Volume 24, Number 12—December 2018
Dispatch
Candidatus Cryptoplasma Associated with Green Lizards and Ixodes ricinus Ticks, Slovakia, 2004–2011
Abstract
During 2004–2011, we collected green lizards and Ixodes ricinus ticks in Slovak Karst National Park in Slovakia; 90% (36/40) of lizards and 37% of ticks removed from lizards were infected with family Anaplasmataceae bacteria. Only Candidatus Cryptoplasma sp. REP (reptile) was identified in these samples. Green lizards transmit this bacterium.
The family Anaplasmataceae (Rickettsiales; Alphaproteobacteria) comprises bacteria that are able to invade and infect their vertebrate host’s blood cells, bone marrow–derived phagocytic cells, and endothelial cells; these bacteria can also infect cells of insects, helminths, and arthropod reproductive tissues (1–3). Tickborne family members include bacteria of Anaplasma, Ehrlichia, Candidatus Neoehrlichia sp., and Candidatus Cryptoplasma californiense (4).
Although reptiles play a role as hosts for ixodid and argasid ticks, their role in maintaining tickborne Anaplasmataceae bacteria in the environment has not been described. Nieto et al. (5) suggested that lizards and snakes in the far western part of the United States could become exposed to Anaplasma phagocytophilum when fed on by infected ticks. Moreover, Rejmanek et al. detected 2 highly dissimilar strains of A. phagocytophilum in the same lizard species (6). In Europe, an undescribed Anaplasma sp. was detected in Ixodes ricinus ticks feeding on sand lizards and sand lizard blood samples (7,8). In our study, we sought to confirm these previous findings by determining whether family Anaplasmataceae bacteria were present in lizards and their feeding ticks in Slovakia.
We conducted this study in the Slovak Karst National Park in Slovakia (48°36′N, 20°52′E) during 2004–2011. We carried out lizard capture and sample collections with official permits (6103/2007-2.1 and 5498/2011-2.2) issued by the Ministry of Environment of the Slovak Republic. We captured 103 green lizards (Lacerta viridis) and collected blood from 40 (30 males and 10 females). We collected 235 I. ricinus ticks (118 larvae and 117 nymphs) from 63 green lizards and 271 questing I. ricinus ticks (132 nymphs, 76 males, and 63 females) from the same area and immediately stored them in 70% ethanol.
We isolated DNA from lizard blood using a DNeasy Blood & Tissue Kit (QIAGEN, Hilden Germany) and isolated DNA from ticks by alkaline hydrolysis. We performed PCR amplification in 25-μL (total) reaction mixtures using the MasterTaq DNA Polymerase Kit (Eppendorf AG, Hamburg, Germany). We amplified sequences using the primer combinations EHR747 plus EHR521 or fD1 plus rP2 (9), which spanned almost the entire 16S rRNA sequence (Table 1). We examined the ≈250-bp gene fragment of 16S rRNA by single-strand conformation polymorphism (SSCP) analysis to determine Anaplasmataceae species type (10). We performed SSCP analysis following the protocol of Derdakova et al. (11). We ran positive control samples A. phagocytophilum, A. ovis, Wolbachia sp., and Candidatus N. mikurensis with each reaction. We purified the PCR products obtained using the GenElute PCR Clean-Up Kit (Sigma-Aldrich, Buchs, Switzerland) and sequenced both strands. We edited variants obtained in this study (1,410 bp) using MEGA 4.0.2 (https://megasoftware.net/) and checked by eye. We made comparisons to sequences in GenBank with BLASTn 2.2.26 (https://pods.iplantcollaborative.org/wiki/display/DEapps/Blastn-2.2.26). For phylogenetic analysis of our variant (GenBank accession no. MG924904), we aligned 17 related sequences obtained from the GenBank database and constructed a phylogenetic tree using the Bayesian inference method (12).
We examined all blood and ticks collected for the presence of family Anaplasmataceae bacteria. Anaplasmataceae family members were present in 36 lizards (28 males and 8 females). Of the ticks removed from lizards, 87 (37%) were infected, and of questing ticks, 18 (6.6%) were infected (Table 2).
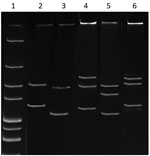
Denatured and electrophoresed PCR products from samples demonstrated several SSCP profiles, of which 1 was clearly distinguishable from the profiles of the Anaplasmataceae species used as controls (Figure 1). We detected this unique profile in all lizard blood samples, all ticks feeding on lizards, and some questing ticks. We sequenced representatives of this unidentified SSCP profile (≈247 bp; GenBank accession nos. KY031322–3) and compared them with DNA fragments in the GenBank database. The closest related (99% identity) 16S rRNA sequences were from uncultured Anaplasma sp. isolates from questing I. ricinus ticks from Morocco (GenBank accession no. AY672415), Tunisia (GenBank accession no. AY672420), and France (GenBank accession no. GU734325). Sequencing of a longer (1,410-bp) fragment of the 16S rRNA gene revealed 99.1% similarity with the Candidatus C. californiense isolate from I. pacificus ticks in California (Figure 2). The 16S rRNA sequence obtained in this study was found to share a maximum of 94% identity with A. phagocytophilum Norway variant 2 (GenBank accession no. CP015376). The phylogenetic tree we constructed using 16S rRNA gene sequences showed that the reptile-associated Candidatus Cryptoplasma sp. REP (reptile) clustered in a separate branch with Candidatus C. californiense, indicating the isolate represents a lineage distinct from other known Anaplasmataceae species (e.g., A. phagocytophilum, A. marginale, A. platys, Ehrlichia muris, E. chaffeensis, and E. ewingii).
The role of ectotherm animals, especially lizards, in the maintenance of vectorborne pathogens is not clear. The interaction between reptiles and Anaplasmataceae family members has only been investigated in a few studies. Our findings expand knowledge on this research topic. Only limited information about the reptile–Anaplasma relationship exists. Ekner et al. suggested that sand lizards could potentially serve as a reservoir host for species of the Anaplasmataceae family when she discovered that ticks collected from these lizards in Poland were infected with Anaplasma-like pathogens (8). Although A. phagocytophilum might be transmitted by reptiles to a limited extent (5), the Anaplasma-like species detected in reptiles could also be a novel species, as suggested by Rejmanek et al. (6).
Despite the fact that lizards are exposed to a number of family Anaplasmataceae bacteria through infected ticks, our findings suggest that, except for Canditatus Cryptoplasma sp. REP, green lizards do not acquire infections with these species. In short, we detected Canditatus Cryptoplasma sp. REP in 90% of examined lizards, 37% of ticks feeding on lizards, and 6.6% of questing ticks in localities with lizards.
On the basis of our results, we cautiously speculate that Canditatus Cryptoplasma sp. REP is selected for and other genospecies selected against in ticks feeding on lizards. The Canditatus Cryptoplasma sp. REP variant had a high homology (100%) with a sequence obtained from an Apodemus agrarius mouse from Slovakia (13), which indicates that rodents or other mammals might also become infected with this bacterium and contribute (to a lesser extent) to the circulation of these bacteria in nature.
In conclusion, we found a yet to be named species of Canditatus Cryptoplasma sp. (Canditatus Cryptoplasma sp. REP) in questing I. ricinus ticks, I. ricinus ticks collected from and feeding on green lizards, and the blood of green lizards in Slovakia. These results indicate that green lizards serve as an intermediate host for this bacterium and that lizards can influence the enzootic maintenance and circulation of bacteria in the environment. However, other hosts besides reptiles could be involved in the Canditatus Cryptoplasma sp. REP lifecycle as well, though probably to a lesser extent.
Dr. Kočíková is a postdoctoral researcher at the Department of Vector-Borne Diseases, Institute of Parasitology, Slovak Academy of Sciences, Košice, Slovakia. Her research interests include ecology and epidemiology of ticks and tickborne pathogens, the role of reptiles in transmission of vectorborne microorganisms, and molecular diversity of emerging parasitic and infectious disease agents in light of global changes.
Acknowledgment
This study was conducted within the framework of the project Environmental Protection against Parasitozoonoses under the Influence of Global Climate and Social Changes (project no. 26220220116), which is supported by the Research and Development Operating Program funded by the European Fund for Regional Development (0.4). Additional support was provided by the Scientific Grant Agency of the Ministry of Education of the Slovak Republic, Slovak Academy of Sciences VEGA 2/0113/18 (to I.M.), and EurNegVec COST Action TD1303. We thank the Fulbright Foundation (USA, to V.A.C.) for support.
References
- Dumler JS. Anaplasma and Ehrlichia infection. Ann N Y Acad Sci. 2005;1063:361–73. DOIPubMedGoogle Scholar
- Rikihisa Y. Mechanisms to create a safe haven by members of the family Anaplasmataceae. Ann N Y Acad Sci. 2003;990:548–55. DOIPubMedGoogle Scholar
- Rar V, Golovljova I. Anaplasma, Ehrlichia, and “Candidatus Neoehrlichia” bacteria: pathogenicity, biodiversity, and molecular genetic characteristics, a review. Infect Genet Evol. 2011;11:1842–61. DOIPubMedGoogle Scholar
- Eshoo MW, Carolan HE, Massire C, Chou DM, Crowder CD, Rounds MA, et al. Survey of Ixodes pacificus ticks in California reveals a diversity of microorganisms and a novel and widespread Anaplasmataceae species. PLoS One. 2015;10:e0135828. DOIPubMedGoogle Scholar
- Nieto NC, Foley JE, Bettaso J, Lane RS. Reptile infection with Anaplasma phagocytophilum, the causative agent of granulocytic anaplasmosis. J Parasitol. 2009;95:1165–70. DOIPubMedGoogle Scholar
- Rejmanek D, Bradburd G, Foley J. Molecular characterization reveals distinct genospecies of Anaplasma phagocytophilum from diverse North American hosts. J Med Microbiol. 2012;61:204–12. DOIPubMedGoogle Scholar
- Tijsse-Klasen E, Fonville M, Reimerink JHJ, Spitzen-van der Sluijs A, Sprong H. Role of sand lizards in the ecology of Lyme and other tick-borne diseases in the Netherlands. Parasit Vectors. 2010;3:42. DOIPubMedGoogle Scholar
- Ekner A, Dudek K, Sajkowska Z, Majláthová V, Majláth I, Tryjanowski P. Anaplasmataceae and Borrelia burgdorferi sensu lato in the sand lizard Lacerta agilis and co-infection of these bacteria in hosted Ixodes ricinus ticks. Parasit Vectors. 2011;4:182. DOIPubMedGoogle Scholar
- Weisburg WG, Barns SM, Pelletier DA, Lane DJ. 16S ribosomal DNA amplification for phylogenetic study. J Bacteriol. 1991;173:697–703. DOIPubMedGoogle Scholar
- Pancholi P, Kolbert CP, Mitchell PD, Reed KD Jr, Dumler JS, Bakken JS, et al. Ixodes dammini as a potential vector of human granulocytic ehrlichiosis. J Infect Dis. 1995;172:1007–12. DOIPubMedGoogle Scholar
- Derdáková M, Beati L, Pet’ko B, Stanko M, Fish D. Genetic variability within Borrelia burgdorferi sensu lato genospecies established by PCR-single-strand conformation polymorphism analysis of the rrfA-rrlB intergenic spacer in ixodes ricinus ticks from the Czech Republic. Appl Environ Microbiol. 2003;69:509–16. DOIPubMedGoogle Scholar
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol. 2013;30:2725–9. DOIPubMedGoogle Scholar
- Stefancíková A, Derdáková M, Lencáková D, Ivanová R, Stanko M, Cisláková L, et al. Serological and molecular detection of Borrelia burgdorferi sensu lato and Anaplasmataceae in rodents. Folia Microbiol (Praha). 2008;53:493–9. DOIPubMedGoogle Scholar
Figures
Tables
Cite This ArticleOriginal Publication Date: October 25, 2018
Table of Contents – Volume 24, Number 12—December 2018
| EID Search Options |
|---|
|
|
|
|
|
|

Please use the form below to submit correspondence to the authors or contact them at the following address:
Viktória Majláthová, Institute of Parasitology, Slovak Academy of Sciences, Hlinkova 3, 040 01 Košice, Slovakia
Top