Volume 26, Number 8—August 2020
Research Letter
Heartland Virus in Lone Star Ticks, Alabama, USA
Abstract
We detected Heartland virus (HRTV) in lone star nymphs collected in 2018 in northern Alabama, USA. Real-time reverse transcription PCR selective for the small segment of the HRTV genome and confirmatory sequencing of positive samples showed high identity with HRTV strains sequenced from Tennessee and Missouri.
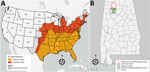
Heartland virus (HRTV) is an emerging pathogenic phlebovirus first identified in the United States in 2009 and now reported in 15 states (1,2). Nymphal lone star ticks (Amblyomma americanum) are considered the primary vectors of HRTV, and a variety of domestic and endemic mammalian species are potential amplification hosts of this virus (2,3). Although A. americanum ticks are well-established throughout the eastern, southeastern, and midwestern United States, their range is expanding northward and westward, most likely because of increased host availability and abundance, changes in environmental and climatic conditions, and adaptive genetic variation (Figure, panel A) (4). We tested for HRTV in A. americanum ticks collected in Alabama, USA, a state within the range of this vector where HRTV has not been documented previously from ticks.
From June 1, 2018, through August 31, 2018, we collected ticks as previously described (5) in the William B. Bankhead National Forest, Alabama (34.2270°N, 87.3461°W; Figure, panel B). In preparation for pathogen screening, we separated ticks into pools. Nymph tick pools ranged from 1 to 5 tick(s) of the same species per pool. We screened adult ticks individually (i.e., 1 adult tick per pool) (Appendix Table). We did not include larvae in pathogen screening. We used molecular methods to extract viral RNA and detect the small (S) segment of the HRTV genome using the HRTV-4 primer and probe set (6) in tick pools (Appendix Table). We sequenced HRTV-4–positive samples using the Ion Torrent Personal Genomic Machine system (Life Technologies, https://www.thermofisher.com) at the Centers for Disease Control and Prevention (CDC; Fort Collins, CO, USA) as described previously (7). We obtained sequences of the HTRV S segment of other HRTV samples and strains from the GenBank database, and aligned sequences using the MUSCLE alignment tool (https://www.ebi.ac.uk/Tools/msa/muscle) in MEGA software (8). We also included a closely related severe fever with thrombocytopenia syndrome virus isolate from the GenBank database as an outgroup for this analysis. We used a maximum-likelihood tree approach with 1,000 bootstrap replications to generate the genetic relationships between the Alabama samples and the other HRTV samples available through the GenBank database.
We collected 964 ticks, of which 921 were A. americanum (872 nymphs, 22 adult males, and 27 adult females) and 43 were Dermacentor variabilis (20 adult males and 23 adult females). We tested the ticks in 337 screened tick pools (Appendix Table). We amplified HRTV-4 from 5 pools that each contained 4 A. americanum nymphs. Therefore, the bias-corrected maximum-likelihood estimate of the infection rate (9) in questing A. americanum nymphs collected from the William B. Bankhead National Forest during 2018 was 0.58 (95% CI 0.21–1.27) and minimum infection rate (9) was 0.57 (95% CI 0.07–1.07) per 100 ticks screened on the basis of 235 nymph pools tested. To confirm results, we randomly selected homogenate from 3 of 5 HRTV-4–positive pools and submitted 3 individual RNA samples for sequencing at CDC. Sequencing RNA directly from tick homogenate confirmed HRTV in each of the 3 pools. Although we did not obtain whole-genome sequences, we identified partial coding sequences of all 3 HRTV segments in each pool. Maximum-likelihood phylogenetic inference of 730 nt of the S segment confirmed the BLAST analysis (https://blast.ncbi.nlm.nih.gov/Blast.cgi) and placed the generated HRTV S segment (submitted under GenBank accession no. MT052710) in a well-supported clade with HRTV strains previously described in Missouri and Tennessee (Appendix Figure).
Our findings of HRTV in A. americanum ticks in Alabama update knowledge of the virus’ distribution in the United States (Figure, panel A). Our findings also suggest A. americanum nymphs are the primary vectors of HRTV. As the geographic range of A. americanum continues to expand, we encourage enhanced surveillance and screening for HRTV to provide a more accurate and up-to-date understanding of where this tickborne virus probably occurs in the United States. Treatment for HRTV infection is limited to supportive care only; clinical data from the southeastern United States show that Heartland virus has a 10% death rate (10). Surveillance of HRTV in tick vector species is necessary to gain a comprehensive understanding of the environmental determinants that may put humans at risk for encountering the vector and to identify the geographic host range (both current and potential) of this emerging pathogen in the United States.
Mr. Newman is a PhD candidate in biological sciences at Tennessee State University, Nashville, Tennessee. His interests are landscape genomics, the function of biodiversity in vectorborne disease ecology, and application of One Health concepts to wildlife parasitology.
Acknowledgments
We thank Allison Cochran for forest stand access and logistical support at the William B. Bankhead National Forest and Stuart Robertson and Alexander Binney for field sampling assistance. We also thank Amy Lambert for enabling collaboration and sample sequencing for this research, Javier Monzón for permission to recreate his A. americanum range map, and Suzanne Wade for help creating the maps.
This study was supported by the US Department Agriculture, National Institute of Food and Agriculture 1890 Institution Teaching, Research and Extension Capacity Building Grants Program.
References
- Centers for Disease Control and Prevention, National Center for Emerging and Zoonotic Infectious Diseases, Division of Vector-Borne Diseases. Heartland virus disease (Heartland): statistics & maps. 2018 Oct 22 [cited 2020 Feb 11]. https://www.cdc.gov/heartland-virus/statistics/index.html
- Riemersma KK, Komar N. Heartland virus neutralizing antibodies in vertebrate wildlife, United States, 2009–2014. Emerg Infect Dis. 2015;21:1830–3. DOIPubMedGoogle Scholar
- Bosco-Lauth AM, Panella NA, Root JJ, Gidlewski T, Lash RR, Harmon JR, et al. Serological investigation of heartland virus (Bunyaviridae: Phlebovirus) exposure in wild and domestic animals adjacent to human case sites in Missouri 2012-2013. Am J Trop Med Hyg. 2015;92:1163–7. DOIPubMedGoogle Scholar
- Monzón JD, Atkinson EG, Henn BM, Benach JL. Population and evolutionary genomics of Amblyomma americanum, an expanding arthropod disease vector. Genome Biol Evol. 2016;8:1351–60. DOIPubMedGoogle Scholar
- Newman BC, Sutton WB, Wang Y, Schweitzer CJ, Moncayo AC, Miller BT. A standardized method for the construction of a tick drag/flag sampling approach and evaluation of sampling efficacy. Exp Appl Acarol. 2019;79:433–46. DOIPubMedGoogle Scholar
- Savage HM, Godsey MS Jr, Lambert A, Panella NA, Burkhalter KL, Harmon JR, et al. First detection of heartland virus (Bunyaviridae: Phlebovirus) from field collected arthropods. Am J Trop Med Hyg. 2013;89:445–52. DOIPubMedGoogle Scholar
- Hughes HR, Russell BJ, Lambert AJ. First complete genome sequences of Anopheles A virus of the genus Orthobunyavirus. Genome Announc. 2017;5:e01331–17. DOIPubMedGoogle Scholar
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 2018;35:1547–9. DOIPubMedGoogle Scholar
- Biggerstaff BJ. PooledInfRate, version 4.0: a Microsoft Office add-in to compute prevalence estimates from pooled samples. Fort Collins (CO): Centers for Disease Control and Prevention; 2009 [cited 2020 Apr 16]. https://www.cdc.gov/westnile/resourcepages/mosqsurvsoft.html
- Hevey MA, O’Halloran JA, Jagger BW, Staples JE, Lambert AJ, Panella AJ, et al. Heartland virus infection in a heart transplant recipient from the Heartland. Transpl Infect Dis. 2019;21:
e13098 . DOIPubMedGoogle Scholar
Figure
Cite This ArticleOriginal Publication Date: July 15, 2020
Table of Contents – Volume 26, Number 8—August 2020
| EID Search Options |
|---|
|
|
|
|
|
|
Please use the form below to submit correspondence to the authors or contact them at the following address:
Brent C. Newman, Department of Agricultural and Environmental Sciences, 3500 John A. Merritt Blvd, Nashville, TN 37209, USA
Top