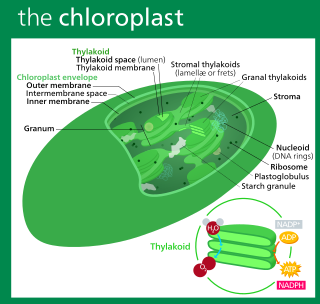
A chloroplast is a type of organelle known as a plastid that conducts photosynthesis mostly in plant and algal cells. Chloroplasts have a high concentration of chlorophyll pigments which capture the energy from sunlight and convert it to chemical energy and release oxygen. The chemical energy created is then used to make sugar and other organic molecules from carbon dioxide in a process called the Calvin cycle. Chloroplasts carry out a number of other functions, including fatty acid synthesis, amino acid synthesis, and the immune response in plants. The number of chloroplasts per cell varies from one, in some unicellular algae, up to 100 in plants like Arabidopsis and wheat.
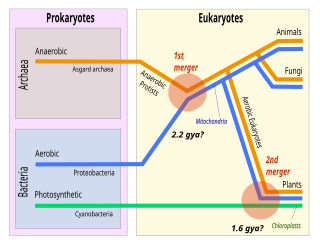
Symbiogenesis is the leading evolutionary theory of the origin of eukaryotic cells from prokaryotic organisms. The theory holds that mitochondria, plastids such as chloroplasts, and possibly other organelles of eukaryotic cells are descended from formerly free-living prokaryotes taken one inside the other in endosymbiosis. Mitochondria appear to be phylogenetically related to Rickettsiales bacteria, while chloroplasts are thought to be related to cyanobacteria.
The cryptomonads are a group of algae, most of which have plastids. They are traditionally considered a division of algae among phycologists, under the name of Cryptophyta. They are common in freshwater, and also occur in marine and brackish habitats. Each cell is around 10–50 μm in size and flattened in shape, with an anterior groove or pocket. At the edge of the pocket there are typically two slightly unequal flagella. Some may exhibit mixotrophy. They are classified as clade Cryptomonada, which is divided into two classes: heterotrophic Goniomonadea and phototrophic Cryptophyceae. The two groups are united under three shared morphological characteristics: presence of a periplast, ejectisomes with secondary scroll, and mitochondrial cristae with flat tubules. Genetic studies as early as 1994 also supported the hypothesis that Goniomonas was sister to Cryptophyceae. A study in 2018 found strong evidence that the common ancestor of Cryptomonada was an autotrophic protist.
A plastid is a membrane-bound organelle found in the cells of plants, algae, and some other eukaryotic organisms. Plastids are considered to be intracellular endosymbiotic cyanobacteria.

Horizontal gene transfer (HGT) or lateral gene transfer (LGT) is the movement of genetic material between organisms other than by the ("vertical") transmission of DNA from parent to offspring (reproduction). HGT is an important factor in the evolution of many organisms. HGT is influencing scientific understanding of higher-order evolution while more significantly shifting perspectives on bacterial evolution.

Nucleomorphs are small, vestigial eukaryotic nuclei found between the inner and outer pairs of membranes in certain plastids. They are thought to be vestiges of red and green algal nuclei that were engulfed by a larger eukaryote. Because the nucleomorph lies between two sets of membranes, nucleomorphs support the endosymbiotic theory and are evidence that the plastids containing them are complex plastids. Having two sets of membranes indicate that the plastid, a prokaryote, was engulfed by a eukaryote, an alga, which was then engulfed by another eukaryote, the host cell, making the plastid an example of secondary endosymbiosis.
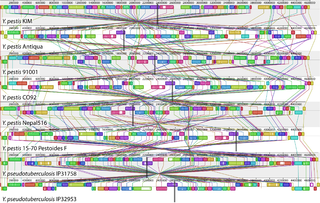
Comparative genomics is a branch of biological research that examines genome sequences across a spectrum of species, spanning from humans and mice to a diverse array of organisms from bacteria to chimpanzees. This large-scale holistic approach compares two or more genomes to discover the similarities and differences between the genomes and to study the biology of the individual genomes. Comparison of whole genome sequences provides a highly detailed view of how organisms are related to each other at the gene level. By comparing whole genome sequences, researchers gain insights into genetic relationships between organisms and study evolutionary changes. The major principle of comparative genomics is that common features of two organisms will often be encoded within the DNA that is evolutionarily conserved between them. Therefore, Comparative genomics provides a powerful tool for studying evolutionary changes among organisms, helping to identify genes that are conserved or common among species, as well as genes that give unique characteristics of each organism. Moreover, these studies can be performed at different levels of the genomes to obtain multiple perspectives about the organisms.

Sequence homology is the biological homology between DNA, RNA, or protein sequences, defined in terms of shared ancestry in the evolutionary history of life. Two segments of DNA can have shared ancestry because of three phenomena: either a speciation event (orthologs), or a duplication event (paralogs), or else a horizontal gene transfer event (xenologs).
Indel (insertion-deletion) is a molecular biology term for an insertion or deletion of bases in the genome of an organism. Indels ≥ 50 bases in length are classified as structural variants.

Hydnoroideae is a subfamily of parasitic flowering plants in the order Piperales. Traditionally, and as recently as the APG III system it given family rank under the name Hydnoraceae. It is now submerged in the Aristolochiaceae. It contains two genera, Hydnora and Prosopanche:

The Archaeplastida are a major group of eukaryotes, comprising the photoautotrophic red algae (Rhodophyta), green algae, land plants, and the minor group glaucophytes. It also includes the non-photosynthetic lineage Rhodelphidia, a predatorial (eukaryotrophic) flagellate that is sister to the Rhodophyta, and probably the microscopic picozoans. The Archaeplastida have chloroplasts that are surrounded by two membranes, suggesting that they were acquired directly through a single endosymbiosis event by phagocytosis of a cyanobacterium. All other groups which have chloroplasts, besides the amoeboid genus Paulinella, have chloroplasts surrounded by three or four membranes, suggesting they were acquired secondarily from red or green algae. Unlike red and green algae, glaucophytes have never been involved in secondary endosymbiosis events.

The Rafflesiaceae are a family of rare parasitic plants comprising 36 species in 3 genera found in the tropical forests of east and southeast Asia, including Rafflesia arnoldii, which has the largest flowers of all plants. The plants are endoparasites of vines in the genus Tetrastigma (Vitaceae) and lack stems, leaves, roots, and any photosynthetic tissue. They rely entirely on their host plants for both water and nutrients, and only then emerge as flowers from the roots or lower stems of the host plants.
The Mesostigmatophyceae are a class of basal green algae found in freshwater. In a narrow circumscription, the class contains a single genus, Mesostigma. AlgaeBase then places the order within its circumscription of Charophyta. A clade containing Chlorokybus and Spirotaenia may either be added, or treated as a sister, with Chlorokybus placed in a separate class, Chlorokybophyceae. When broadly circumscribed, Mesostigmatophyceae may be placed as sister to all other green algae, or as sister to all Streptophyta.

The UTC clade is a grouping of Chlorophyta.

Chloroplast DNA (cpDNA), also known as plastid DNA (ptDNA) is the DNA located in chloroplasts, which are photosynthetic organelles located within the cells of some eukaryotic organisms. Chloroplasts, like other types of plastid, contain a genome separate from that in the cell nucleus. The existence of chloroplast DNA was identified biochemically in 1959, and confirmed by electron microscopy in 1962. The discoveries that the chloroplast contains ribosomes and performs protein synthesis revealed that the chloroplast is genetically semi-autonomous. The first complete chloroplast genome sequences were published in 1986, Nicotiana tabacum (tobacco) by Sugiura and colleagues and Marchantia polymorpha (liverwort) by Ozeki et al. Since then, tens of thousands of chloroplast genomes from various species have been sequenced.
Jeffrey Donald Palmer is a Distinguished Professor of Biology at Indiana University Bloomington.
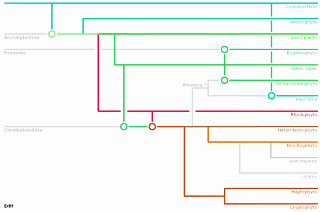
A plastid is a membrane-bound organelle found in plants, algae and other eukaryotic organisms that contribute to the production of pigment molecules. Most plastids are photosynthetic, thus leading to color production and energy storage or production. There are many types of plastids in plants alone, but all plastids can be separated based on the number of times they have undergone endosymbiotic events. Currently there are three types of plastids; primary, secondary and tertiary. Endosymbiosis is reputed to have led to the evolution of eukaryotic organisms today, although the timeline is highly debated.
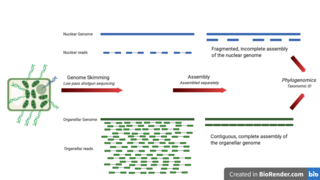
Genome skimming is a sequencing approach that uses low-pass, shallow sequencing of a genome, to generate fragments of DNA, known as genome skims. These genome skims contain information about the high-copy fraction of the genome. The high-copy fraction of the genome consists of the ribosomal DNA, plastid genome (plastome), mitochondrial genome (mitogenome), and nuclear repeats such as microsatellites and transposable elements. It employs high-throughput, next generation sequencing technology to generate these skims. Although these skims are merely 'the tip of the genomic iceberg', phylogenomic analysis of them can still provide insights on evolutionary history and biodiversity at a lower cost and larger scale than traditional methods. Due to the small amount of DNA required for genome skimming, its methodology can be applied in other fields other than genomics. Tasks like this include determining the traceability of products in the food industry, enforcing international regulations regarding biodiversity and biological resources, and forensics.

















