
Microbial DNA barcoding is the use of DNA metabarcoding to characterize a mixture of microorganisms. DNA metabarcoding is a method of DNA barcoding that uses universal genetic markers to identify DNA of a mixture of organisms. [1]

Microbial DNA barcoding is the use of DNA metabarcoding to characterize a mixture of microorganisms. DNA metabarcoding is a method of DNA barcoding that uses universal genetic markers to identify DNA of a mixture of organisms. [1]
| Part of a series on |
| DNA barcoding |
|---|
 |
| By taxa |
| Other |
Using metabarcoding to assess microbial communities has a long history. Back in 1972, Carl Woese, Mitchell Sogin and Stephen Sogin first tried to detect several families within bacteria using the 5S rRNA gene. [2] Only a few years later, a new tree of life with three domains was proposed by again Woese and colleagues, who were the first to use the small subunit of the ribosomal RNA (SSU rRNA) gene to distinguish between bacteria, archaea and eukaryotes. [3] Out of this approach, the SSU rRNA gene made its way to be the most frequently used genetic marker for both prokaryotes (16S rRNA) and eukaryotes (18S rRNA). The tedious process of cloning those DNA fragments for sequencing got fastened up by the steady improvement of sequencing technologies. With the development of HTS (High-Throughput-Sequencing) in the early 2000s and the ability to deal with this massive data using modern bioinformatics and cluster algorithms, investigating microbial life got much easier.
Genetic diversity is varying from species to species. Therefore, it is possible to identify distinct species by the recovery of a short DNA sequence from a standard part of the genome. This short sequence is defined as barcode sequence. Requirements for a specific part of the genome to serve as barcode should be a high variation between two different species, but not much differences in the gene between two individuals of the same species to make differentiating individual species easier. [4] [5] For both bacteria and archaea the 16S rRNA/rDNA gene is used. It is a common housekeeping gene in all prokaryotic organisms and therefore is used as a standard barcode to assess prokaryotic diversity. For protists, the corresponding 18S rRNA/rDNA gene is used. [6] To distinguish different species of fungi, the ITS (Internal Transcribed Spacer) region of the ribosomal cistron is used. [7]
The existing diversity of the microbial world is not unraveled completely yet, although we know that it is mainly composed by bacteria, fungi and unicellular eukaryotes. [4] Taxonomic identification of microbial eukaryotes requires exceedingly skillful expertise and is often difficult due to small sizes of the organisms, fragmented individuals, hidden diversity and cryptic species. [8] [9] Further, prokaryotes can simply not be taxonomically assigned using traditional methods like microscopy, because they are too small and morphologically indistinguishable. Therefore, via the use of DNA metabarcoding, it is possible to identify organisms without taxonomic expertise by matching short High Throughput Sequences (HTS)-derived gene fragments to a reference sequence database, e.g. NCBI. [10] These mentioned qualities make DNA barcoding a cost-effective, reliable and less time-consuming method, compared to the traditional ones, to meet the increasing need for large-scale environmental assessments.
A lot of studies followed the first usage of Woese et al., and are now covering a variety of applications. Not only in biological or ecological research metabarcoding is used. Also in medicine and human biology bacterial barcodes are used, e.g. to investigate the microbiome and bacterial colonization of the human gut in normal and obese twins [11] or comparison studies of newborn, child and adult gut bacteria composition. [12] Additionally, barcoding plays a major role in biomonitoring of e.g. rivers and streams [13] and grassland restoration. [14] Conservation parasitology, environmental parasitology and paleoparasitology rely on barcoding as a useful tool in disease investigating and management, too. [15]
Cyanobacteria are a group of photosynthetic prokaryotes. Similar as in other prokaryotes, taxonomy of cyanobacteria using DNA sequences is mostly based on similarity within the 16S ribosomal gene. [16] Thus, the most common barcode used for identification of cyanobacteria is 16S rDNA marker. While it is difficult to define species within prokaryotic organisms, 16S marker can be used for determining individual operational taxonomic units (OTUs). In some cases, these OTUs can also be linked to traditionally defined species and can therefore be considered a reliable representation of the evolutionary relationships. [17]

However, when analyzing a taxonomic structure or biodiversity of a whole cyanobacterial community (see DNA metabarcoding), it is more informative to use markers specific for cyanobacteria. Universal 16S bacterial primers have been used successfully to isolate cyanobacterial rDNA from environmental samples, but they also recover many bacterial sequences. [18] [19] The use of cyanobacteria-specific [20] or phyto-specific 16S markers is commonly used for focusing on cyanobacteria only. [21] A few sets of such primers have been tested for barcoding or metabarcoding of environmental samples and gave good results, screening out majority of non-photosynthetic or non-cyanobacterial organisms. [22] [21] [23] [24]
Number of sequenced cyanobacterial genomes available in databases is increasing. [25] Besides 16S marker, phylogenetic studies could therefore include also more variable sequences, such as sequences of protein-coding genes (gyrB, rpoC, rpoD, [26] rbcL, hetR, [27] psbA, [28] [29] rnpB, [30] nifH, [31] nifD [32] ), internal transcribed spacer of ribosomal RNA genes (16S-23S rRNA-ITS) [33] [25] or phycocyanin intergenic spacer (PC-IGS). [33] However, nifD and nifH can only be used for identification of nitrogen-fixing cyanobacterial strains.
DNA barcoding of cyanobacteria can be applied in various ecological, evolutionary and taxonomical studies. Some examples include assessment of cyanobacterial diversity and community structure, [34] identification of harmful cyanobacteria in ecologically and economically important waterbodies [35] and assessment of cyanobacterial symbionts in marine invertebrates. [24] It has a potential to serve as a part of routine monitoring programs for occurrence of cyanobacteria, as well as early detection of potentially toxic species in waterbodies. This might help us detect harmful species before they start to form blooms and thus improve our water management strategies. Species identification based on environmental DNA could be particularly useful for cyanobacteria, as traditional identification using microscopy is challenging. Their morphological characteristics which are the basis for species delimitation vary in different growth conditions. [20] [36] Identification under microscope is also time-consuming and therefore relatively costly. Molecular methods can detect much lower concentration of cyanobacterial cells in the sample than traditional identification methods.
The reference database is a collection of DNA sequences, which are assigned to either a species or a function. It can be used to link molecular obtained sequences of an organism to pre-existing taxonomy. General databases like the NCBI platform include all kind of sequences, either whole genomes or specific marker genes of all organisms. There are also different platforms where only sequences from a distinct group of organisms are stored, e.g. UNITE database [37] exclusively for fungi sequences or the PR2 database solely for protist ribosomal sequences. [38] Some databases are curated, which allows a taxonomic assignment with higher accuracy than using uncurated databases as a reference.

Nanoarchaeota is a proposed phylum in the domain Archaea that currently has only one representative, Nanoarchaeum equitans, which was discovered in a submarine hydrothermal vent and first described in 2002.

The Thermoproteota are prokaryotes that have been classified as a phylum of the domain Archaea. Initially, the Thermoproteota were thought to be sulfur-dependent extremophiles but recent studies have identified characteristic Thermoproteota environmental rRNA indicating the organisms may be the most abundant archaea in the marine environment. Originally, they were separated from the other archaea based on rRNA sequences; other physiological features, such as lack of histones, have supported this division, although some crenarchaea were found to have histones. Until 2005 all cultured Thermoproteota had been thermophilic or hyperthermophilic organisms, some of which have the ability to grow at up to 113 °C. These organisms stain Gram negative and are morphologically diverse, having rod, cocci, filamentous and oddly-shaped cells. Recent evidence shows that some members of the Thermoproteota are methanogens.

The Korarchaeota is a proposed phylum within the Archaea. The name is derived from the Greek noun koros or kore, meaning young man or young woman, and the Greek adjective archaios which means ancient. They are also known as Xenarchaeota. The name is equivalent to Candidatus Korarchaeota, and they go by the name Xenarchaeota or Xenarchaea as well.
In molecular biology, a hybridization probe (HP) is a fragment of DNA or RNA, usually 15–10000 nucleotides long, which can be radioactively or fluorescently labeled. HPs can be used to detect the presence of nucleotide sequences in analyzed RNA or DNA that are complementary to the sequence in the probe. The labeled probe is first denatured into single stranded DNA (ssDNA) and then hybridized to the target ssDNA or RNA immobilized on a membrane or in situ.
Metagenomics is the study of genetic material recovered directly from environmental or clinical samples by a method called sequencing. The broad field may also be referred to as environmental genomics, ecogenomics, community genomics or microbiomics.

George Edward Fox is an astrobiologist, a Professor Emeritus and researcher at the University of Houston. He is an elected fellow of the American Academy of Microbiology, the American Association for the Advancement of Science, American Institute for Medical and Biological Engineering and the International Astrobiology Society. Fox received his B.A. degree in 1967, and completed his Ph.D. degree in 1974; both in chemical engineering at Syracuse University.

16S ribosomal RNA is the RNA component of the 30S subunit of a prokaryotic ribosome. It binds to the Shine-Dalgarno sequence and provides most of the SSU structure.
Synechocystis sp. PCC6803 is a strain of unicellular, freshwater cyanobacteria. Synechocystis sp. PCC6803 is capable of both phototrophic growth by oxygenic photosynthesis during light periods and heterotrophic growth by glycolysis and oxidative phosphorylation during dark periods. Gene expression is regulated by a circadian clock and the organism can effectively anticipate transitions between the light and dark phases.
An operational taxonomic unit (OTU) is an operational definition used to classify groups of closely related individuals. The term was originally introduced in 1963 by Robert R. Sokal and Peter H. A. Sneath in the context of numerical taxonomy, where an "operational taxonomic unit" is simply the group of organisms currently being studied. Numerical taxonomy is a method in biological systematics that involves using numerical techniques to classify taxonomic units based on the states of their characteristics. In this sense, an OTU is a pragmatic definition to group individuals by similarity, equivalent to but not necessarily in line with classical Linnaean taxonomy or modern evolutionary taxonomy.
In molecular biology, and more importantly high-throughput DNA sequencing, a chimera is a single DNA sequence originating when multiple transcripts or DNA sequences get joined. Chimeras can be considered artifacts and be filtered out from the data during processing to prevent spurious inferences of biological variation. However, chimeras should not be confused with chimeric reads, who are generally used by structural variant callers to detect structural variation events and are not always an indication of the presence of a chimeric transcript or gene.

DNA barcoding is a method of species identification using a short section of DNA from a specific gene or genes. The premise of DNA barcoding is that by comparison with a reference library of such DNA sections, an individual sequence can be used to uniquely identify an organism to species, just as a supermarket scanner uses the familiar black stripes of the UPC barcode to identify an item in its stock against its reference database. These "barcodes" are sometimes used in an effort to identify unknown species or parts of an organism, simply to catalog as many taxa as possible, or to compare with traditional taxonomy in an effort to determine species boundaries.

Bacterial taxonomy is subfield of taxonomy devoted to the classification of bacteria specimens into taxonomic ranks.
Community fingerprinting is a set of molecular biology techniques that can be used to quickly profile the diversity of a microbial community. Rather than directly identifying or counting individual cells in an environmental sample, these techniques show how many variants of a gene are present. In general, it is assumed that each different gene variant represents a different type of microbe. Community fingerprinting is used by microbiologists studying a variety of microbial systems to measure biodiversity or track changes in community structure over time. The method analyzes environmental samples by assaying genomic DNA. This approach offers an alternative to microbial culturing, which is important because most microbes cannot be cultured in the laboratory. Community fingerprinting does not result in identification of individual microbe species; instead, it presents an overall picture of a microbial community. These methods are now largely being replaced by high throughput sequencing, such as targeted microbiome analysis and metagenomics.
Microbial phylogenetics is the study of the manner in which various groups of microorganisms are genetically related. This helps to trace their evolution. To study these relationships biologists rely on comparative genomics, as physiology and comparative anatomy are not possible methods.

Mitchell Sogin is an American microbiologist. He is a distinguished senior scientist at the Marine Biological Laboratory in Woods Hole, Massachusetts. His research investigates the evolution and diversity of single-celled organisms.

DNA barcoding is an alternative method to the traditional morphological taxonomic classification, and has frequently been used to identify species of aquatic macroinvertebrates. Many are crucial indicator organisms in the bioassessment of freshwater and marine ecosystems.

DNA barcoding of algae is commonly used for species identification and phylogenetic studies. Algae form a phylogenetically heterogeneous group, meaning that the application of a single universal barcode/marker for species delimitation is unfeasible, thus different markers/barcodes are applied for this aim in different algal groups.

DNA barcoding methods for fish are used to identify groups of fish based on DNA sequences within selected regions of a genome. These methods can be used to study fish, as genetic material, in the form of environmental DNA (eDNA) or cells, is freely diffused in the water. This allows researchers to identify which species are present in a body of water by collecting a water sample, extracting DNA from the sample and isolating DNA sequences that are specific for the species of interest. Barcoding methods can also be used for biomonitoring and food safety validation, animal diet assessment, assessment of food webs and species distribution, and for detection of invasive species.

Fungal DNA barcoding is the process of identifying species of the biological kingdom Fungi through the amplification and sequencing of specific DNA sequences and their comparison with sequences deposited in a DNA barcode database such as the ISHAM reference database, or the Barcode of Life Data System (BOLD). In this attempt, DNA barcoding relies on universal genes that are ideally present in all fungi with the same degree of sequence variation. The interspecific variation, i.e., the variation between species, in the chosen DNA barcode gene should exceed the intraspecific (within-species) variation.
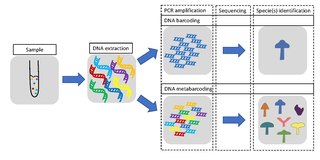
Metabarcoding is the barcoding of DNA/RNA in a manner that allows for the simultaneous identification of many taxa within the same sample. The main difference between barcoding and metabarcoding is that metabarcoding does not focus on one specific organism, but instead aims to determine species composition within a sample.