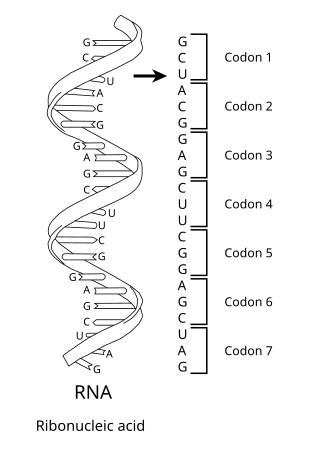
The genetic code is the set of rules used by living cells to translate information encoded within genetic material into proteins. Translation is accomplished by the ribosome, which links proteinogenic amino acids in an order specified by messenger RNA (mRNA), using transfer RNA (tRNA) molecules to carry amino acids and to read the mRNA three nucleotides at a time. The genetic code is highly similar among all organisms and can be expressed in a simple table with 64 entries.
In molecular biology, a stop codon is a codon that signals the termination of the translation process of the current protein. Most codons in messenger RNA correspond to the addition of an amino acid to a growing polypeptide chain, which may ultimately become a protein; stop codons signal the termination of this process by binding release factors, which cause the ribosomal subunits to disassociate, releasing the amino acid chain.
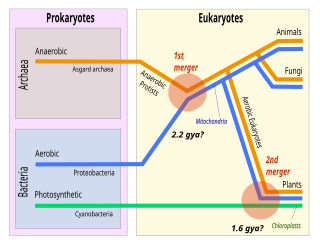
Symbiogenesis is the leading evolutionary theory of the origin of eukaryotic cells from prokaryotic organisms. The theory holds that mitochondria, plastids such as chloroplasts, and possibly other organelles of eukaryotic cells are descended from formerly free-living prokaryotes taken one inside the other in endosymbiosis. Mitochondria appear to be phylogenetically related to Rickettsiales bacteria, while chloroplasts are thought to be related to cyanobacteria.
In biology, translation is the process in living cells in which proteins are produced using RNA molecules as templates. The generated protein is a sequence of amino acids. This sequence is determined by the sequence of nucleotides in the RNA. The nucleotides are considered three at a time. Each such triple results in addition of one specific amino acid to the protein being generated. The matching from nucleotide triple to amino acid is called the genetic code. The translation is performed by a large complex of functional RNA and proteins called ribosomes. The entire process is called gene expression.

Transfer RNA is an adaptor molecule composed of RNA, typically 76 to 90 nucleotides in length. In a cell, it provides the physical link between the genetic code in messenger RNA (mRNA) and the amino acid sequence of proteins, carrying the correct sequence of amino acids to be combined by the protein-synthesizing machinery, the ribosome. Each three-nucleotide codon in mRNA is complemented by a three-nucleotide anticodon in tRNA. As such, tRNAs are a necessary component of translation, the biological synthesis of new proteins in accordance with the genetic code.

The start codon is the first codon of a messenger RNA (mRNA) transcript translated by a ribosome. The start codon always codes for methionine in eukaryotes and archaea and a N-formylmethionine (fMet) in bacteria, mitochondria and plastids.
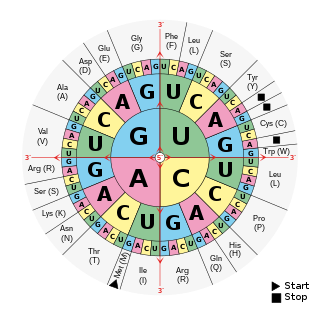
A codon table can be used to translate a genetic code into a sequence of amino acids. The standard genetic code is traditionally represented as an RNA codon table, because when proteins are made in a cell by ribosomes, it is messenger RNA (mRNA) that directs protein synthesis. The mRNA sequence is determined by the sequence of genomic DNA. In this context, the standard genetic code is referred to as translation table 1. It can also be represented in a DNA codon table. The DNA codons in such tables occur on the sense DNA strand and are arranged in a 5′-to-3′ direction. Different tables with alternate codons are used depending on the source of the genetic code, such as from a cell nucleus, mitochondrion, plastid, or hydrogenosome.
The pterobranchia mitochondrial code is a genetic code used by the mitochondrial genome of Rhabdopleura compacta (Pterobranchia). The Pterobranchia are one of the two groups in the Hemichordata which together with the Echinodermata and Chordata form the three major lineages of deuterostomes. AUA translates to isoleucine in Rhabdopleura as it does in the Echinodermata and Enteropneusta while AUA encodes methionine in the Chordata. The assignment of AGG to lysine is not found elsewhere in deuterostome mitochondria but it occurs in some taxa of Arthropoda. This code shares with many other mitochondrial codes the reassignment of the UGA STOP to tryptophan, and AGG and AGA to an amino acid other than arginine. The initiation codons in Rhabdopleura compacta are ATG and GTG.
The vertebrate mitochondrial code is the genetic code found in the mitochondria of all vertebrata.
The yeast mitochondrial code is a genetic code used by the mitochondrial genome of yeasts, notably Saccharomyces cerevisiae, Candida glabrata, Hansenula saturnus, and Kluyveromyces thermotolerans.
The bacterial, archaeal and plant plastid code is the DNA code used by bacteria, archaea, prokaryotic viruses and chloroplast proteins. It is essentially the same as the standard code, however there are some variations in alternative start codons.
The mold, protozoan, and coelenterate mitochondrial code and the mycoplasma/spiroplasma code is the genetic code used by various organisms, in some cases with slight variations, notably the use of UGA as a tryptophan codon rather than a stop codon.
The invertebrate mitochondrial code is a genetic code used by the mitochondrial genome of invertebrates. Mitochondria contain their own DNA and reproduce independently from their host cell. Variation in translation of the mitochondrial genetic code occurs when DNA codons result in non-standard amino acids has been identified in invertebrates, most notably arthropods. This variation has been helpful as a tool to improve upon the phylogenetic tree of invertebrates, like flatworms.
The echinoderm and flatworm mitochondrial code is a genetic code used by the mitochondria of certain echinoderm and flatworm species.
The alternative yeast nuclear code is a genetic code found in certain yeasts. However, other yeast, including Saccharomyces cerevisiae, Candida azyma, Candida diversa, Candida magnoliae, Candida rugopelliculosa, Yarrowia lipolytica, and Zygoascus hellenicus, definitely use the standard (nuclear) code.
The ascidian mitochondrial code is a genetic code found in the mitochondria of Ascidia.
The alternative flatworm mitochondrial code is a genetic code found in the mitochondria of Platyhelminthes and Nematodes.
The pachysolen tannophilus nuclear code is a genetic code found in the ascomycete fungus Pachysolen tannophilus.
The Condylostoma nuclear code is a genetic code used by the nuclear genome of the heterotrich ciliate Condylostoma magnum. This code, along with translation tables 27 and 31, is remarkable in that every one of the 64 possible codons can be a sense codon. Experimental evidence suggests that translation termination relies on context, specifically proximity to the poly(A) tail. Near such a tail, PABP could help terminate the protein by recruiting eRF1 and eRF3 to prevent the cognate tRNA from binding.
The Cephalodiscidae mitochondrial code is a genetic code used by the mitochondrial genome of Cephalodiscidae (Pterobranchia). The Pterobranchia are one of the two groups in the Hemichordata which together with the Echinodermata and Chordata form the major clades of deuterostomes.





