Related Research Articles

Early modern human (EMH), or anatomically modern human (AMH), are terms used to distinguish Homo sapiens that are anatomically consistent with the range of phenotypes seen in contemporary humans, from extinct archaic human species. This distinction is useful especially for times and regions where anatomically modern and archaic humans co-existed, for example, in Paleolithic Europe. Among the oldest known remains of Homo sapiens are those found at the Omo-Kibish I archaeological site in south-western Ethiopia, dating to about 233,000 to 196,000 years ago, the Florisbad site in South Africa, dating to about 259,000 years ago, and the Jebel Irhoud site in Morocco, dated about 315,000 years ago.

The indigenous peoples of Western New Guinea in Indonesia and Papua New Guinea, commonly called Papuans, are Melanesians. There is genetic evidence for two major historical lineages in New Guinea and neighboring islands: a first wave from the Malay Archipelago perhaps 50,000 years ago when New Guinea and Australia were a single landmass called Sahul and, much later, a wave of Austronesian people from the north who introduced Austronesian languages and pigs about 3,500 years ago. They also left a small but significant genetic trace in many coastal Papuan peoples.

The Bacho Kiro cave is situated 5 km (3.1 mi) west of the town Dryanovo, Bulgaria, only 300 m (980 ft) away from the Dryanovo Monastery. It is embedded in the canyons of the Andaka and Dryanovo River. It was opened in 1890 and the first recreational visitors entered the cave in 1938, two years before it was renamed in honor of Bulgarian National Revival leader, teacher and revolutionary Bacho Kiro. The cave is a four-storey labyrinth of galleries and corridors with a total length of 3,600 m (11,800 ft), 700 m (2,300 ft) of which are maintained for public access and equipped with electrical lights since 1964. An underground river has over time carved out the many galleries that contain countless stalactone, stalactite, and stalagmite speleothem formations of great beauty. Galleries and caverns of a 1,200 m (3,900 ft) long section have been musingly named as a popular description of this fairy-tale underground world. The formations succession: Bacho Kiro’s Throne, The Dwarfs, The Sleeping Princess, The Throne Hall, The Reception Hall, The Haidouti Meeting-Ground, The Fountain and the Sacrificial Altar.

Human genetic variation is the genetic differences in and among populations. There may be multiple variants of any given gene in the human population (alleles), a situation called polymorphism.
David Emil Reich is an American geneticist known for his research into the population genetics of ancient humans, including their migrations and the mixing of populations, discovered by analysis of genome-wide patterns of mutations. He is professor in the department of genetics at the Harvard Medical School, and an associate of the Broad Institute. Reich was highlighted as one of Nature's 10 for his contributions to science in 2015. He received the Dan David Prize in 2017, the NAS Award in Molecular Biology, the Wiley Prize, and the Darwin–Wallace Medal in 2019. In 2021 he was awarded the Massry Prize.

The genetic history of Europe includes information around the formation, ethnogenesis, and other DNA-specific information about populations indigenous, or living in Europe.

Peștera cu Oase is a system of 12 karstic galleries and chambers located near the city Anina, in the Caraș-Severin county, southwestern Romania, where some of the oldest European early modern human (EEMH) remains, between 42,000 and 37,000 years old, have been found.

Early human migrations are the earliest migrations and expansions of archaic and modern humans across continents. They are believed to have begun approximately 2 million years ago with the early expansions out of Africa by Homo erectus. This initial migration was followed by other archaic humans including H. heidelbergensis, which lived around 500,000 years ago and was the likely ancestor of Denisovans and Neanderthals as well as modern humans. Early hominids had likely crossed land bridges that have now sunk.

In human mitochondrial genetics, L is the mitochondrial DNA macro-haplogroup that is at the root of the anatomically modern human mtDNA phylogenetic tree. As such, it represents the most ancestral mitochondrial lineage of all currently living modern humans, also dubbed "Mitochondrial Eve".

In paleoanthropology, the recent African origin of modern humans or the "Out of Africa" theory (OOA) is the most widely accepted model of the geographic origin and early migration of anatomically modern humans. It follows the early expansions of hominins out of Africa, accomplished by Homo erectus and then Homo neanderthalensis.
The multiregional hypothesis, multiregional evolution (MRE), or polycentric hypothesis, is a scientific model that provides an alternative explanation to the more widely accepted "Out of Africa" model of monogenesis for the pattern of human evolution.

The Denisovans or Denisova hominins are an extinct species or subspecies of archaic human that ranged across Asia during the Lower and Middle Paleolithic, and lived, based on current evidence, from 285 to 25 thousand years ago. Denisovans are known from few physical remains; consequently, most of what is known about them comes from DNA evidence. No formal species name has been established pending more complete fossil material.

Interbreeding between archaic and modern humans occurred during the Middle Paleolithic and early Upper Paleolithic. The interbreeding happened in several independent events that included Neanderthals and Denisovans, as well as several unidentified hominins.

The term Eurasian backflow, or Eurasian back-migrations, has been used to describe several pre-Neolithic and Neolithic migration events of humans from western Eurasia back to Africa.
Genetic studies on Neanderthal ancient DNA became possible in the late 1990s. The Neanderthal genome project, established in 2006, presented the first fully sequenced Neanderthal genome in 2013.

Denny is an ~90,000 year old fossil specimen belonging to a ~13-year-old Neanderthal-Denisovan hybrid girl. To date, she is the only first-generation hybrid hominin ever discovered. Denny’s remains consist of a single fossilized fragment of a long bone discovered among over 2,000 visually unidentifiable fragments excavated at the Denisova Cave in the Altai Mountains, Russia in 2012.
Basal Eurasian is a proposed lineage of anatomically modern humans with reduced, or zero, Neanderthal admixture (ancestry) compared to other ancient non-Africans. Basal Eurasians represent a sister lineage to other Eurasians and may have originated from the Southern Middle East, specifically the Arabian Peninsula, or North Africa, and are said to have contributed ancestry to various West Eurasian, South Asian, and Central Asian as well as African groups. This Basal Eurasian component is also proposed to explain the lower archaic admixture among modern West Eurasians compared with East Eurasians, although alternatives without the need of such Basal admixture exist as well. Basal Eurasian ancestry had likely admixed into West Eurasian groups present in West Asia as early as 26,000 years ago, prior to the Last Glacial Maximum, with this ancestry being subsequently spread by later migrations, such as those of the Anatolian Neolithic Farmers into Europe during the Holocene.
Eukaryote hybrid genomes result from interspecific hybridization, where closely related species mate and produce offspring with admixed genomes. The advent of large-scale genomic sequencing has shown that hybridization is common, and that it may represent an important source of novel variation. Although most interspecific hybrids are sterile or less fit than their parents, some may survive and reproduce, enabling the transfer of adaptive variants across the species boundary, and even result in the formation of novel evolutionary lineages. There are two main variants of hybrid species genomes: allopolyploid, which have one full chromosome set from each parent species, and homoploid, which are a mosaic of the parent species genomes with no increase in chromosome number.
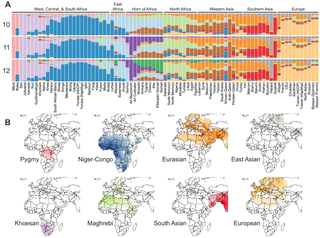
The genetic history of Africa summarizes the genetic makeup and population history of African populations in Africa, composed of the overall genetic history, including the regional genetic histories of North Africa, West Africa, East Africa, Central Africa, and Southern Africa, as well as the recent origin of modern humans in Africa. The Sahara served as a trans-regional passageway and place of dwelling for people in Africa during various humid phases and periods throughout the history of Africa. It also served as a biological barrier that restricted geneflow between the northern and central parts of Africa since its desertification, contributing to the diverse and distinct population structures on the continent. Nonetheless, this did not stop contact between peoples north and south of the Sahara at various points, especially in prehistoric times when the climate conditions were warmer and wetter.
Basal West African is a hypothetical line of descent that is no longer extant.
References
- 1 2 Beerli, P (2004). "Effect of unsampled populations on the estimation of population sizes and migration rates between sampled populations". Molecular Ecology. 13 (4): 827–836. doi: 10.1111/j.1365-294x.2004.02101.x . PMID 15012758. S2CID 18326408.
- ↑ Skatkin, M (2005). "Seeing ghosts: the effect of unsampled populations on migration rates estimated for sampled populations". Molecular Ecology. 14 (1): 67–73. doi:10.1111/j.1365-294X.2004.02393.x. PMID 15643951. S2CID 17600283.
- ↑ Patterson, N (2012). "Ancient admixture in human history". Genetics. 192 (3): 1065–93. doi:10.1534/genetics.112.145037. PMC 3522152 . PMID 22960212.
- ↑ Raghavan, M (2013). "Upper Palaeolithic Siberian genome reveals dual ancestry of Native Americans". Nature. 505 (7481): 87–91. Bibcode:2014Natur.505...87R. doi:10.1038/nature12736. PMC 4105016 . PMID 24256729.
- ↑ Callaway, E (2015). ""Ghost population" hints at long-lost migration to the Americas". Nature. doi:10.1038/nature.2015.18029. S2CID 181337948.
- 1 2 Skoglund, Pontus; et al. (2017). "Reconstructing Prehistoric African Population Structure". Cell. 171 (1): 59–71.e21. doi:10.1016/j.cell.2017.08.049. ISSN 0092-8674. OCLC 7144495602. PMC 5679310 . PMID 28938123. S2CID 1257429.
- ↑ Arun Durvasula; Sriram Sankararaman (2020). "Recovering signals of ghost archaic introgression in African populations". Science Advances. 6 (7): eaax5097. Bibcode:2020SciA....6.5097D. doi: 10.1126/sciadv.aax5097 . PMC 7015685 . PMID 32095519. "Non-African populations (Han Chinese in Beijing and Utah residents with northern and western European ancestry) also show analogous patterns in the CSFS, suggesting that a component of archaic ancestry was shared before the split of African and non-African populations...One interpretation of the recent time of introgression that we document is that archaic forms persisted in Africa until fairly recently. Alternately, the archaic population could have introgressed earlier into a modern human population, which then subsequently interbred with the ancestors of the populations that we have analyzed here. The models that we have explored here are not mutually exclusive, and it is plausible that the history of African populations includes genetic contributions from multiple divergent populations, as evidenced by the large effective population size associated with the introgressing archaic population...Given the uncertainty in our estimates of the time of introgression, we wondered whether jointly analyzing the CSFS from both the CEU (Utah residents with Northern and Western European ancestry) and YRI genomes could provide additional resolution. Under model C, we simulated introgression before and after the split between African and non-African populations and observed qualitative differences between the two models in the high-frequency–derived allele bins of the CSFS in African and non-African populations (fig. S40). Using ABC to jointly fit the high-frequency–derived allele bins of the CSFS in CEU and YRI (defined as greater than 50% frequency), we find that the lower limit on the 95% credible interval of the introgression time is older than the simulated split between CEU and YRI (2800 versus 2155 generations B.P.), indicating that at least part of the archaic lineages seen in the YRI are also shared with the CEU..."
- ↑ Supplementary Materials for Recovering signals of ghost archaic introgression in African populations", section "S8.2" "We simulated data using the same priors in Section S5.2, but computed the spectrum for both YRI [West African Yoruba] and CEU [a population of European origin] . We found that the best fitting parameters were an archaic split time of 27,000 generations ago (95% HPD: 26,000-28,000), admixture fraction of 0.09 (95% HPD: 0.04-0.17), admixture time of 3,000 generations ago (95% HPD: 2,800-3,400), and an effective population size of 19,700 individuals (95% HPD: 19,300-20,200). We find that the lower bound of the admixture time is further back than the simulated split between CEU and YRI (2155 generations ago), providing some evidence in favor of a pre-Out-of-Africa event. This model suggests that many populations outside of Africa should also contain haplotypes from this introgression event, though detection is difficult because many methods use unadmixed outgroups to detect introgressed haplotypes [Browning et al., 2018, Skov et al., 2018, Durvasula and Sankararaman, 2019] (5, 53, 22). It is also possible that some of these haplotypes were lost during the Out-of-Africa bottleneck."
- ↑ Durvasula, Arun; Sankararaman, Sriram (2020). "Recovering signals of ghost archaic introgression in African populations". Science Advances. 6 (7): eaax5097. Bibcode:2020SciA....6.5097D. doi:10.1126/sciadv.aax5097. PMC 7015685 . PMID 32095519. S2CID 211472946.
- ↑ Bergström, A; McCarthy, S; Hui, R; Almarri, M; Ayub, Q (2020). "Insights into human genetic variation and population history from 929 diverse genomes". Science . 367 (6484): eaay5012. doi:10.1126/science.aay5012. PMC 7115999 . PMID 32193295. "An analysis of archaic sequences in modern populations identifies ancestral genetic variation in African populations that likely predates modern humans and has been lost in most non-African populations...We found small amounts of Neanderthal ancestry in West African genomes, most likely reflecting Eurasian admixture. Despite their very low levels or absence of archaic ancestry, African populations share many Neanderthal and Denisovan variants that are absent from Eurasia, reflecting how a larger proportion of the ancestral human variation has been maintained in Africa."
- ↑ Frantz, L (2015). "Evidence of long-term gene flow and selection during domestication from analyses of Eurasian wild and domestic pig genomes". Nature Genetics. 47 (10): 1141–1148. doi:10.1038/ng.3394. PMID 26323058. S2CID 205350534.
- ↑ Gopalakrishnan, Shyam; Sinding, Mikkel-Holger S.; Ramos-Madrigal, Jazmín; Niemann, Jonas; Samaniego Castruita, Jose A.; Vieira, Filipe G.; Carøe, Christian; Montero, Marc de Manuel; Kuderna, Lukas; Serres, Aitor; González-Basallote, Víctor Manuel; Liu, Yan-Hu; Wang, Guo-Dong; Marques-Bonet, Tomas; Mirarab, Siavash; Fernandes, Carlos; Gaubert, Philippe; Koepfli, Klaus-Peter; Budd, Jane; Rueness, Eli Knispel; Heide-Jørgensen, Mads Peter; Petersen, Bent; Sicheritz-Ponten, Thomas; Bachmann, Lutz; Wiig, Øystein; Hansen, Anders J.; Gilbert, M. Thomas P. (2018). "Interspecific Gene Flow Shaped the Evolution of the Genus Canis". Current Biology. 28 (21): 3441–3449.e5. doi:10.1016/j.cub.2018.08.041. PMC 6224481 . PMID 30344120.