sequence and Phylogenetic Analyses
KASV sequence data were imported into Geneious 11.1.2 (Biomatters, https://www.geneious.com). After removing KASV-specific primers from the sequences (2013 tick pools), we used BBDuk to trim adaptors and low-quality reads from both sequence ends (minimum quality 30). Long sequence reads were retained (>93% of maximum read length) and normalized to a target coverage level of 40 with a minimum depth of 2. Merged reads were mapped to the concatenated genome sequence of the KASV Z-52963 isolate using the Geneious mapper (minimum mapping quality 30), and consensus sequences were then extracted and parsed according to gene.
We used the MUSCLE algorithm (https://www.ebi.ac.uk/Tools/msa/muscle) to generate N, GP, and RdRp nucleotide and deduced amino acid alignments from the new KASV sequences and existing KASV, YOGV, and LPHV sequences. We constructed maximum-likelihood phylogenies using the PhyML 3.0 algorithm (20) in conjunction with the best-fit nucleotide substitution model (21) on the ATGC Montpellier Bioinformatics Platform (http://www.atgc-montpellier.fr). We visualized phylogenies using TreeGraph 2 (http://treegraph.bioinfweb.info) (22). We estimated the global ratio of the rate of nonsynonymous (dN) nucleotide substitutions to the rate of synonymous (dS) nucleotide substitutions (dN:dS) across the KASV nucleotide alignments using the fixed effects likelihood method with the HyPhy version 1.8.2 software (https://www.hyphy.org).
Description of the O. (R.) faini Tick Collections
We collected 3,125 O. (R.) faini ticks (625 pools of 5 each) from the rock crevices within R. aegyptiacus bat roosting sites in Python Cave, Uganda, in 2013 and 975 O. (R.) faini ticks (195 pools of 5 each) from the same location in 2017. We confirmed the O. (R.) faini species designation by comparative genetic analysis of the 16S rRNA gene of a set of ticks and by morphologic examination (12). Screening the O. (R.) faini tick pools for the tick-specific 16S rRNA gene revealed that 4.3% (27/625) of the 2013 pools and 3.6% (7/195) of the 2017 pools were negative, indicating that these samples contained RNA inhibitors and were unsuitable for downstream KASV qRT-PCR analysis.
Detection of KASV RNA in Ticks
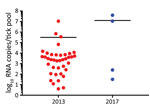
We detected KASV RNA in 39/598 of the 2013 O. (R.) faini tick pools and 4/188 of the 2017 O. (R.) faini tick pools, resulting in maximum-likelihood estimates of KASV infection prevalence at the individual tick level of 1.34% (95% CI 0.94%–1.83%) and 0.43% (95% CI 0.12%–1.10%), respectively. Based off a standard curve using synthetic KASV RNA, the mean KASV load of the positive 2013 tick pools was 5.5 (range 0.6–7.0) log10 RNA copies, and the mean KASV load of the positive 2017 tick pools was 7.1 (range 1.5–7.6) log10 RNA copies (Figure 1).
Isolation of Infectious KASV from Ticks
We isolated infectious KASV from 50% (2/4) of the 2017 KASV RNA-positive tick pools. As expected, the 2 KASV isolates were derived from the tick pools with the highest KASV RNA loads (UGA-Tick-20170048: 7.6 log10 RNA copies and UGA-Tick-20170128: 7.0 log10RNA copies) (Figure 1). A qRT-PCR targeting the eukaryotic 18S rRNA gene was used to screen the KASV-isolation positive tick pools for R. aegyptiacus blood. We found no trace of R. aegyptiacus blood in tick pool UGA-Tick-20170048, indicating that the 5 ticks in this pool had not recently taken a blood meal from an actively KASV-infected R. aegyptiacus bat. This finding suggests that KASV in this tick pool resulted from active virus replication in >1 tick.
Circulation of Genetically Diverse KASVs
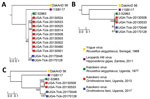
We attempted genomic sequencing on 9 of the 2013 KASV RNA-positive tick pools with KASV loads >3.6 log10 RNA copies and on the two 2017 KASV tick isolates. We obtained complete sequence coverage for the N (1,545 nt) for 11 of the KASV-positive tick pools, the GP (4,314 nt) for 4 of the KASV-positive tick pools, and the RdRp (11,919 nt) genes for 4 of the KASV-positive tick pools. Maximum-likelihood phylogenies constructed from N, GP, and RdRp gene sequences from the 2013 and 2017 KASV-positive tick pools, as well as prototype KASV (Z-52963, R. aegyptiacus, Uganda, 1977), YOGV (DakAnD 56, R. aegyptiacus, Senegal, 1968), and LPHV (11SB17, H. gigas, Zambia, 2011) isolate sequences, had similar topologies and virus species groupings (Figure 2). Consistent with previous results (2,23,24), YOGV diverged first followed by LPHV, and finally the KASV lineage. The N phylogeny showed 2 distinct KASV lineages defined by >9.2% interlineage nucleotide divergences, and the RdRp phylogeny showed 2 distinct KASV lineages defined by >12.0% interlineage nucleotide divergences; the first lineage included the prototype Z-52963 sequence plus the 2013 tick sequences and the second lineage included the 2017 tick sequences (Appendix Tables 3, 4). Deduced amino acid alignments of the N and RdRp proteins revealed that most KASV lineage-defining nucleotide substitutions were synonymous with interlineage amino acid divergences ranging from 0.6% to 1.0% for the N protein and 2.4% to 2.6% for the RdRp protein. In contrast to the N and RdRp phylogenies, the GP phylogeny shows that the Z-52963 sequence diverged first, followed by the divergence of the 2013 and the 2017 tick sequence groups. Furthermore, the KASV sequences in the GP phylogeny are considerably more similar to one another at the nucleotide level (<2.2% nt divergence and <0.9% aa divergence) and do not form 2 distinct lineages (Appendix Table 5). Consistent with the phylogenetic and KASV gene/protein divergence data, dN:dS estimates demonstrated that the N gene was under the strongest purifying selection (0.0110), followed by the RdRp (0.0264) and GP (0.0650) genes.
We detected KASV RNA in 43 O. (R.) faini tick pools collected from a large R. aegyptiacus bat colony at Python Cave, Uganda, over a 4-year span. The mean KASV RNA load for the 39 positive 2013 tick pools stored in MagMax Lysis Binding Buffer was lower (5.5 [range 0.6–7.0] log10 RNA copies) than the 4 positive 2017 tick pools stored in sterile media supplemented with FBS and antimicrobial drugs (7.1 [range 1.5–7.6] log10 RNA copies). Although this difference might have resulted from the choice of sample preservation buffer, it could also be attributed to the type of storage vial (internally threaded for 2017 ticks vs. externally threaded for 2013 ticks), number of RNA freeze-thaw cycles (0 for 2017 ticks vs. 1 for 2013 ticks), month of tick collection in relation to the natural history of KASV infection in O. (R.) faini ticks (September for ticks collected in 2017 vs. April for ticks collected in 2013), or sample size effect (39 KASV-positive tick pools in 2013 vs. 4 KASV-positive tick pools in 2017). We could not attempt virus isolation on the 2013 KASV-positive tick pools because they were placed directly in virucidal buffer; however, we isolated infectious KASV from 2 of the 2017 KASV RNA-positive tick pools. Importantly, molecular evidence of R. aegyptiacus blood was not detected in 1 of the KASV isolation-positive tick pools. This finding indicates that the presence of infectious KASV in this tick pool resulted from active virus replication in >1 tick and not from ingestion of a recent blood meal by a tick feeding on a viremic R. aegyptiacus bat. Although we did not assess whether KASV can disseminate to the salivary glands of O. (R.) faini ticks and then be successfully transmitted to R. aegyptiacus bats, our data coupled with the results of a previous study demonstrating a 2.7% prevalence of active KASV infection and a 67.6% KASV seroprevalence in R. aegyptiacus captured at Kasokero Cave, Uganda (6), suggest that this virus is maintained in an enzootic transmission cycle involving R. aegyptiacus bats and O. (R.) faini ticks. The isolation of KASV from O. (R.) faini ticks collected from a R. aegyptiacus bat roost in Lanner Gorge Cave, South Africa, in 1994–1995 (R. Swanepoel, unpub. data) supports this notion and suggests that KASV has a widespread geographic distribution. Tick transmission of KASV also is consistent with our knowledge of the vector status of the orthonairoviruses (2). Of the 14 currently recognized orthonairovirus species (1), 13 have now been associated with a tick host.
Genetic analysis of the KASV nucleotide alignment showed that the N and RdRp gene sequences were highly divergent, whereas the GP gene sequences were highly conserved. However, the low dN:dS estimates, together with the high level of conservation between the deduced amino acid sequences for the N, GP, and RdRp proteins, suggest that strong purifying selection purged deleterious mutations. This finding is consistent with findings of previous studies demonstrating that arbovirus evolution is constrained to enable alternating infection of disparate vertebrate and arthropod hosts (25–27).
Additional work is needed to fully understand the roles that R. aegyptiacus bats and O. (R.) faini ticks play in maintaining KASV over time (9). Collection and separation of O. (R.) faini ticks according to life history stage, as well as experimental tick work, will be important in determining whether these ticks serve as amplification or reservoir hosts for KASV. Detection of infectious KASV in larval, nymphal, and adult stages of O. (R.) faini ticks would suggest that the virus is transtadially transmitted, and ticks serve as virus amplification hosts. Likewise, detection of KASV in nymphs and adults originating from KASV-artificially infected O. (R.) faini larvae would suggest the virus is transtadially transmitted from one generation to the next. Isolation of KASV in O. (R.) faini eggs found in nature or in larvae originating from KASV-artificially infected female ticks would indicate that the virus is transovarially transmitted and that ticks are reservoir hosts for the virus because a vertebrate host is not required for long-term virus survival. Similarly, a longitudinal ecologic investigation of KASV infection in R. aegyptiacus bats, as well as experimental KASV infection of captive bats, is critical in defining the relationship this virus has with its vertebrate host. Although KASV has previously been isolated from 2 wild-caught R. aegyptiacus bats (6), the isolation of actively replicating KASV over several days in experimentally infected R. aegyptiacus bats will confirm the ability of this bat species to serve as an amplification host for the virus. The detection of KASV in oral, rectal, or urogenital shedding collected from experimentally infected or wild-caught R. aegyptiacus bats will not only provide evidence that these bats are reservoir hosts of the virus but also indicate that they are capable of transmitting the virus to humans that encroach upon their habitat.
Although no human cases of KASV infection have been reported since the initial UVRI-associated cases described in 1977 (6), surveillance of populations at risk for KASV infection has never been conducted. Miners, herders, tourists, and researchers often frequent mines and caves occupied by large colonies of R. aegyptiacus bats. Entry into R. aegyptiacus bat–inhabited environments has been linked to the spillover of several pathogenic agents into the human population, including Marburg virus (11,28,29), Sosuga virus (30,31), and Borrelia spirochetes (13). Similarly, humans who enter environments occupied by R. aegyptiacus bats and O. (R.) faini ticks are likely to be at risk for KASV infection. Surveillance of these at-risk populations for evidence of active or past infection KASV infection is needed to determine the true burden of KASV infection in humans.
Dr. Schuh is a research scientist with the Viral Special Pathogens Branch, Division of High-Consequence Pathogens and Pathology, National Center for Emerging and Zoonotic Infectious Diseases, Centers for Disease Control and Prevention. Her research interests include investigating the enzootic transmission dynamics of tickborne and batborne viruses and identifying ecologic drivers underlying the spillover of zoonotic viruses into the human population.
Acknowledgments
We thank Margret Driciru and the Uganda Wildlife Authority rangers at Python Cave in Queen Elizabeth National Park for providing support during the tick collections and Shannon Whitmer for providing Illumina sequencing advice. We are grateful to Kenneth Plante, Scott Weaver, Nikos Vasilakis, Robert Tesh, and Thomas Ksiazek for providing the KASV Z-52963 isolate and KASV mouse immune ascitic fluid from the World Reference Center for Emerging Viruses and Arboviruses.
This work was funded in part through an interagency agreement with the Defense Threat Reduction Agency, HDTRA16-025-33037.
References
- Maes P, Adkins S, Alkhovsky SV, Avšič-Županc T, Ballinger MJ, Bente DA, et al. Taxonomy of the order Bunyavirales: second update 2018. Arch Virol. 2019;164:927–41. DOIPubMedGoogle Scholar
- Walker PJ, Widen SG, Firth C, Blasdell KR, Wood TG, Travassos da Rosa AP, et al. Genomic characterization of Yogue, Kasokero, Issyk-Kul, Keterah, Gossas, and Thiafora viruses: nairoviruses naturally infecting bats, shrews, and ticks. Am J Trop Med Hyg. 2015;93:1041–51. DOIPubMedGoogle Scholar
- Springer MS. Phylogenetics: bats united, microbats divided. Curr Biol. 2013;23:R999–1001. DOIPubMedGoogle Scholar
- Lei M, Dong D. Phylogenomic analyses of bat subordinal relationships based on transcriptome data. Sci Rep. 2016;6:27726. DOIPubMedGoogle Scholar
- Hutcheon JM, Kirsch JAW. A moveable face: deconstructing the Microchiroptera and a new classification of extant bats. Acta Chiropt. 2006;8:1–10. DOIGoogle Scholar
- Kalunda M, Mukwaya LG, Mukuye A, Lule M, Sekyalo E, Wright J, et al. Kasokero virus: a new human pathogen from bats (Rousettus aegyptiacus) in Uganda. Am J Trop Med Hyg. 1986;35:387–92. DOIPubMedGoogle Scholar
- Ishii A, Ueno K, Orba Y, Sasaki M, Moonga L, Hang’ombe BM, et al. A nairovirus isolated from African bats causes haemorrhagic gastroenteritis and severe hepatic disease in mice. Nat Commun. 2014;5:5651. DOIPubMedGoogle Scholar
- Mans BJ, Featherston J, Kvas M, Pillay K-A, de Klerk DG, Pienaar R, et al. Argasid and ixodid systematics: Implications for soft tick evolution and systematics, with a new argasid species list. Ticks Tick Borne Dis. 2019;10:219–40. DOIPubMedGoogle Scholar
- Hoogstraal H. Argasid and nuttalliellid ticks as parasites and vectors. Adv Parasitol. 1985;24:135–238. DOIPubMedGoogle Scholar
- Braack LEO. Arthropod inhabitants of a tropical cave ‘island’ environment provisioned by bats. Biol Conserv. 1989;48:77–84. DOIGoogle Scholar
- Amman BR, Carroll SA, Reed ZD, Sealy TK, Balinandi S, Swanepoel R, et al. Seasonal pulses of Marburg virus circulation in juvenile Rousettus aegyptiacus bats coincide with periods of increased risk of human infection. PLoS Pathog. 2012;8:
e1002877 . DOIPubMedGoogle Scholar - Schuh AJ, Amman BR, Apanaskevich DA, Sealy TK, Nichol ST, Towner JS. No evidence for the involvement of the argasid tick Ornithodoros faini in the enzootic maintenance of marburgvirus within Egyptian rousette bats Rousettus aegyptiacus. Parasit Vectors. 2016;9:128. DOIPubMedGoogle Scholar
- Qiu Y, Nakao R, Hang’ombe BM, Sato K, Kajihara M, Kanchela S, et al. Human borreliosis caused by a New World relapsing fever Borrelia-like organism in the Old World. Clin Infect Dis. 2019;69:107–12. DOIPubMedGoogle Scholar
- Sándor AD, Corduneanu A, Péter Á, Mihalca AD, Barti L, Csősz I, et al. Bats and ticks: host selection and seasonality of bat-specialist ticks in eastern Europe. Parasit Vectors. 2019;12:605. DOIPubMedGoogle Scholar
- Shope RE, Sather GE. Diagnostic procedures for viral, rickettsial and chlamydial infections. In: Schmidt NJ, Lennette EH, editors. Arboviruses. 5th ed. Washington (DC): American Public Health Association; 1979. p. 767–814.
- Shone SM, Dillon HJ, Hom SS, Delgado N. A novel real-time PCR assay for the speciation of medically important ticks. Vector Borne Zoonotic Dis. 2006;6:152–60. DOIPubMedGoogle Scholar
- Cowling DW, Gardner IA, Johnson WO. Comparison of methods for estimation of individual-level prevalence based on pooled samples. Prev Vet Med. 1999;39:211–25. DOIPubMedGoogle Scholar
- Andreassen A, Jore S, Cuber P, Dudman S, Tengs T, Isaksen K, et al. Prevalence of tick borne encephalitis virus in tick nymphs in relation to climatic factors on the southern coast of Norway. Parasit Vectors. 2012;5:177. DOIPubMedGoogle Scholar
- Quick J, Grubaugh ND, Pullan ST, Claro IM, Smith AD, Gangavarapu K, et al. Multiplex PCR method for MinION and Illumina sequencing of Zika and other virus genomes directly from clinical samples. Nat Protoc. 2017;12:1261–76. DOIPubMedGoogle Scholar
- Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 2010;59:307–21. DOIPubMedGoogle Scholar
- Lefort V, Longueville JE, Gascuel O. SMS: Smart Model Selection in PhyML. Mol Biol Evol. 2017;34:2422–4. DOIPubMedGoogle Scholar
- Stöver BC, Müller KF. TreeGraph 2: combining and visualizing evidence from different phylogenetic analyses. BMC Bioinformatics. 2010;11:7. DOIPubMedGoogle Scholar
- Walker PJ, Widen SG, Wood TG, Guzman H, Tesh RB, Vasilakis N. A global genomic characterization of nairoviruses identifies nine discrete genogroups with distinctive structural characteristics and host-vector associations. Am J Trop Med Hyg. 2016;94:1107–22. DOIPubMedGoogle Scholar
- Aguilar PV, Marciel de Souza W, Silvas JA, Wood T, Widen S, Fumagalli MJ, et al. Genetic characterization of the Patois serogroup (genus Orthobunyavirus; family Peribunyaviridae) and evidence that Estero Real virus is a member of the genus Orthonairovirus. Am J Trop Med Hyg. 2018;99:451–7. DOIPubMedGoogle Scholar
- Coffey LL, Vasilakis N, Brault AC, Powers AM, Tripet F, Weaver SC. Arbovirus evolution in vivo is constrained by host alternation. Proc Natl Acad Sci U S A. 2008;105:6970–5. DOIPubMedGoogle Scholar
- Vasilakis N, Deardorff ER, Kenney JL, Rossi SL, Hanley KA, Weaver SC. Mosquitoes put the brake on arbovirus evolution: experimental evolution reveals slower mutation accumulation in mosquito than vertebrate cells. PLoS Pathog. 2009;5:
e1000467 . DOIPubMedGoogle Scholar - Grubaugh ND, Rückert C, Armstrong PM, Bransfield A, Anderson JF, Ebel GD, et al. Transmission bottlenecks and RNAi collectively influence tick-borne flavivirus evolution. Virus Evol. 2016;2:
vew033 . DOIPubMedGoogle Scholar - Swanepoel R, Smit SB, Rollin PE, Formenty P, Leman PA, Kemp A, et al.; International Scientific and Technical Committee for Marburg Hemorrhagic Fever Control in the Democratic Republic of Congo. Studies of reservoir hosts for Marburg virus. Emerg Infect Dis. 2007;13:1847–51. DOIPubMedGoogle Scholar
- Towner JS, Amman BR, Sealy TK, Carroll SA, Comer JA, Kemp A, et al. Isolation of genetically diverse Marburg viruses from Egyptian fruit bats. PLoS Pathog. 2009;5:
e1000536 . DOIPubMedGoogle Scholar - Amman BR, Albariño CG, Bird BH, Nyakarahuka L, Sealy TK, Balinandi S, et al. A recently discovered pathogenic paramyxovirus, Sosuga virus, is present in Rousettus aegyptiacus fruit bats at multiple locations in Uganda. J Wildl Dis. 2015;51:774–9. DOIPubMedGoogle Scholar
- Albariño CG, Foltzer M, Towner JS, Rowe LA, Campbell S, Jaramillo CM, et al. Novel paramyxovirus associated with severe acute febrile disease, South Sudan and Uganda, 2012. Emerg Infect Dis. 2014;20:211–6. DOIPubMedGoogle Scholar
Figures
Cite This ArticleOriginal Publication Date: November 12, 2020
Table of Contents – Volume 26, Number 12—December 2020
| EID Search Options |
|---|
|
|
|
|
|
|
Please use the form below to submit correspondence to the authors or contact them at the following address:
Jonathan S. Towner, Centers for Disease Control and Prevention, 1600 Clifton Rd NE, Mailstop H18-B, Atlanta, GA 30329-4027, USA
Top