
Protein primary structure is the linear sequence of amino acids in a peptide or protein. By convention, the primary structure of a protein is reported starting from the amino-terminal (N) end to the carboxyl-terminal (C) end. Protein biosynthesis is most commonly performed by ribosomes in cells. Peptides can also be synthesized in the laboratory. Protein primary structures can be directly sequenced, or inferred from DNA sequences.

In molecular biology, post-translational modification (PTM) is the covalent process of changing proteins following protein biosynthesis. PTMs may involve enzymes or occur spontaneously. Proteins are created by ribosomes, which translate mRNA into polypeptide chains, which may then change to form the mature protein product. PTMs are important components in cell signalling, as for example when prohormones are converted to hormones.

Lipid-anchored proteins are proteins located on the surface of the cell membrane that are covalently attached to lipids embedded within the cell membrane. These proteins insert and assume a place in the bilayer structure of the membrane alongside the similar fatty acid tails. The lipid-anchored protein can be located on either side of the cell membrane. Thus, the lipid serves to anchor the protein to the cell membrane. They are a type of proteolipids.
A signal peptide is a short peptide present at the N-terminus of most newly synthesized proteins that are destined toward the secretory pathway. These proteins include those that reside either inside certain organelles, secreted from the cell, or inserted into most cellular membranes. Although most type I membrane-bound proteins have signal peptides, most type II and multi-spanning membrane-bound proteins are targeted to the secretory pathway by their first transmembrane domain, which biochemically resembles a signal sequence except that it is not cleaved. They are a kind of target peptide.
The N-terminus (also known as the amino-terminus, NH2-terminus, N-terminal end or amine-terminus) is the start of a protein or polypeptide, referring to the free amine group (-NH2) located at the end of a polypeptide. Within a peptide, the amine group is bonded to the carboxylic group of another amino acid, making it a chain. That leaves a free carboxylic group at one end of the peptide, called the C-terminus, and a free amine group on the other end called the N-terminus. By convention, peptide sequences are written N-terminus to C-terminus, left to right (in LTR writing systems). This correlates the translation direction to the text direction, because when a protein is translated from messenger RNA, it is created from the N-terminus to the C-terminus, as amino acids are added to the carboxyl end of the protein.
In chemistry, acylation is a broad class of chemical reactions in which an acyl group is added to a substrate. The compound providing the acyl group is called the acylating agent. The substrate to be acylated and the product include the following:

Prenylation is the addition of hydrophobic molecules to a protein or a biomolecule. It is usually assumed that prenyl groups (3-methylbut-2-en-1-yl) facilitate attachment to cell membranes, similar to lipid anchors like the GPI anchor, though direct evidence of this has not been observed. Prenyl groups have been shown to be important for protein–protein binding through specialized prenyl-binding domains.
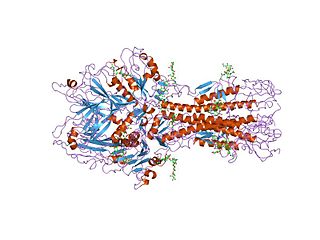
Hemagglutinin esterase (HEs) is a glycoprotein that certain enveloped viruses possess and use as an invading mechanism. HEs helps in the attachment and destruction of certain sialic acid receptors that are found on the host cell surface. Viruses that possess HEs include influenza C virus, toroviruses, and coronaviruses of the subgenus Embecovirus. HEs is a dimer transmembrane protein consisting of two monomers, each monomer is made of three domains. The three domains are: membrane fusion, esterase, and receptor binding domains.
Chemical modification refers to a number of various processes involving the alteration of the chemical constitution or structure of molecules.
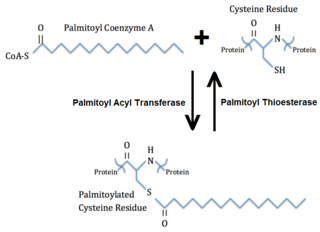
In molecular biology, palmitoylation is the covalent attachment of fatty acids, such as palmitic acid, to cysteine (S-palmitoylation) and less frequently to serine and threonine (O-palmitoylation) residues of proteins, which are typically membrane proteins. The precise function of palmitoylation depends on the particular protein being considered. Palmitoylation enhances the hydrophobicity of proteins and contributes to their membrane association. Palmitoylation also appears to play a significant role in subcellular trafficking of proteins between membrane compartments, as well as in modulating protein–protein interactions.
Group-specific antigen, or gag, is the polyprotein that contains the core structural proteins of an Ortervirus. It was named as such because scientists used to believe it was antigenic. Now it is known that it makes up the inner shell, not the envelope exposed outside. It makes up all the structural units of viral conformation and provides supportive framework for mature virion.

Glycylpeptide N-tetradecanoyltransferase 2 known also as N-myristoyltransferase, is an enzyme that in humans is encoded by the NMT2 gene.

In enzymology, a glycylpeptide N-tetradecanoyltransferase is an enzyme that catalyzes the chemical reaction

Glycylpeptide N-tetradecanoyltransferase 1 also known as myristoyl-CoA:protein N-myristoyltransferase 1 (NMT-1) is an enzyme that in humans is encoded by the NMT1 gene. It belongs to the protein N-terminal methyltransferase and glycylpeptide N-tetradecanoyltransferase family of enzymes.

Myristoylated alanine-rich C-kinase substrate is a protein that in humans is encoded by the MARCKS gene. It plays important roles in cell shape, cell motility, secretion, transmembrane transport, regulation of the cell cycle, and neural development. Recently, MARCKS has been implicated in the exocytosis of a number of vesicles and granules such as mucin and chromaffin. It is also the name of a protein family, of which MARCKS is the most studied member. They are intrinsically disordered proteins, with an acidic pH, with high proportions of alanine, glycine, proline, and glutamic acid. They are membrane-bound through a lipid anchor at the N-terminus, and a polybasic domain in the middle. They are regulated by Ca2+/calmodulin and protein kinase C. In their unphosphorylated form, they bind to actin filaments, causing them to crosslink, and sequester acidic membrane phospholipids such as PIP2.
BAALC is a gene that codes for the brain and acute leukemia cytoplasmic protein. The official symbol (BAALC) and official name is maintained by the HGNC. The function of BAALC is not fully understood yet, but has been suggested to have synaptic roles involving the post synaptic lipid raft. Lipid rafts are microdomains that are enriched with cholesterol and sphingolipids, lipid raft functions include membrane trafficking, signal processing, and regulation of the actin cytoskeleton. The postsynaptic lipid raft possesses many proteins and is one of the major sites for signal processing and postsynaptic density (PSD). Along with its involvement in the post synaptic lipid rafts, BAALC expression has been associated with Acute Lymphoblastic Leukemia and Acute Myeloid Leukemia.
Src kinase family is a family of non-receptor tyrosine kinases that includes nine members: Src, Yes, Fyn, and Fgr, forming the SrcA subfamily, Lck, Hck, Blk, and Lyn in the SrcB subfamily, and Frk in its own subfamily. Frk has homologs in invertebrates such as flies and worms, and Src homologs exist in organisms as diverse as unicellular choanoflagellates, but the SrcA and SrcB subfamilies are specific to vertebrates. Src family kinases contain six conserved domains: a N-terminal myristoylated segment, a SH2 domain, a SH3 domain, a linker region, a tyrosine kinase domain, and C-terminal tail.
Protein methylation is a type of post-translational modification featuring the addition of methyl groups to proteins. It can occur on the nitrogen-containing side-chains of arginine and lysine, but also at the amino- and carboxy-termini of a number of different proteins. In biology, methyltransferases catalyze the methylation process, activated primarily by S-adenosylmethionine. Protein methylation has been most studied in histones, where the transfer of methyl groups from S-adenosyl methionine is catalyzed by histone methyltransferases. Histones that are methylated on certain residues can act epigenetically to repress or activate gene expression.
A proteolipid is a protein covalently linked to lipid molecules, which can be fatty acids, isoprenoids or sterols. The process of such a linkage is known as protein lipidation, and falls into the wider category of acylation and post-translational modification. Proteolipids are abundant in brain tissue, and are also present in many other animal and plant tissues. They include ghrelin, a peptide hormone associated with feeding. Many proteolipids have bound fatty acid chains, which often provide an interface for interacting with biological membranes and act as lipidons that direct proteins to specific zones.

O-Octadecylhydroxylamine (ODHA) is a white solid organic compound with the formula C18H39NO. ODHA is a noncanonical lipid, which contains a saturated alkyl tail and an aminooxy headgroup. This noncanonical lipid can be site selectively appended to the N-terminal of desired biopolymers such as peptides. ODHA drives the supramolecular assembly of modified protein, presumably through the hydrophobic collapse of ODHA chains.

















