
Amino acids are organic compounds that contain both amino and carboxylic acid functional groups. Although over 500 amino acids exist in nature, by far the most important are the 22 α-amino acids incorporated into proteins. Only these 22 appear in the genetic code of life.
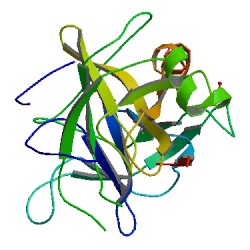
Chymotrypsin (EC 3.4.21.1, chymotrypsins A and B, alpha-chymar ophth, avazyme, chymar, chymotest, enzeon, quimar, quimotrase, alpha-chymar, alpha-chymotrypsin A, alpha-chymotrypsin) is a digestive enzyme component of pancreatic juice acting in the duodenum, where it performs proteolysis, the breakdown of proteins and polypeptides. Chymotrypsin preferentially cleaves peptide amide bonds where the side chain of the amino acid N-terminal to the scissile amide bond (the P1 position) is a large hydrophobic amino acid (tyrosine, tryptophan, and phenylalanine). These amino acids contain an aromatic ring in their side chain that fits into a hydrophobic pocket (the S1 position) of the enzyme. It is activated in the presence of trypsin. The hydrophobic and shape complementarity between the peptide substrate P1 side chain and the enzyme S1 binding cavity accounts for the substrate specificity of this enzyme. Chymotrypsin also hydrolyzes other amide bonds in peptides at slower rates, particularly those containing leucine at the P1 position.
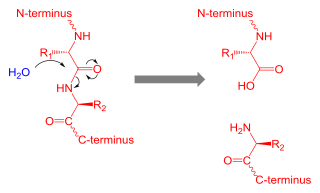
Proteolysis is the breakdown of proteins into smaller polypeptides or amino acids. Uncatalysed, the hydrolysis of peptide bonds is extremely slow, taking hundreds of years. Proteolysis is typically catalysed by cellular enzymes called proteases, but may also occur by intra-molecular digestion.

Trypsin is an enzyme in the first section of the small intestine that starts the digestion of protein molecules by cutting long chains of amino acids into smaller pieces. It is a serine protease from the PA clan superfamily, found in the digestive system of many vertebrates, where it hydrolyzes proteins. Trypsin is formed in the small intestine when its proenzyme form, the trypsinogen produced by the pancreas, is activated. Trypsin cuts peptide chains mainly at the carboxyl side of the amino acids lysine or arginine. It is used for numerous biotechnological processes. The process is commonly referred to as trypsinogen proteolysis or trypsinization, and proteins that have been digested/treated with trypsin are said to have been trypsinized.
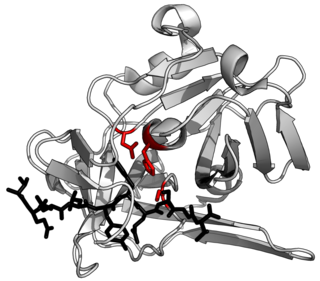
A protease is an enzyme that catalyzes proteolysis, breaking down proteins into smaller polypeptides or single amino acids, and spurring the formation of new protein products. They do this by cleaving the peptide bonds within proteins by hydrolysis, a reaction where water breaks bonds. Proteases are involved in numerous biological pathways, including digestion of ingested proteins, protein catabolism, and cell signaling.
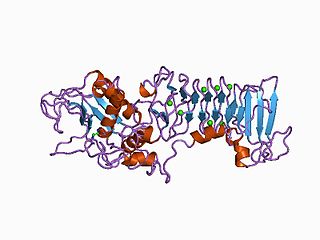
Matrix metalloproteinases (MMPs), also known as matrix metallopeptidases or matrixins, are metalloproteinases that are calcium-dependent zinc-containing endopeptidases; other family members are adamalysins, serralysins, and astacins. The MMPs belong to a larger family of proteases known as the metzincin superfamily.
The C-terminus is the end of an amino acid chain, terminated by a free carboxyl group (-COOH). When the protein is translated from messenger RNA, it is created from N-terminus to C-terminus. The convention for writing peptide sequences is to put the C-terminal end on the right and write the sequence from N- to C-terminus.
The N-terminus (also known as the amino-terminus, NH2-terminus, N-terminal end or amine-terminus) is the start of a protein or polypeptide, referring to the free amine group (-NH2) located at the end of a polypeptide. Within a peptide, the amine group is bonded to the carboxylic group of another amino acid, making it a chain. That leaves a free carboxylic group at one end of the peptide, called the C-terminus, and a free amine group on the other end called the N-terminus. By convention, peptide sequences are written N-terminus to C-terminus, left to right (in LTR writing systems). This correlates the translation direction to the text direction, because when a protein is translated from messenger RNA, it is created from the N-terminus to the C-terminus, as amino acids are added to the carboxyl end of the protein.
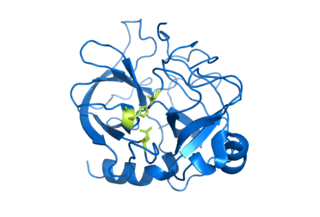
Serine proteases are enzymes that cleave peptide bonds in proteins. Serine serves as the nucleophilic amino acid at the (enzyme's) active site. They are found ubiquitously in both eukaryotes and prokaryotes. Serine proteases fall into two broad categories based on their structure: chymotrypsin-like (trypsin-like) or subtilisin-like.
A metalloproteinase, or metalloprotease, is any protease enzyme whose catalytic mechanism involves a metal. An example is ADAM12 which plays a significant role in the fusion of muscle cells during embryo development, in a process known as myogenesis.

Digestive enzymes take part in the chemical process of digestion, which follows the mechanical process of digestion. Food consists of macromolecules of proteins, carbohydrates, and fats that need to be broken down chemically by digestive enzymes in the mouth, stomach, pancreas, and duodenum, before being able to be absorbed into the bloodstream. Initial breakdown is achieved by chewing (mastication) and the use of digestive enzymes of saliva. Once in the stomach further mechanical churning takes place mixing the food with secreted gastric acid. Digestive gastric enzymes take part in some of the chemical process needed for absorption. Most of the enzymatic activity, and hence absorption takes place in the duodenum.
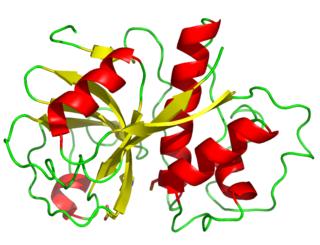
Papain, also known as papaya proteinase I, is a cysteine protease enzyme present in papaya and mountain papaya. It is the namesake member of the papain-like protease family.
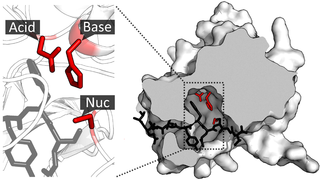
A catalytic triad is a set of three coordinated amino acids that can be found in the active site of some enzymes. Catalytic triads are most commonly found in hydrolase and transferase enzymes. An acid-base-nucleophile triad is a common motif for generating a nucleophilic residue for covalent catalysis. The residues form a charge-relay network to polarise and activate the nucleophile, which attacks the substrate, forming a covalent intermediate which is then hydrolysed to release the product and regenerate free enzyme. The nucleophile is most commonly a serine or cysteine amino acid, but occasionally threonine or even selenocysteine. The 3D structure of the enzyme brings together the triad residues in a precise orientation, even though they may be far apart in the sequence.
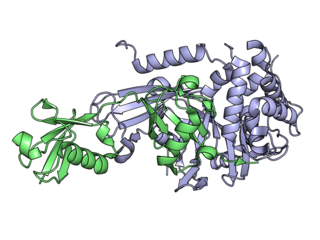
Deubiquitinating enzymes (DUBs), also known as deubiquitinating peptidases, deubiquitinating isopeptidases, deubiquitinases, ubiquitin proteases, ubiquitin hydrolases, or ubiquitin isopeptidases, are a large group of proteases that cleave ubiquitin from proteins. Ubiquitin is attached to proteins in order to regulate the degradation of proteins via the proteasome and lysosome; coordinate the cellular localisation of proteins; activate and inactivate proteins; and modulate protein-protein interactions. DUBs can reverse these effects by cleaving the peptide or isopeptide bond between ubiquitin and its substrate protein. In humans there are nearly 100 DUB genes, which can be classified into two main classes: cysteine proteases and metalloproteases. The cysteine proteases comprise ubiquitin-specific proteases (USPs), ubiquitin C-terminal hydrolases (UCHs), Machado-Josephin domain proteases (MJDs) and ovarian tumour proteases (OTU). The metalloprotease group contains only the Jab1/Mov34/Mpr1 Pad1 N-terminal+ (MPN+) (JAMM) domain proteases.
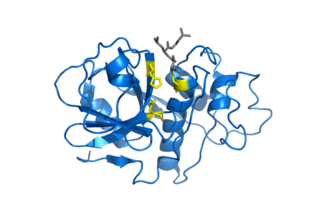
Cysteine proteases, also known as thiol proteases, are hydrolase enzymes that degrade proteins. These proteases share a common catalytic mechanism that involves a nucleophilic cysteine thiol in a catalytic triad or dyad.
Gelatinases are enzymes capable of degrading gelatin through hydrolysis, playing a major role in degradation of extracellular matrix and tissue remodeling. Gelatinases are a type of matrix metalloproteinase (MMP), a family of enzymes that depend on zinc as a cofactor and can break down parts of the extracellular matrix. MMPs have multiple subgroups, including gelatinase A and gelatinase B. Gelatinases are assigned a variety of Enzyme Commission numbers: gelatinase A uses 3.4.24.24, and gelatinase B uses 3.4.24.35, in which the first three numbers are same. The first digit, 3, is the class. Class 3 enzymes are hydrolases, enzymes that catalyze hydrolysis reactions, that is, they cleave bonds in presence of water. The next digit represents sub-class 4, or proteases, which are enzymes who hydrolyze peptide bonds in proteins. The next number is the sub-subclass of 24, which consists of metalloendopeptidases which contain metal ions in their active sites, in this case zinc, which help in cleaving peptide bonds. The last part of the EC number is the serial number, identifying specific enzymes within a sub-subclass. 24 represents gelatinase A, which is a metalloproteinase that breaks down gelatin and collagen, while 35 represents gelatinase B, which hydrolyzes peptide bonds.

A carboxypeptidase is a protease enzyme that hydrolyzes (cleaves) a peptide bond at the carboxy-terminal (C-terminal) end of a protein or peptide. This is in contrast to an aminopeptidases, which cleave peptide bonds at the N-terminus of proteins. Humans, animals, bacteria and plants contain several types of carboxypeptidases that have diverse functions ranging from catabolism to protein maturation. At least two mechanisms have been discussed.
Protein metabolism denotes the various biochemical processes responsible for the synthesis of proteins and amino acids (anabolism), and the breakdown of proteins by catabolism.
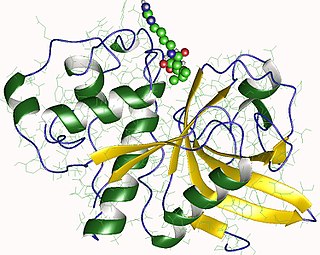
Actinidain is a type of cysteine protease enzyme found in fruits including kiwifruit, pineapple, mango, banana, figs, and papaya. This enzyme is part of the peptidase C1 family of papain-like proteases.
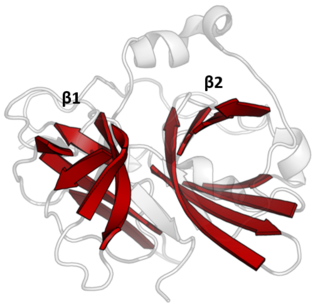
The PA clan is the largest group of proteases with common ancestry as identified by structural homology. Members have a chymotrypsin-like fold and similar proteolysis mechanisms but can have identity of <10%. The clan contains both cysteine and serine proteases. PA clan proteases can be found in plants, animals, fungi, eubacteria, archaea and viruses.















