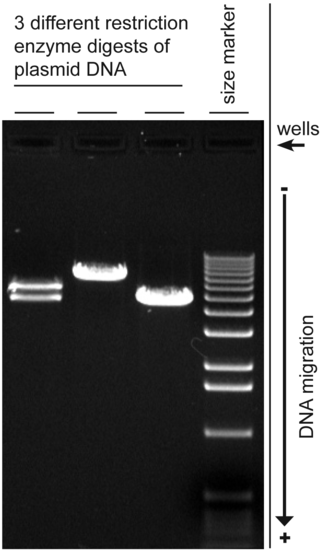
Agarose gel electrophoresis is a method of gel electrophoresis used in biochemistry, molecular biology, genetics, and clinical chemistry to separate a mixed population of macromolecules such as DNA or proteins in a matrix of agarose, one of the two main components of agar. The proteins may be separated by charge and/or size, and the DNA and RNA fragments by length. Biomolecules are separated by applying an electric field to move the charged molecules through an agarose matrix, and the biomolecules are separated by size in the agarose gel matrix.

Agarose is a heteropolysaccharide, generally extracted from certain red algae. It is a linear polymer made up of the repeating unit of agarobiose, which is a disaccharide made up of D-galactose and 3,6-anhydro-L-galactopyranose. Agarose is one of the two principal components of agar, and is purified from agar by removing agar's other component, agaropectin.

Gel electrophoresis is an electrophoresis method for separation and analysis of biomacromolecules and their fragments, based on their size and charge through a supportive medium. It is used in clinical chemistry to separate proteins by charge or size and in biochemistry and molecular biology to separate a mixed population of DNA and RNA fragments by length, to estimate the size of DNA and RNA fragments or to separate proteins by charge.

Polyacrylamide gel electrophoresis (PAGE) is a technique widely used in biochemistry, forensic chemistry, genetics, molecular biology and biotechnology to separate biological macromolecules, usually proteins or nucleic acids, according to their electrophoretic mobility. Electrophoretic mobility is a function of the length, conformation, and charge of the molecule. Polyacrylamide gel electrophoresis is a powerful tool used to analyze RNA samples. When polyacrylamide gel is denatured after electrophoresis, it provides information on the sample composition of the RNA species.

Gel electrophoresis of nucleic acids is an analytical technique to separate DNA or RNA fragments by size and reactivity. Nucleic acid molecules are placed on a gel, where an electric field induces the nucleic acids to migrate toward the positively charged anode. The molecules separate as they travel through the gel based on the each molecule's size and shape. Longer molecules move more slowly because the gel resists their movement more forcefully than it resists shorter molecules. After some time, the electricity is turned off and the positions of the different molecules are analyzed.
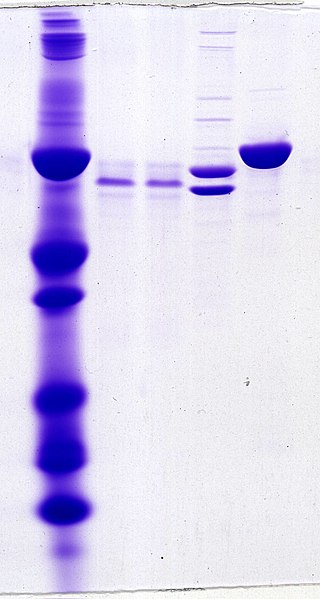
Protein electrophoresis is a method for analysing the proteins in a fluid or an extract. The electrophoresis may be performed with a small volume of sample in a number of alternative ways with or without a supporting medium, namely agarose or polyacrylamide. Variants of gel electrophoresis include SDS-PAGE, free-flow electrophoresis, electrofocusing, isotachophoresis, affinity electrophoresis, immunoelectrophoresis, counterelectrophoresis, and capillary electrophoresis. Each variant has many subtypes with individual advantages and limitations. Gel electrophoresis is often performed in combination with electroblotting or immunoblotting to give additional information about a specific protein.
The first isolation of deoxyribonucleic acid (DNA) was done in 1869 by Friedrich Miescher. DNA extraction is the process of isolating DNA from the cells of an organism isolated from a sample, typically a biological sample such as blood, saliva, or tissue. It involves breaking open the cells, removing proteins and other contaminants, and purifying the DNA so that it is free of other cellular components. The purified DNA can then be used for downstream applications such as PCR, sequencing, or cloning. Currently, it is a routine procedure in molecular biology or forensic analyses.
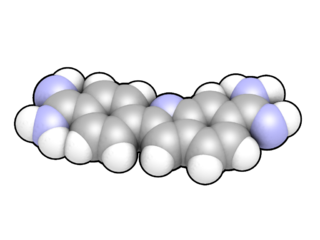
DAPI, or 4′,6-diamidino-2-phenylindole, is a fluorescent stain that binds strongly to adenine–thymine-rich regions in DNA. It is used extensively in fluorescence microscopy. As DAPI can pass through an intact cell membrane, it can be used to stain both live and fixed cells, though it passes through the membrane less efficiently in live cells and therefore provides a marker for membrane viability.
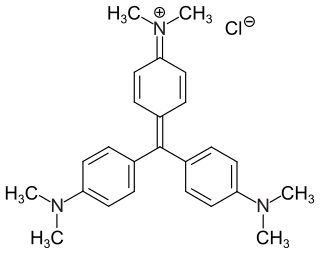
Crystal violet or gentian violet, also known as methyl violet 10B or hexamethyl pararosaniline chloride, is a triarylmethane dye used as a histological stain and in Gram's method of classifying bacteria. Crystal violet has antibacterial, antifungal, and anthelmintic (vermicide) properties and was formerly important as a topical antiseptic. The medical use of the dye has been largely superseded by more modern drugs, although it is still listed by the World Health Organization.
In molecular biology, gel extraction or gel isolation is a technique used to isolate a desired fragment of intact DNA from an agarose gel following agarose gel electrophoresis. After extraction, fragments of interest can be mixed, precipitated, and enzymatically ligated together in several simple steps. This process, usually performed on plasmids, is the basis for rudimentary genetic engineering.
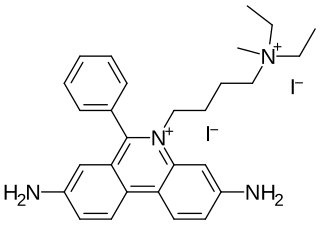
Propidium iodide is a fluorescent intercalating agent that can be used to stain cells and nucleic acids. PI binds to DNA by intercalating between the bases with little or no sequence preference. When in an aqueous solution, PI has a fluorescent excitation maximum of 493 nm (blue-green), and an emission maximum of 636 nm (red). After binding DNA, the quantum yield of PI is enhanced 20-30 fold, and the excitation/emission maximum of PI is shifted to 535 nm (green) / 617 nm (orange-red). Propidium iodide is used as a DNA stain in flow cytometry to evaluate cell viability or DNA content in cell cycle analysis, or in microscopy to visualize the nucleus and other DNA-containing organelles. Propidium Iodide is not membrane-permeable, making it useful to differentiate necrotic, apoptotic and healthy cells based on membrane integrity. PI also binds to RNA, necessitating treatment with nucleases to distinguish between RNA and DNA staining. PI is widely used in fluorescence staining and visualization of the plant cell wall.
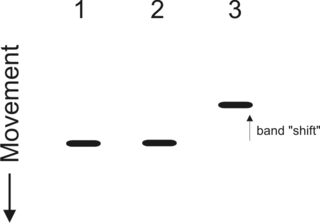
An electrophoretic mobility shift assay (EMSA) or mobility shift electrophoresis, also referred as a gel shift assay, gel mobility shift assay, band shift assay, or gel retardation assay, is a common affinity electrophoresis technique used to study protein–DNA or protein–RNA interactions. This procedure can determine if a protein or mixture of proteins is capable of binding to a given DNA or RNA sequence, and can sometimes indicate if more than one protein molecule is involved in the binding complex. Gel shift assays are often performed in vitro concurrently with DNase footprinting, primer extension, and promoter-probe experiments when studying transcription initiation, DNA gang replication, DNA repair or RNA processing and maturation, as well as pre-mRNA splicing. Although precursors can be found in earlier literature, most current assays are based on methods described by Garner and Revzin and Fried and Crothers.
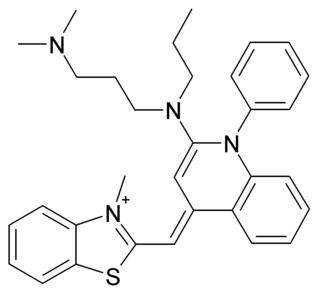
SYBR Green I (SG) is an asymmetrical cyanine dye used as a nucleic acid stain in molecular biology. The SYBR family of dyes is produced by Molecular Probes Inc., now owned by Thermo Fisher Scientific. SYBR Green I binds to DNA. The resulting DNA-dye-complex best absorbs 497 nanometer blue light and emits green light. The stain preferentially binds to double-stranded DNA, but will stain single-stranded (ss) DNA with lower performance. SYBR Green can also stain RNA with a lower performance than ssDNA.

An electrophoretic color marker is a chemical used to monitor the progress of agarose gel electrophoresis and polyacrylamide gel electrophoresis (PAGE) since DNA, RNA, and most proteins are colourless. The color markers are made up of a mixture of dyes that migrate through the gel matrix alongside the sample of interest. They are typically designed to have different mobilities from the sample components and to generate colored bands that can be used to assess the migration and separation of sample components.
Fluorescence is used in the life sciences generally as a non-destructive way of tracking or analysing biological molecules. Some proteins or small molecules in cells are naturally fluorescent, which is called intrinsic fluorescence or autofluorescence. The intrinsic DNA fluorescence is very weak.Alternatively, specific or general proteins, nucleic acids, lipids or small molecules can be "labelled" with an extrinsic fluorophore, a fluorescent dye which can be a small molecule, protein or quantum dot. Several techniques exist to exploit additional properties of fluorophores, such as fluorescence resonance energy transfer, where the energy is passed non-radiatively to a particular neighbouring dye, allowing proximity or protein activation to be detected; another is the change in properties, such as intensity, of certain dyes depending on their environment allowing their use in structural studies.

A gel doc, also known as a gel documentation system, gel image system or gel imager, refers to equipment widely used in molecular biology laboratories for the imaging and documentation of nucleic acid and protein suspended within polyacrylamide or agarose gels. Genetic information is stored in DNA. Polyacrylamide or agarose gel electrophoresis procedures are carried out to examine nucleic acids or proteins in order to analyze the genetic data. For protein analysis, two-dimensional gel electrophoresis is employed (2-DGE) which is one of the methods most frequently used in comparative proteomic investigations that can distinguish thousands of proteins in a single run. Proteins are separated using 2-DGE first, based on their isoelectric points (pIs) in one dimension and then based on their molecular mass in the other. After that, a thorough qualitative and quantitative analysis of the proteomes is performed using gel documentation with software image assessment methods on the 2-DGE gels stained for protein visibility. Gels are typically stained with Ethidium bromide or other nucleic acid stains such as GelGreen.
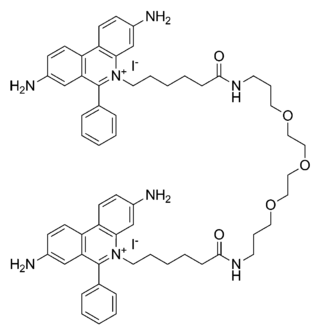
GelRed is an intercalating nucleic acid stain used in molecular genetics for agarose gel DNA electrophoresis. GelRed structurally consists of two ethidium subunits that are bridged by a linear oxygenated spacer.
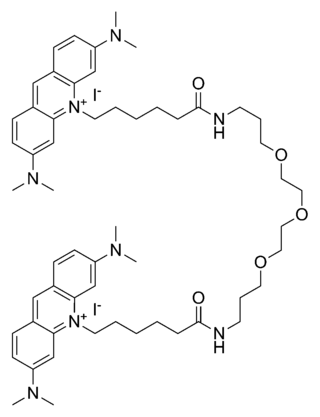
GelGreen is an intercalating nucleic acid stain used in molecular genetics for agarose gel DNA electrophoresis. GelGreen consists of two acridine orange subunits that are bridged by a linear oxygenated spacer.
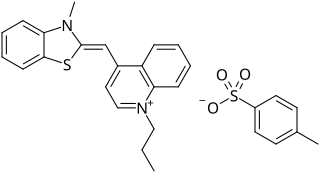
SYBR Safe is a cyanine dye used as a nucleic acid stain in molecular biology. SYBR Safe is one of a number of SYBR dyes made by the Life Technologies Corporation. SYBR Safe binds to DNA. The resulting DNA-dye-complex absorbs blue light and emits green light.
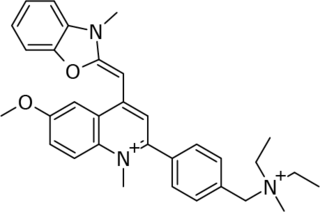
SYBR Gold is an asymmetrical cyanine dye. It can be used as a stain for double-stranded DNA, single-stranded DNA, and RNA. SYBR Gold is the most sensitive fluorescent stain of the SYBR family of dyes for the detection of nucleic acids. The SYBR family of dyes is produced by Molecular Probes Inc., now owned by Thermo Fisher Scientific


























