
A bacteriophage, also known informally as a phage, is a duplodnaviria virus that infects and replicates within bacteria and archaea. The term was derived from "bacteria" and the Greek φαγεῖν, meaning "to devour". Bacteriophages are composed of proteins that encapsulate a DNA or RNA genome, and may have structures that are either simple or elaborate. Their genomes may encode as few as four genes and as many as hundreds of genes. Phages replicate within the bacterium following the injection of their genome into its cytoplasm.
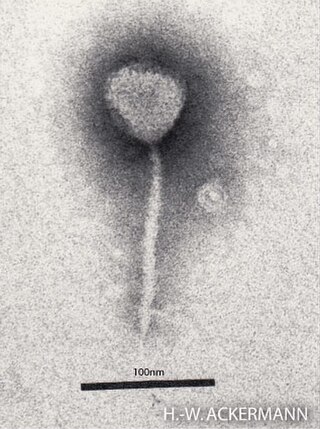
Enterobacteria phage λ is a bacterial virus, or bacteriophage, that infects the bacterial species Escherichia coli. It was discovered by Esther Lederberg in 1950. The wild type of this virus has a temperate life cycle that allows it to either reside within the genome of its host through lysogeny or enter into a lytic phase, during which it kills and lyses the cell to produce offspring. Lambda strains, mutated at specific sites, are unable to lysogenize cells; instead, they grow and enter the lytic cycle after superinfecting an already lysogenized cell.
A provirus is a virus genome that is integrated into the DNA of a host cell. In the case of bacterial viruses (bacteriophages), proviruses are often referred to as prophages. However, proviruses are distinctly different from prophages and these terms should not be used interchangeably. Unlike prophages, proviruses do not excise themselves from the host genome when the host cell is stressed.

A cloning vector is a small piece of DNA that can be stably maintained in an organism, and into which a foreign DNA fragment can be inserted for cloning purposes. The cloning vector may be DNA taken from a virus, the cell of a higher organism, or it may be the plasmid of a bacterium. The vector contains features that allow for the convenient insertion of a DNA fragment into the vector or its removal from the vector, for example through the presence of restriction sites. The vector and the foreign DNA may be treated with a restriction enzyme that cuts the DNA, and DNA fragments thus generated contain either blunt ends or overhangs known as sticky ends, and vector DNA and foreign DNA with compatible ends can then be joined together by molecular ligation. After a DNA fragment has been cloned into a cloning vector, it may be further subcloned into another vector designed for more specific use.

A prophage is a bacteriophage genome that is integrated into the circular bacterial chromosome or exists as an extrachromosomal plasmid within the bacterial cell. Integration of prophages into the bacterial host is the characteristic step of the lysogenic cycle of temperate phages. Prophages remain latent in the genome through multiple cell divisions until activation by an external factor, such as UV light, leading to production of new phage particles that will lyse the cell and spread. As ubiquitous mobile genetic elements, prophages play important roles in bacterial genetics and evolution, such as in the acquisition of virulence factors.
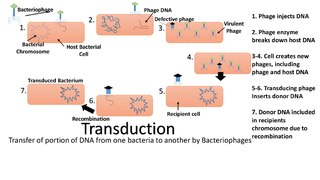
Transduction is the process by which foreign DNA is introduced into a cell by a virus or viral vector. An example is the viral transfer of DNA from one bacterium to another and hence an example of horizontal gene transfer. Transduction does not require physical contact between the cell donating the DNA and the cell receiving the DNA, and it is DNase resistant. Transduction is a common tool used by molecular biologists to stably introduce a foreign gene into a host cell's genome.
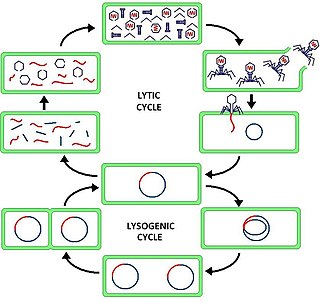
The lytic cycle is one of the two cycles of viral reproduction, the other being the lysogenic cycle. The lytic cycle results in the destruction of the infected cell and its membrane. Bacteriophages that only use the lytic cycle are called virulent phages.

Escherichia virus T4 is a species of bacteriophages that infect Escherichia coli bacteria. It is a double-stranded DNA virus in the subfamily Tevenvirinae from the family Myoviridae. T4 is capable of undergoing only a lytic lifecycle and not the lysogenic lifecycle. The species was formerly named T-even bacteriophage, a name which also encompasses, among other strains, Enterobacteria phage T2, Enterobacteria phage T4 and Enterobacteria phage T6.
In virology, temperate refers to the ability of some bacteriophages to display a lysogenic life cycle. Many temperate phages can integrate their genomes into their host bacterium's chromosome, together becoming a lysogen as the phage genome becomes a prophage. A temperate phage is also able to undergo a productive, typically lytic life cycle, where the prophage is expressed, replicates the phage genome, and produces phage progeny, which then leave the bacterium. With phage the term virulent is often used as an antonym to temperate, but more strictly a virulent phage is one that has lost its ability to display lysogeny through mutation rather than a phage lineage with no genetic potential to ever display lysogeny.

Lysogeny, or the lysogenic cycle, is one of two cycles of viral reproduction. Lysogeny is characterized by integration of the bacteriophage nucleic acid into the host bacterium's genome or formation of a circular replicon in the bacterial cytoplasm. In this condition the bacterium continues to live and reproduce normally, while the bacteriophage lies in a dormant state in the host cell. The genetic material of the bacteriophage, called a prophage, can be transmitted to daughter cells at each subsequent cell division, and later events can release it, causing proliferation of new phages via the lytic cycle. Lysogenic cycles can also occur in eukaryotes, although the method of DNA incorporation is not fully understood. For instance the AIDS viruses can either infect humans lytically, or lay dormant (lysogenic) as part of the infected cells' genome, keeping the ability to return to lysis at a later time. The rest of this article is about lysogeny in bacterial hosts.

Bacteriophage T7 is a bacteriophage, a virus that infects bacteria. It infects most strains of Escherichia coli and relies on these hosts to propagate. Bacteriophage T7 has a lytic life cycle, meaning that it destroys the cell it infects. It also possesses several properties that make it an ideal phage for experimentation: its purification and concentration have produced consistent values in chemical analyses; it can be rendered noninfectious by exposure to UV light; and it can be used in phage display to clone RNA binding proteins.
Salmonella virus P22 is a bacteriophage in the Podoviridae family that infects Salmonella typhimurium. Like many phages, it has been used in molecular biology to induce mutations in cultured bacteria and to introduce foreign genetic material. P22 has been used in generalized transduction and is an important tool for investigating Salmonella genetics.
P1 is a temperate bacteriophage that infects Escherichia coli and some other bacteria. When undergoing a lysogenic cycle the phage genome exists as a plasmid in the bacterium unlike other phages that integrate into the host DNA. P1 has an icosahedral head containing the DNA attached to a contractile tail with six tail fibers. The P1 phage has gained research interest because it can be used to transfer DNA from one bacterial cell to another in a process known as transduction. As it replicates during its lytic cycle it captures fragments of the host chromosome. If the resulting viral particles are used to infect a different host the captured DNA fragments can be integrated into the new host's genome. This method of in vivo genetic engineering was widely used for many years and is still used today, though to a lesser extent. P1 can also be used to create the P1-derived artificial chromosome cloning vector which can carry relatively large fragments of DNA. P1 encodes a site-specific recombinase, Cre, that is widely used to carry out cell-specific or time-specific DNA recombination by flanking the target DNA with loxP sites.
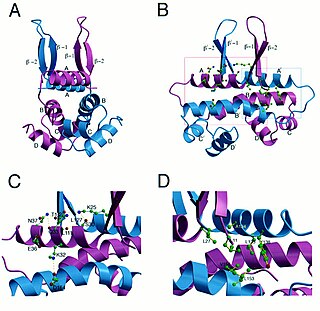
fis is an E. coli gene encoding the Fis protein. The regulation of this gene is more complex than most other genes in the E. coli genome, as Fis is an important protein which regulates expression of other genes. It is supposed that fis is regulated by H-NS, IHF and CRP. It also regulates its own expression (autoregulation). Fis is one of the most abundant DNA binding proteins in Escherichia coli under nutrient-rich growth conditions.
A P1-derived artificial chromosome, or PAC, is a DNA construct derived from the DNA of P1 bacteriophages and Bacterial artificial chromosome. It can carry large amounts of other sequences for a variety of bioengineering purposes in bacteria. It is one type of the efficient cloning vector used to clone DNA fragments in Escherichia coli cells.
Zygotic induction occurs when a bacterial cell carrying the silenced DNA of a bacterial virus in its chromosome transfers the viral DNA along with its own DNA to another bacterial cell lacking the virus, causing the recipient of the DNA to break open. In the donor cell, a repressor protein encoded by the prophage keeps the viral genes turned off so that virus is not produced. When DNA is transferred to the recipient cell by conjugation, the viral genes in the transferred DNA are immediately turned on because the recipient cell lacks the repressor. As a result, many virus are made in the recipient cell, and lysis eventually occurs to release the new virus.
Escherichia virus 186 is a virus of the family Myoviridae, genus Eganvirus.
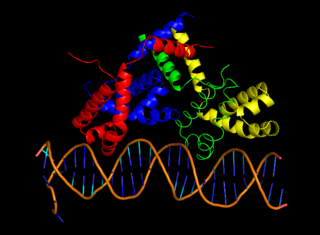
cII or transcriptional activator II is a DNA-binding protein and important transcription factor in the life cycle of lambda phage. It is encoded in the lambda phage genome by the 291 base pair cII gene. cII plays a key role in determining whether the bacteriophage will incorporate its genome into its host and lie dormant (lysogeny), or replicate and kill the host (lysis).
Escherichia virus CC31, formerly known as Enterobacter virus CC31, is a dsDNA bacteriophage of the subfamily Tevenvirinae responsible for infecting the bacteria family of Enterobacteriaceae. It is one of two discovered viruses of the genus Karamvirus, diverging away from the previously discovered T4virus, as a clonal complex (CC). CC31 was first isolated from Escherichia coli B strain S/6/4 and is primarily associated with Escherichia, even though is named after Enterobacter.
The T7 expression system is used in the field of microbiology to clone recombinant DNA using strains of E. coli. It is the most popular system for expressing recombinant proteins in E. coli.
2. Bertani, G., STUDIES ON LYSOGENESIS I.: The Mode of Phage Liberation by Lysogenic Escherichia coli1. Journal of Bacteriology, 1951. 62(3): p. 293.









