A provirus is a virus genome that is integrated into the DNA of a host cell. In the case of bacterial viruses (bacteriophages), proviruses are often referred to as prophages. However, proviruses are distinctly different from prophages and these terms should not be used interchangeably. Unlike prophages, proviruses do not excise themselves from the host genome when the host cell is stressed.

A reverse transcriptase (RT) is an enzyme used to convert RNA genome to DNA, a process termed reverse transcription. Reverse transcriptases are used by viruses such as HIV and hepatitis B to replicate their genomes, by retrotransposon mobile genetic elements to proliferate within the host genome, and by eukaryotic cells to extend the telomeres at the ends of their linear chromosomes. Contrary to a widely held belief, the process does not violate the flows of genetic information as described by the classical central dogma, as transfers of information from RNA to DNA are explicitly held possible.
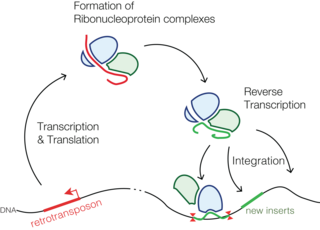
Retrotransposons are mobile elements which move in the host genome by converting their transcribed RNA into DNA through the reverse transcription. Thus, they differ from Class II transposable elements, or DNA transposons, in utilizing an RNA intermediate for the transposition and leaving the transposition donor site unchanged.
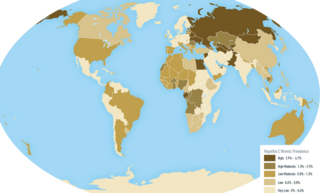
An oncovirus or oncogenic virus is a virus that can cause cancer. This term originated from studies of acutely transforming retroviruses in the 1950–60s, when the term "oncornaviruses" was used to denote their RNA virus origin. With the letters "RNA" removed, it now refers to any virus with a DNA or RNA genome causing cancer and is synonymous with "tumor virus" or "cancer virus". The vast majority of human and animal viruses do not cause cancer, probably because of longstanding co-evolution between the virus and its host. Oncoviruses have been important not only in epidemiology, but also in investigations of cell cycle control mechanisms such as the retinoblastoma protein.
Metaviridae is a family of viruses which exist as Ty3-gypsy LTR retrotransposons in a eukaryotic host's genome. They are closely related to retroviruses: members of the family Metaviridae share many genomic elements with retroviruses, including length, organization, and genes themselves. This includes genes that encode reverse transcriptase, integrase, and capsid proteins. The reverse transcriptase and integrase proteins are needed for the retrotransposon activity of the virus. In some cases, virus-like particles can be formed from capsid proteins.
Lentivirus is a genus of retroviruses that cause chronic and deadly diseases characterized by long incubation periods, in humans and other mammalian species. The genus includes the human immunodeficiency virus (HIV), which causes AIDS. Lentiviruses are distributed worldwide, and are known to be hosted in apes, cows, goats, horses, cats, and sheep as well as several other mammals.
Gammaretrovirus is a genus in the Retroviridae family. Example species are the murine leukemia virus and the feline leukemia virus. They cause various sarcomas, leukemias and immune deficiencies in mammals, reptiles and birds.
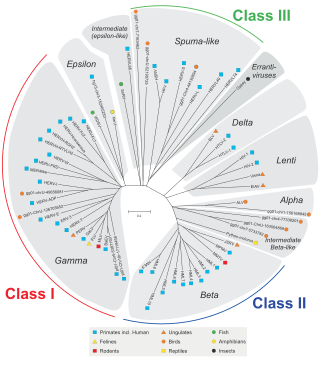
Endogenous retroviruses (ERVs) are endogenous viral elements in the genome that closely resemble and can be derived from retroviruses. They are abundant in the genomes of jawed vertebrates, and they comprise up to 5–8% of the human genome.
The genome and proteins of HIV (human immunodeficiency virus) have been the subject of extensive research since the discovery of the virus in 1983. "In the search for the causative agent, it was initially believed that the virus was a form of the Human T-cell leukemia virus (HTLV), which was known at the time to affect the human immune system and cause certain leukemias. However, researchers at the Pasteur Institute in Paris isolated a previously unknown and genetically distinct retrovirus in patients with AIDS which was later named HIV." Each virion comprises a viral envelope and associated matrix enclosing a capsid, which itself encloses two copies of the single-stranded RNA genome and several enzymes. The discovery of the virus itself occurred two years following the report of the first major cases of AIDS-associated illnesses.
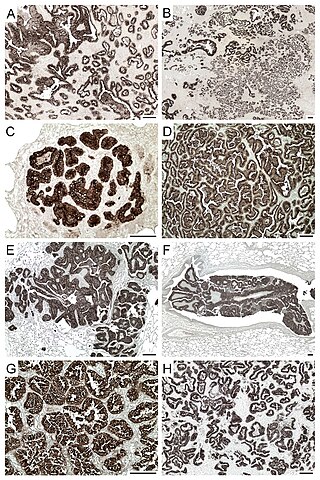
Jaagsiekte sheep retrovirus (JSRV) is a betaretrovirus which is the causative agent of a contagious lung cancer in sheep, called ovine pulmonary adenocarcinoma.
Rous sarcoma virus (RSV) is a retrovirus and is the first oncovirus to have been described. It causes sarcoma in chickens.
The murine leukemia viruses are retroviruses named for their ability to cause cancer in murine (mouse) hosts. Some MLVs may infect other vertebrates. MLVs include both exogenous and endogenous viruses. Replicating MLVs have a positive sense, single-stranded RNA (ssRNA) genome that replicates through a DNA intermediate via the process of reverse transcription.
Human foamy virus (HFV) is a retrovirus in the genus Spumavirus. The spumaviruses are complex and significantly different from the other six genera of retroviruses in several ways. The foamy viruses derive their name from the characteristic ‘foamy’ appearance of the cytopathic effect (CPE) induced in the cells. Foamy virus in humans occurs only as a result of zoonotic infection.
Pseudodiploid or pseudoploid refers to one of the essential components in viral reproduction. It means having two RNA genomes per virion but giving rise to only one DNA copy in infected cells.
Simian foamy virus (SFV) is a species of the genus Spumavirus that belongs to the family of Retroviridae. It has been identified in a wide variety of primates, including prosimians, New World and Old World monkeys, as well as apes, and each species has been shown to harbor a unique (species-specific) strain of SFV, including African green monkeys, baboons, macaques, and chimpanzees. As it is related to the more well-known retrovirus human immunodeficiency virus (HIV), its discovery in primates has led to some speculation that HIV may have been spread to the human species in Africa through contact with blood from apes, monkeys, and other primates, most likely through bushmeat-hunting practices.
Bovine immunodeficiency virus (BIV) is a retrovirus belonging to the genus Lentivirus. It is similar to the human immunodeficiency virus (HIV) and infects cattle. The cells primarily infected are lymphocytes and monocytes/macrophages.
Mason-Pfizer monkey virus (M-PMV), formerly Simian retrovirus (SRV), is a species of retroviruses that usually infect and cause a fatal immune deficiency in Asian macaques. The ssRNA virus appears sporadically in mammary carcinoma of captive macaques at breeding facilities which expected as the natural host, but the prevalence of this virus in feral macaques remains unknown. M-PMV was transmitted naturally by virus-containing body fluids, via biting, scratching, grooming, and fighting. Cross contaminated instruments or equipment (fomite) can also spread this virus among animals.
Human Endogenous Retrovirus-W (HERV-W) is a family of Human Endogenous Retroviruses (HERVs).
Bovine foamy virus (BFV) is a ss(+)RNA retrovirus that belongs to the genus spumaviridae. Spumaviruses differ from the other six members of family retroviridae, both structurally and in pathogenic nature. Spumaviruses derive their name from spuma the latin for "foam". The 'foam' aspect of 'foamy virus' comes from syncytium formation and the rapid vacuolization of infected cells, creating a 'foamy' appearance.
Feline foamy virus or Feline syncytial virus is a retrovirus and belongs to the family Retroviridae and the subfamily Spumaretrovirinae. It shares the genus Felispumavirus with only Puma feline foamy virus. There has been controversy on whether FeFV is nonpathogenic as the virus is generally asymptomatic in affected cats and does not cause disease. However, some changes in kidney and lung tissue have been observed over time in cats affected with FeFV, which may or may not be directly affiliated. This virus is fairly common and infection rates gradually increase with a cat's age. Study results from antibody examinations and PCR analysis have shown that over 70% of felines over 9 years old were seropositive for Feline foamy virus. Viral infections are similar between male and female domesticated cats whereas in the wild, more feral females cats are affected with FeFV.










