
In organic chemistry, a sulfide or thioether is an organosulfur functional group with the connectivity R−S−R' as shown on right. Like many other sulfur-containing compounds, volatile sulfides have foul odors. A sulfide is similar to an ether except that it contains a sulfur atom in place of the oxygen. The grouping of oxygen and sulfur in the periodic table suggests that the chemical properties of ethers and sulfides are somewhat similar, though the extent to which this is true in practice varies depending on the application.
Methanogens are anaerobic archaea that produce methane as a byproduct of their energy metabolism, i.e., catabolism. Methane production, or methanogenesis, is the only biochemical pathway for ATP generation in methanogens. All known methanogens belong exclusively to the domain Archaea, although some bacteria, plants, and animal cells are also known to produce methane. However, the biochemical pathway for methane production in these organisms differs from that in methanogens and does not contribute to ATP formation. Methanogens belong to various phyla within the domain Archaea. Previous studies placed all known methanogens into the superphylum Euryarchaeota. However, recent phylogenomic data have led to their reclassification into several different phyla. Methanogens are common in various anoxic environments, such as marine and freshwater sediments, wetlands, the digestive tracts of animals, wastewater treatment plants, rice paddy soil, and landfills. While some methanogens are extremophiles, such as Methanopyrus kandleri, which grows between 84 and 110°C, or Methanonatronarchaeum thermophilum, which grows at a pH range of 8.2 to 10.2 and a Na+ concentration of 3 to 4.8 M, most of the isolates are mesophilic and grow around neutral pH.
Methanogenesis or biomethanation is the formation of methane coupled to energy conservation by microbes known as methanogens. It is the fourth and final stage of anaerobic digestion. Organisms capable of producing methane for energy conservation have been identified only from the domain Archaea, a group phylogenetically distinct from both eukaryotes and bacteria, although many live in close association with anaerobic bacteria. The production of methane is an important and widespread form of microbial metabolism. In anoxic environments, it is the final step in the decomposition of biomass. Methanogenesis is responsible for significant amounts of natural gas accumulations, the remainder being thermogenic.
Methanotrophs are prokaryotes that metabolize methane as their source of carbon and chemical energy. They are bacteria or archaea, can grow aerobically or anaerobically, and require single-carbon compounds to survive.
Microbial metabolism is the means by which a microbe obtains the energy and nutrients it needs to live and reproduce. Microbes use many different types of metabolic strategies and species can often be differentiated from each other based on metabolic characteristics. The specific metabolic properties of a microbe are the major factors in determining that microbe's ecological niche, and often allow for that microbe to be useful in industrial processes or responsible for biogeochemical cycles.

Coenzyme B is a coenzyme required for redox reactions in methanogens. The full chemical name of coenzyme B is 7-mercaptoheptanoylthreoninephosphate. The molecule contains a thiol, which is its principal site of reaction.
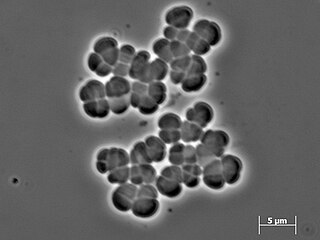
In taxonomy, the Methanosarcinaceae are a family of the Methanosarcinales.

The Wood–Ljungdahl pathway is a set of biochemical reactions used by some bacteria. It is also known as the reductive acetyl-coenzyme A (acetyl-CoA) pathway. This pathway enables these organisms to use hydrogen as an electron donor, and carbon dioxide as an electron acceptor and as a building block for biosynthesis.
Anaerobic oxidation of methane (AOM) is a methane-consuming microbial process occurring in anoxic marine and freshwater sediments. AOM is known to occur among mesophiles, but also in psychrophiles, thermophiles, halophiles, acidophiles, and alkophiles. During AOM, methane is oxidized with different terminal electron acceptors such as sulfate, nitrate, nitrite and metals, either alone or in syntrophy with a partner organism.
In enzymology, a 2-oxopropyl-CoM reductase (carboxylating) (EC 1.8.1.5) is an enzyme that catalyzes the chemical reaction

In enzymology, coenzyme-B sulfoethylthiotransferase, also known as methyl-coenzyme M reductase (MCR) or most systematically as 2-(methylthio)ethanesulfonate:N-(7-thioheptanoyl)-3-O-phosphothreonine S-(2-sulfoethyl)thiotransferase is an enzyme that catalyzes the final step in the formation of methane. It does so by combining the hydrogen donor coenzyme B and the methyl donor coenzyme M. Via this enzyme, most of the natural gas on earth was produced. Ruminants produce methane because their rumens contain methanogenic prokaryotes (Archaea) that encode and express the set of genes of this enzymatic complex.

Coenzyme F420 is a family of coenzymes involved in redox reactions in a number of bacteria and archaea. It is derived from coenzyme FO (7,8-didemethyl-8-hydroxy-5-deazariboflavin) and differs by having a oligoglutamyl tail attached via a 2-phospho-L-lactate bridge. F420 is so named because it is a flavin derivative with an absorption maximum at 420 nm.

Methanobrevibacter smithii is the predominant methanogenic archaeon in the microbiota of the human gut. M. smithii has a coccobacillus shape. It plays an important role in the efficient digestion of polysaccharides (complex sugars) by consuming the end products of bacterial fermentation (H2, CO2, acetate, and formate). M. smithii is a hydrogenotrophic methanogen that utilizes hydrogen by combining it with carbon dioxide to form methane. The removal of hydrogen by M. smithii is thought to allow an increase in the extraction of energy from nutrients by shifting bacterial fermentation to more oxidized end products.

F430 is the cofactor (sometimes called the coenzyme) of the enzyme methyl coenzyme M reductase (MCR). MCR catalyzes the reaction EC 2.8.4.1 that releases methane in the final step of methanogenesis:
(Methyl-Co methylamine-specific corrinoid protein):coenzyme M methyltransferase is an enzyme with systematic name methylated monomethylamine-specific corrinoid protein:coenzyme M methyltransferase. This enzyme catalyses the following chemical reaction
Methylamine-corrinoid protein Co-methyltransferase is an enzyme with the systematic name monomethylamine:5-hydroxybenzimidazolylcobamide Co-methyltransferase. This enzyme catalyzes the following chemical reaction:

Methanococcus maripaludis is a species of methanogenic archaea found in marine environments, predominantly salt marshes. M. maripaludis is a non-pathogenic, gram-negative, weakly motile, non-spore-forming, and strictly anaerobic mesophile. It is classified as a chemolithoautotroph. This archaeon has a pleomorphic coccoid-rod shape of 1.2 by 1.6 μm, in average size, and has many unique metabolic processes that aid in survival. M. maripaludis also has a sequenced genome consisting of around 1.7 Mbp with over 1,700 identified protein-coding genes. In ideal conditions, M. maripaludis grows quickly and can double every two hours.
The sulfate-methane transition zone (SMTZ) is a zone in oceans, lakes, and rivers typically found below the sediment surface in which sulfate and methane coexist. The formation of a SMTZ is driven by the diffusion of sulfate down the sediment column and the diffusion of methane up the sediments. At the SMTZ, their diffusion profiles meet and sulfate and methane react with one another, which allows the SMTZ to harbor a unique microbial community whose main form of metabolism is anaerobic oxidation of methane (AOM). The presence of AOM marks the transition from dissimilatory sulfate reduction to methanogenesis as the main metabolism utilized by organisms.

C1 chemistry is the chemistry of one-carbon molecules. Although many compounds and ions contain only one carbon, stable and abundant C-1 feedstocks are the focus of research. Four compounds are of major industrial importance: methane, carbon monoxide, carbon dioxide, and methanol. Technologies that interconvert these species are often used massively to match supply to demand.

Hydroxyarchaeol is a core lipid unique to archaea, similar to archaeol, with a hydroxide functional group at the carbon-3 position of one of its ether side chains. It is found exclusively in certain taxa of methanogenic archaea, and is a common biomarker for methanogenesis and methane-oxidation. Isotopic analysis of hydroxyarchaeol can be informative about the environment and substrates for methanogenesis.












