WO2022159687A1 - Phenotypic assay to identify protein degraders - Google Patents
Phenotypic assay to identify protein degraders Download PDFInfo
- Publication number
- WO2022159687A1 WO2022159687A1 PCT/US2022/013294 US2022013294W WO2022159687A1 WO 2022159687 A1 WO2022159687 A1 WO 2022159687A1 US 2022013294 W US2022013294 W US 2022013294W WO 2022159687 A1 WO2022159687 A1 WO 2022159687A1
- Authority
- WO
- WIPO (PCT)
- Prior art keywords
- cells
- test compound
- inhibitor
- small molecule
- neddylation
- Prior art date
Links
- 239000001064 degrader Substances 0.000 title claims abstract description 20
- 108090000623 proteins and genes Proteins 0.000 title claims description 30
- 102000004169 proteins and genes Human genes 0.000 title claims description 21
- 238000012247 phenotypical assay Methods 0.000 title description 2
- 238000000034 method Methods 0.000 claims abstract description 48
- 150000003384 small molecules Chemical class 0.000 claims abstract description 31
- 102000052581 Cullin Human genes 0.000 claims abstract description 14
- 108700020475 Cullin Proteins 0.000 claims abstract description 14
- 230000001419 dependent effect Effects 0.000 claims abstract description 13
- 210000004027 cell Anatomy 0.000 claims description 111
- 150000001875 compounds Chemical class 0.000 claims description 71
- 230000009527 neddylation Effects 0.000 claims description 57
- 239000003112 inhibitor Substances 0.000 claims description 51
- 238000012360 testing method Methods 0.000 claims description 47
- MPUQHZXIXSTTDU-QXGSTGNESA-N sulfamic acid [(1S,2S,4R)-4-[4-[[(1S)-2,3-dihydro-1H-inden-1-yl]amino]-7-pyrrolo[2,3-d]pyrimidinyl]-2-hydroxycyclopentyl]methyl ester Chemical group C1[C@H](O)[C@H](COS(=O)(=O)N)C[C@H]1N1C2=NC=NC(N[C@@H]3C4=CC=CC=C4CC3)=C2C=C1 MPUQHZXIXSTTDU-QXGSTGNESA-N 0.000 claims description 36
- -1 small molecule tyrosine kinase inhibitor Chemical class 0.000 claims description 21
- 238000003556 assay Methods 0.000 claims description 18
- 230000014509 gene expression Effects 0.000 claims description 17
- SETFNECMODOHTO-UHFFFAOYSA-N indisulam Chemical compound C1=CC(S(=O)(=O)N)=CC=C1S(=O)(=O)NC1=CC=CC2=C1NC=C2Cl SETFNECMODOHTO-UHFFFAOYSA-N 0.000 claims description 16
- 210000004962 mammalian cell Anatomy 0.000 claims description 16
- 229950009881 indisulam Drugs 0.000 claims description 15
- 230000003247 decreasing effect Effects 0.000 claims description 14
- DOEVCIHTTTYVCC-UHFFFAOYSA-N 1-(3-chloro-4-methylphenyl)-3-[[2-(2,6-dioxopiperidin-3-yl)-1-oxo-3h-isoindol-5-yl]methyl]urea Chemical compound C1=C(Cl)C(C)=CC=C1NC(=O)NCC1=CC=C(C(=O)N(C2)C3C(NC(=O)CC3)=O)C2=C1 DOEVCIHTTTYVCC-UHFFFAOYSA-N 0.000 claims description 13
- 230000001965 increasing effect Effects 0.000 claims description 13
- 230000003833 cell viability Effects 0.000 claims description 12
- 239000003814 drug Substances 0.000 claims description 10
- 238000003559 RNA-seq method Methods 0.000 claims description 9
- 239000011324 bead Substances 0.000 claims description 9
- 229940121358 tyrosine kinase inhibitor Drugs 0.000 claims description 9
- 230000015556 catabolic process Effects 0.000 claims description 8
- 238000006731 degradation reaction Methods 0.000 claims description 8
- 238000003753 real-time PCR Methods 0.000 claims description 8
- 239000005483 tyrosine kinase inhibitor Substances 0.000 claims description 8
- 206010028980 Neoplasm Diseases 0.000 claims description 7
- 230000008859 change Effects 0.000 claims description 7
- 210000005260 human cell Anatomy 0.000 claims description 7
- TZTRUHFXPVXWRD-QTQZEZTPSA-N 4-amino-7-[(2R,3R,4S,5R)-3,4-dihydroxy-5-[(sulfamoylamino)methyl]oxolan-2-yl]-5-[2-(2-ethoxy-6-fluorophenyl)ethynyl]pyrrolo[2,3-d]pyrimidine Chemical group NC=1C2=C(N=CN=1)N(C=C2C#CC1=C(C=CC=C1F)OCC)[C@@H]1O[C@@H]([C@H]([C@H]1O)O)CNS(N)(=O)=O TZTRUHFXPVXWRD-QTQZEZTPSA-N 0.000 claims description 6
- 201000011510 cancer Diseases 0.000 claims description 6
- 238000001943 fluorescence-activated cell sorting Methods 0.000 claims description 6
- 238000011534 incubation Methods 0.000 claims description 6
- 231100000053 low toxicity Toxicity 0.000 claims description 6
- 210000004698 lymphocyte Anatomy 0.000 claims description 6
- 238000007481 next generation sequencing Methods 0.000 claims description 6
- 230000004952 protein activity Effects 0.000 claims description 6
- 230000026447 protein localization Effects 0.000 claims description 6
- 210000000130 stem cell Anatomy 0.000 claims description 6
- 229940124597 therapeutic agent Drugs 0.000 claims description 6
- 108020004999 messenger RNA Proteins 0.000 claims description 5
- 229930014626 natural product Natural products 0.000 claims description 5
- 230000001413 cellular effect Effects 0.000 claims description 4
- 238000000684 flow cytometry Methods 0.000 claims description 4
- 231100000252 nontoxic Toxicity 0.000 claims description 4
- 230000003000 nontoxic effect Effects 0.000 claims description 4
- 210000001519 tissue Anatomy 0.000 claims description 4
- 108091033409 CRISPR Proteins 0.000 claims description 3
- 239000002246 antineoplastic agent Substances 0.000 claims description 3
- 238000001574 biopsy Methods 0.000 claims description 3
- 230000004663 cell proliferation Effects 0.000 claims description 3
- 210000002889 endothelial cell Anatomy 0.000 claims description 3
- 210000002919 epithelial cell Anatomy 0.000 claims description 3
- 210000002950 fibroblast Anatomy 0.000 claims description 3
- 210000004602 germ cell Anatomy 0.000 claims description 3
- 210000003494 hepatocyte Anatomy 0.000 claims description 3
- 238000003119 immunoblot Methods 0.000 claims description 3
- 210000002510 keratinocyte Anatomy 0.000 claims description 3
- 229940043355 kinase inhibitor Drugs 0.000 claims description 3
- 238000012083 mass cytometry Methods 0.000 claims description 3
- 210000002752 melanocyte Anatomy 0.000 claims description 3
- 210000005033 mesothelial cell Anatomy 0.000 claims description 3
- 210000000663 muscle cell Anatomy 0.000 claims description 3
- 210000000066 myeloid cell Anatomy 0.000 claims description 3
- 210000003098 myoblast Anatomy 0.000 claims description 3
- 210000003061 neural cell Anatomy 0.000 claims description 3
- 210000000963 osteoblast Anatomy 0.000 claims description 3
- 210000002997 osteoclast Anatomy 0.000 claims description 3
- 239000003757 phosphotransferase inhibitor Substances 0.000 claims description 3
- 229940126586 small molecule drug Drugs 0.000 claims description 2
- 238000012216 screening Methods 0.000 abstract description 2
- IAZDPXIOMUYVGZ-UHFFFAOYSA-N Dimethylsulphoxide Chemical compound CS(C)=O IAZDPXIOMUYVGZ-UHFFFAOYSA-N 0.000 description 15
- 231100000419 toxicity Toxicity 0.000 description 13
- 230000001988 toxicity Effects 0.000 description 13
- 238000013537 high throughput screening Methods 0.000 description 9
- 102000004190 Enzymes Human genes 0.000 description 7
- 108090000790 Enzymes Proteins 0.000 description 7
- 102000006275 Ubiquitin-Protein Ligases Human genes 0.000 description 7
- 108010083111 Ubiquitin-Protein Ligases Proteins 0.000 description 7
- 229940079593 drug Drugs 0.000 description 7
- 239000000758 substrate Substances 0.000 description 7
- 101000941994 Homo sapiens Protein cereblon Proteins 0.000 description 6
- 239000003292 glue Substances 0.000 description 6
- 238000003018 immunoassay Methods 0.000 description 5
- 230000017854 proteolysis Effects 0.000 description 5
- 238000003734 CellTiter-Glo Luminescent Cell Viability Assay Methods 0.000 description 4
- TZYWCYJVHRLUCT-VABKMULXSA-N N-benzyloxycarbonyl-L-leucyl-L-leucyl-L-leucinal Chemical compound CC(C)C[C@@H](C=O)NC(=O)[C@H](CC(C)C)NC(=O)[C@H](CC(C)C)NC(=O)OCC1=CC=CC=C1 TZYWCYJVHRLUCT-VABKMULXSA-N 0.000 description 4
- KJDAGXLMHXUAGV-DGWLBADLSA-N [(1r,2r,3s,4r)-2,3-dihydroxy-4-[[2-[3-(trifluoromethylsulfanyl)phenyl]pyrazolo[1,5-a]pyrimidin-7-yl]amino]cyclopentyl]methyl sulfamate Chemical compound O[C@@H]1[C@H](O)[C@@H](COS(=O)(=O)N)C[C@H]1NC1=CC=NC2=CC(C=3C=C(SC(F)(F)F)C=CC=3)=NN12 KJDAGXLMHXUAGV-DGWLBADLSA-N 0.000 description 4
- 239000003795 chemical substances by application Substances 0.000 description 4
- 231100000673 dose–response relationship Toxicity 0.000 description 4
- 230000000694 effects Effects 0.000 description 4
- 239000000203 mixture Substances 0.000 description 4
- 102000036364 Cullin Ring E3 Ligases Human genes 0.000 description 3
- 108091007045 Cullin Ring E3 Ligases Proteins 0.000 description 3
- 102100039768 DDB1- and CUL4-associated factor 15 Human genes 0.000 description 3
- 101000885463 Homo sapiens DDB1- and CUL4-associated factor 15 Proteins 0.000 description 3
- POJZIZBONPAWIV-UHFFFAOYSA-N N-methyl-N-[2-[[[2-[(2-oxo-1,3-dihydroindol-5-yl)amino]-5-(trifluoromethyl)-4-pyrimidinyl]amino]methyl]phenyl]methanesulfonamide Chemical compound CS(=O)(=O)N(C)C1=CC=CC=C1CNC1=NC(NC=2C=C3CC(=O)NC3=CC=2)=NC=C1C(F)(F)F POJZIZBONPAWIV-UHFFFAOYSA-N 0.000 description 3
- 102000038427 NEDD8-activating enzyme E1 Human genes 0.000 description 3
- 108091007790 NEDD8-activating enzyme E1 Proteins 0.000 description 3
- 102100032783 Protein cereblon Human genes 0.000 description 3
- 102100025858 RNA-binding protein 39 Human genes 0.000 description 3
- 101710205955 RNA-binding protein 39 Proteins 0.000 description 3
- 230000006907 apoptotic process Effects 0.000 description 3
- 230000027455 binding Effects 0.000 description 3
- 230000030833 cell death Effects 0.000 description 3
- 230000005754 cellular signaling Effects 0.000 description 3
- 238000011278 co-treatment Methods 0.000 description 3
- 230000005764 inhibitory process Effects 0.000 description 3
- 230000003993 interaction Effects 0.000 description 3
- 239000003550 marker Substances 0.000 description 3
- 230000001404 mediated effect Effects 0.000 description 3
- 231100000331 toxic Toxicity 0.000 description 3
- 230000002588 toxic effect Effects 0.000 description 3
- 238000011282 treatment Methods 0.000 description 3
- YBLWOZUPHDKFOT-UHFFFAOYSA-N 1-(5-chloro-2-methoxyphenyl)-3-(2-methyl-4-quinolinyl)urea Chemical compound COC1=CC=C(Cl)C=C1NC(=O)NC1=CC(C)=NC2=CC=CC=C12 YBLWOZUPHDKFOT-UHFFFAOYSA-N 0.000 description 2
- UEJJHQNACJXSKW-UHFFFAOYSA-N 2-(2,6-dioxopiperidin-3-yl)-1H-isoindole-1,3(2H)-dione Chemical compound O=C1C2=CC=CC=C2C(=O)N1C1CCC(=O)NC1=O UEJJHQNACJXSKW-UHFFFAOYSA-N 0.000 description 2
- CBIAKDAYHRWZCU-UHFFFAOYSA-N 2-bromo-4-[(6,7-dimethoxyquinazolin-4-yl)amino]phenol Chemical compound C=12C=C(OC)C(OC)=CC2=NC=NC=1NC1=CC=C(O)C(Br)=C1 CBIAKDAYHRWZCU-UHFFFAOYSA-N 0.000 description 2
- IPDMWUNUULAXLU-UHFFFAOYSA-N 3-hydroxy-1-methoxy-9,10-dioxo-2-anthracenecarboxaldehyde Chemical compound O=C1C2=CC=CC=C2C(=O)C2=C1C=C(O)C(C=O)=C2OC IPDMWUNUULAXLU-UHFFFAOYSA-N 0.000 description 2
- 102100036876 Cyclin-K Human genes 0.000 description 2
- 108090000266 Cyclin-dependent kinases Proteins 0.000 description 2
- 102000003903 Cyclin-dependent kinases Human genes 0.000 description 2
- 102000012698 DDB1 Human genes 0.000 description 2
- 238000002965 ELISA Methods 0.000 description 2
- 102100036816 Eukaryotic peptide chain release factor GTP-binding subunit ERF3A Human genes 0.000 description 2
- 101000713127 Homo sapiens Cyclin-K Proteins 0.000 description 2
- 101000851788 Homo sapiens Eukaryotic peptide chain release factor GTP-binding subunit ERF3A Proteins 0.000 description 2
- 241000124008 Mammalia Species 0.000 description 2
- PBBRWFOVCUAONR-UHFFFAOYSA-N PP2 Chemical compound C12=C(N)N=CN=C2N(C(C)(C)C)N=C1C1=CC=C(Cl)C=C1 PBBRWFOVCUAONR-UHFFFAOYSA-N 0.000 description 2
- 108700008625 Reporter Genes Proteins 0.000 description 2
- JANPYFTYAGTSIN-FYJGNVAPSA-N SU1498 Chemical compound CC(C)C1=C(O)C(C(C)C)=CC(\C=C(/C#N)C(=O)NCCCC=2C=CC=CC=2)=C1 JANPYFTYAGTSIN-FYJGNVAPSA-N 0.000 description 2
- 102000044159 Ubiquitin Human genes 0.000 description 2
- 108090000848 Ubiquitin Proteins 0.000 description 2
- 230000004075 alteration Effects 0.000 description 2
- 230000022534 cell killing Effects 0.000 description 2
- 230000026374 cyclin catabolic process Effects 0.000 description 2
- 101150077768 ddb1 gene Proteins 0.000 description 2
- 239000002532 enzyme inhibitor Substances 0.000 description 2
- 238000002866 fluorescence resonance energy transfer Methods 0.000 description 2
- 230000012010 growth Effects 0.000 description 2
- WJEOLQLKVOPQFV-UHFFFAOYSA-N masitinib Chemical compound C1CN(C)CCN1CC1=CC=C(C(=O)NC=2C=C(NC=3SC=C(N=3)C=3C=NC=CC=3)C(C)=CC=2)C=C1 WJEOLQLKVOPQFV-UHFFFAOYSA-N 0.000 description 2
- 238000005259 measurement Methods 0.000 description 2
- WIMITXDBYLKRKB-UHFFFAOYSA-N n-[3-chloro-4-[(3-fluorophenyl)methoxy]phenyl]-6-[2-[(2-methylsulfonylethylamino)methyl]-1,3-thiazol-4-yl]quinazolin-4-amine;dihydrochloride Chemical compound Cl.Cl.S1C(CNCCS(=O)(=O)C)=NC(C=2C=C3C(NC=4C=C(Cl)C(OCC=5C=C(F)C=CC=5)=CC=4)=NC=NC3=CC=2)=C1 WIMITXDBYLKRKB-UHFFFAOYSA-N 0.000 description 2
- 230000037361 pathway Effects 0.000 description 2
- 229940124823 proteolysis targeting chimeric molecule Drugs 0.000 description 2
- 230000004063 proteosomal degradation Effects 0.000 description 2
- 238000003127 radioimmunoassay Methods 0.000 description 2
- 229950003647 semaxanib Drugs 0.000 description 2
- WUWDLXZGHZSWQZ-WQLSENKSSA-N semaxanib Chemical compound N1C(C)=CC(C)=C1\C=C/1C2=CC=CC=C2NC\1=O WUWDLXZGHZSWQZ-WQLSENKSSA-N 0.000 description 2
- 230000009758 senescence Effects 0.000 description 2
- 239000000126 substance Substances 0.000 description 2
- YJRLSCDUYLRBIZ-UHFFFAOYSA-N sulochrin Chemical compound COC(=O)C1=CC(O)=CC(OC)=C1C(=O)C1=C(O)C=C(C)C=C1O YJRLSCDUYLRBIZ-UHFFFAOYSA-N 0.000 description 2
- 238000003786 synthesis reaction Methods 0.000 description 2
- 229960001350 tofacitinib Drugs 0.000 description 2
- UJLAWZDWDVHWOW-YPMHNXCESA-N tofacitinib Chemical compound C[C@@H]1CCN(C(=O)CC#N)C[C@@H]1N(C)C1=NC=NC2=C1C=CN2 UJLAWZDWDVHWOW-YPMHNXCESA-N 0.000 description 2
- 230000034512 ubiquitination Effects 0.000 description 2
- 238000010798 ubiquitination Methods 0.000 description 2
- 230000035899 viability Effects 0.000 description 2
- CGEORJKFOZSMEZ-MBZVMHRFSA-N (+)-sesamin monocatechol Chemical compound C1=C(O)C(O)=CC=C1[C@@H]1[C@@H](CO[C@@H]2C=3C=C4OCOC4=CC=3)[C@@H]2CO1 CGEORJKFOZSMEZ-MBZVMHRFSA-N 0.000 description 1
- ZESGNAJSBDILTB-OXVOKJAASA-N (2e,4s,5s,6e,8e)-10-[(2s,3r,6s,8r,9s)-3-butyl-2-[(1e,3e)-4-carboxy-3-methylbuta-1,3-dienyl]-3-(3-carboxypropanoyloxy)-9-methyl-1,7-dioxaspiro[5.5]undecan-8-yl]-5-hydroxy-4,8-dimethyldeca-2,6,8-trienoic acid Chemical compound O1[C@@H](\C=C\C(\C)=C\C(O)=O)[C@](CCCC)(OC(=O)CCC(O)=O)CC[C@@]11O[C@H](C\C=C(/C)\C=C\[C@H](O)[C@@H](C)\C=C\C(O)=O)[C@@H](C)CC1 ZESGNAJSBDILTB-OXVOKJAASA-N 0.000 description 1
- HOCBJBNQIQQQGT-LJQANCHMSA-N (2r)-2-({9-(1-methylethyl)-6-[(4-pyridin-2-ylbenzyl)amino]-9h-purin-2-yl}amino)butan-1-ol Chemical compound C=12N=CN(C(C)C)C2=NC(N[C@@H](CO)CC)=NC=1NCC(C=C1)=CC=C1C1=CC=CC=N1 HOCBJBNQIQQQGT-LJQANCHMSA-N 0.000 description 1
- YOVVNQKCSKSHKT-HNNXBMFYSA-N (2s)-1-[4-[[2-(2-aminopyrimidin-5-yl)-7-methyl-4-morpholin-4-ylthieno[3,2-d]pyrimidin-6-yl]methyl]piperazin-1-yl]-2-hydroxypropan-1-one Chemical compound C1CN(C(=O)[C@@H](O)C)CCN1CC1=C(C)C2=NC(C=3C=NC(N)=NC=3)=NC(N3CCOCC3)=C2S1 YOVVNQKCSKSHKT-HNNXBMFYSA-N 0.000 description 1
- JKNOYWVMHPMBEL-UNXLUWIOSA-N (3z)-2-amino-4-(3,4,5-trihydroxyphenyl)buta-1,3-diene-1,1,3-tricarbonitrile Chemical compound N#CC(C#N)=C(N)\C(C#N)=C\C1=CC(O)=C(O)C(O)=C1 JKNOYWVMHPMBEL-UNXLUWIOSA-N 0.000 description 1
- NXNQLECPAXXYTR-LCYFTJDESA-N (3z)-3-[(1-methylindol-3-yl)methylidene]-1h-pyrrolo[3,2-b]pyridin-2-one Chemical compound C12=CC=CC=C2N(C)C=C1\C=C/1C2=NC=CC=C2NC\1=O NXNQLECPAXXYTR-LCYFTJDESA-N 0.000 description 1
- QDITZBLZQQZVEE-YBEGLDIGSA-N (5z)-5-[(4-pyridin-4-ylquinolin-6-yl)methylidene]-1,3-thiazolidine-2,4-dione Chemical compound S1C(=O)NC(=O)\C1=C\C1=CC=C(N=CC=C2C=3C=CN=CC=3)C2=C1 QDITZBLZQQZVEE-YBEGLDIGSA-N 0.000 description 1
- MYKCTDWWIWGLHW-NTEUORMPSA-N (e)-2-(2,3-dihydroindole-1-carbonyl)-3-(3,4-dihydroxyphenyl)prop-2-enenitrile Chemical compound C1=C(O)C(O)=CC=C1\C=C(/C#N)C(=O)N1C2=CC=CC=C2CC1 MYKCTDWWIWGLHW-NTEUORMPSA-N 0.000 description 1
- HSRMHXWCTRFVHK-NYYWCZLTSA-N (e)-2-(3,4-dihydroxybenzoyl)-3-(4-hydroxy-3-iodo-5-methoxyphenyl)prop-2-enenitrile Chemical compound IC1=C(O)C(OC)=CC(\C=C(/C#N)C(=O)C=2C=C(O)C(O)=CC=2)=C1 HSRMHXWCTRFVHK-NYYWCZLTSA-N 0.000 description 1
- UMGQVUWXNOJOSJ-DGGAMASNSA-N (e)-2-cyano-3-(3,4-dihydroxyphenyl)-n-[(1s)-1-phenylethyl]prop-2-enamide Chemical compound N([C@@H](C)C=1C=CC=CC=1)C(=O)C(\C#N)=C\C1=CC=C(O)C(O)=C1 UMGQVUWXNOJOSJ-DGGAMASNSA-N 0.000 description 1
- FOGXCBUSKWTDKV-NTUHNPAUSA-N (e)-2-cyano-3-[4-hydroxy-3-methoxy-5-(phenylsulfanylmethyl)phenyl]prop-2-enamide Chemical compound COC1=CC(\C=C(/C#N)C(N)=O)=CC(CSC=2C=CC=CC=2)=C1O FOGXCBUSKWTDKV-NTUHNPAUSA-N 0.000 description 1
- DBGZNJVTHYFQJI-ZSOIEALJSA-N (e)-3-(3,4-dimethoxyphenyl)-2-pyridin-3-ylprop-2-enenitrile Chemical compound C1=C(OC)C(OC)=CC=C1\C=C(\C#N)C1=CC=CN=C1 DBGZNJVTHYFQJI-ZSOIEALJSA-N 0.000 description 1
- TYXIVBJQPBWBHO-QCDXTXTGSA-N (e)-3-(3,5-dichlorophenyl)-2-pyridin-3-ylprop-2-enenitrile Chemical compound ClC1=CC(Cl)=CC(\C=C(\C#N)C=2C=NC=CC=2)=C1 TYXIVBJQPBWBHO-QCDXTXTGSA-N 0.000 description 1
- NERXPXBELDBEPZ-RMKNXTFCSA-N (e)-n-[4-[3-chloro-4-[(3-fluorophenyl)methoxy]anilino]-3-cyano-7-ethoxyquinolin-6-yl]-4-(dimethylamino)but-2-enamide Chemical compound C=12C=C(NC(=O)\C=C\CN(C)C)C(OCC)=CC2=NC=C(C#N)C=1NC(C=C1Cl)=CC=C1OCC1=CC=CC(F)=C1 NERXPXBELDBEPZ-RMKNXTFCSA-N 0.000 description 1
- YKLMGKWXBLSKPK-ZSOIEALJSA-N (z)-2-cyano-3-[3-ethoxy-4-hydroxy-5-(phenylsulfanylmethyl)phenyl]prop-2-enamide Chemical compound CCOC1=CC(\C=C(\C#N)C(N)=O)=CC(CSC=2C=CC=CC=2)=C1O YKLMGKWXBLSKPK-ZSOIEALJSA-N 0.000 description 1
- JLOXTZFYJNCPIS-FYWRMAATSA-N (z)-3-amino-3-(4-aminophenyl)sulfanyl-2-[2-(trifluoromethyl)phenyl]prop-2-enenitrile Chemical compound C=1C=CC=C(C(F)(F)F)C=1C(\C#N)=C(/N)SC1=CC=C(N)C=C1 JLOXTZFYJNCPIS-FYWRMAATSA-N 0.000 description 1
- MGUDDBRJHXFTEY-UHFFFAOYSA-N 1-(3-amino-1,2-benzoxazol-5-yl)-6-[4-[2-[(dimethylamino)methyl]imidazol-1-yl]-2-fluorophenyl]-7-fluoroindazole-3-carboxamide Chemical compound CN(C)CC1=NC=CN1C1=CC=C(C=2C(=C3N(C=4C=C5C(N)=NOC5=CC=4)N=C(C3=CC=2)C(N)=O)F)C(F)=C1 MGUDDBRJHXFTEY-UHFFFAOYSA-N 0.000 description 1
- RZUOCXOYPYGSKL-GOSISDBHSA-N 1-[(1s)-1-(4-chloro-3-fluorophenyl)-2-hydroxyethyl]-4-[2-[(2-methylpyrazol-3-yl)amino]pyrimidin-4-yl]pyridin-2-one Chemical compound CN1N=CC=C1NC1=NC=CC(C2=CC(=O)N([C@H](CO)C=3C=C(F)C(Cl)=CC=3)C=C2)=N1 RZUOCXOYPYGSKL-GOSISDBHSA-N 0.000 description 1
- NQHFSECYQAQZBN-LSYPWIJNSA-M 1-[(3s)-3-(3,4-dichlorophenyl)-3-[2-(4-phenyl-1-azoniabicyclo[2.2.2]octan-1-yl)ethyl]piperidin-1-yl]-2-(3-propan-2-yloxyphenyl)ethanone;chloride Chemical compound [Cl-].CC(C)OC1=CC=CC(CC(=O)N2C[C@](CC[N+]34CCC(CC3)(CC4)C=3C=CC=CC=3)(CCC2)C=2C=C(Cl)C(Cl)=CC=2)=C1 NQHFSECYQAQZBN-LSYPWIJNSA-M 0.000 description 1
- VPBYZLCHOKSGRX-UHFFFAOYSA-N 1-[2-chloro-4-(6,7-dimethoxyquinazolin-4-yl)oxyphenyl]-3-propylurea Chemical compound C1=C(Cl)C(NC(=O)NCCC)=CC=C1OC1=NC=NC2=CC(OC)=C(OC)C=C12 VPBYZLCHOKSGRX-UHFFFAOYSA-N 0.000 description 1
- YYDUWLSETXNJJT-MTJSOVHGSA-N 1-[2-fluoro-5-(trifluoromethyl)phenyl]-3-[4-methyl-3-[[(3z)-2-oxo-3-(1h-pyrrol-2-ylmethylidene)-1h-indol-6-yl]amino]phenyl]urea Chemical compound C1=C(NC=2C=C3NC(=O)C(=C\C=4NC=CC=4)/C3=CC=2)C(C)=CC=C1NC(=O)NC1=CC(C(F)(F)F)=CC=C1F YYDUWLSETXNJJT-MTJSOVHGSA-N 0.000 description 1
- QLHHRYZMBGPBJG-UHFFFAOYSA-N 1-[4-[1-(1,4-dioxaspiro[4.5]decan-8-yl)-4-(8-oxa-3-azabicyclo[3.2.1]octan-3-yl)-6-pyrazolo[3,4-d]pyrimidinyl]phenyl]-3-methylurea Chemical compound C1=CC(NC(=O)NC)=CC=C1C1=NC(N2CC3CCC(O3)C2)=C(C=NN2C3CCC4(CC3)OCCO4)C2=N1 QLHHRYZMBGPBJG-UHFFFAOYSA-N 0.000 description 1
- YJZSUCFGHXQWDM-UHFFFAOYSA-N 1-adamantyl 4-[(2,5-dihydroxyphenyl)methylamino]benzoate Chemical compound OC1=CC=C(O)C(CNC=2C=CC(=CC=2)C(=O)OC23CC4CC(CC(C4)C2)C3)=C1 YJZSUCFGHXQWDM-UHFFFAOYSA-N 0.000 description 1
- RGJOJUGRHPQXGF-INIZCTEOSA-N 1-ethyl-3-[4-[4-[(3s)-3-methylmorpholin-4-yl]-7-(oxetan-3-yl)-6,8-dihydro-5h-pyrido[3,4-d]pyrimidin-2-yl]phenyl]urea Chemical compound C1=CC(NC(=O)NCC)=CC=C1C(N=C1N2[C@H](COCC2)C)=NC2=C1CCN(C1COC1)C2 RGJOJUGRHPQXGF-INIZCTEOSA-N 0.000 description 1
- SEERCQIVLBZZJL-UHFFFAOYSA-N 1-n'-[3-fluoro-4-[2-[3-(methylsulfonylmethyl)anilino]pyrimidin-4-yl]oxyphenyl]-1-n-(4-fluorophenyl)cyclopropane-1,1-dicarboxamide Chemical compound CS(=O)(=O)CC1=CC=CC(NC=2N=C(OC=3C(=CC(NC(=O)C4(CC4)C(=O)NC=4C=CC(F)=CC=4)=CC=3)F)C=CN=2)=C1 SEERCQIVLBZZJL-UHFFFAOYSA-N 0.000 description 1
- 125000001637 1-naphthyl group Chemical group [H]C1=C([H])C([H])=C2C(*)=C([H])C([H])=C([H])C2=C1[H] 0.000 description 1
- ZVPDNRVYHLRXLX-UHFFFAOYSA-N 1-ter-butyl-3-p-tolyl-1h-pyrazolo[3,4-d]pyrimidin-4-ylamine Chemical compound C1=CC(C)=CC=C1C1=NN(C(C)(C)C)C2=NC=NC(N)=C12 ZVPDNRVYHLRXLX-UHFFFAOYSA-N 0.000 description 1
- CUNDRHORZHFPLY-UHFFFAOYSA-N 138154-39-9 Chemical compound O=C1C2=CC(O)=CC=C2N2C=NC3=CC=C(NCCN(CC)CC)C1=C32 CUNDRHORZHFPLY-UHFFFAOYSA-N 0.000 description 1
- FPYJSJDOHRDAMT-KQWNVCNZSA-N 1h-indole-5-sulfonamide, n-(3-chlorophenyl)-3-[[3,5-dimethyl-4-[(4-methyl-1-piperazinyl)carbonyl]-1h-pyrrol-2-yl]methylene]-2,3-dihydro-n-methyl-2-oxo-, (3z)- Chemical compound C=1C=C2NC(=O)\C(=C/C3=C(C(C(=O)N4CCN(C)CC4)=C(C)N3)C)C2=CC=1S(=O)(=O)N(C)C1=CC=CC(Cl)=C1 FPYJSJDOHRDAMT-KQWNVCNZSA-N 0.000 description 1
- QFWCYNPOPKQOKV-UHFFFAOYSA-N 2-(2-amino-3-methoxyphenyl)chromen-4-one Chemical compound COC1=CC=CC(C=2OC3=CC=CC=C3C(=O)C=2)=C1N QFWCYNPOPKQOKV-UHFFFAOYSA-N 0.000 description 1
- GFMMXOIFOQCCGU-UHFFFAOYSA-N 2-(2-chloro-4-iodoanilino)-N-(cyclopropylmethoxy)-3,4-difluorobenzamide Chemical compound C=1C=C(I)C=C(Cl)C=1NC1=C(F)C(F)=CC=C1C(=O)NOCC1CC1 GFMMXOIFOQCCGU-UHFFFAOYSA-N 0.000 description 1
- RWEVIPRMPFNTLO-UHFFFAOYSA-N 2-(2-fluoro-4-iodoanilino)-N-(2-hydroxyethoxy)-1,5-dimethyl-6-oxo-3-pyridinecarboxamide Chemical compound CN1C(=O)C(C)=CC(C(=O)NOCCO)=C1NC1=CC=C(I)C=C1F RWEVIPRMPFNTLO-UHFFFAOYSA-N 0.000 description 1
- RQVGFDBMONQTBC-UHFFFAOYSA-N 2-(6,7-dimethoxyquinazolin-4-yl)sulfanyl-5-methyl-1,3,4-thiadiazole Chemical compound C=12C=C(OC)C(OC)=CC2=NC=NC=1SC1=NN=C(C)S1 RQVGFDBMONQTBC-UHFFFAOYSA-N 0.000 description 1
- NPVXOWLPOFYACO-UHFFFAOYSA-N 2-(butylamino)-4-[(4-hydroxycyclohexyl)amino]-n-[(4-imidazol-1-ylphenyl)methyl]pyrimidine-5-carboxamide Chemical compound C1CC(O)CCC1NC1=NC(NCCCC)=NC=C1C(=O)NCC(C=C1)=CC=C1N1C=CN=C1 NPVXOWLPOFYACO-UHFFFAOYSA-N 0.000 description 1
- QFNJFVBKASKGEU-OCEACIFDSA-N 2-(diethylaminomethyl)-4-[(e)-4-[3-(diethylaminomethyl)-4-hydroxyphenyl]hex-3-en-3-yl]phenol Chemical compound C1=C(O)C(CN(CC)CC)=CC(C(\CC)=C(/CC)C=2C=C(CN(CC)CC)C(O)=CC=2)=C1 QFNJFVBKASKGEU-OCEACIFDSA-N 0.000 description 1
- WPXSPHPYZHPIPO-UHFFFAOYSA-N 2-[(3,4-dihydroxy-5-iodophenyl)methylidene]propanedinitrile Chemical compound OC1=CC(C=C(C#N)C#N)=CC(I)=C1O WPXSPHPYZHPIPO-UHFFFAOYSA-N 0.000 description 1
- DWZFIFZYMHNRRV-UHFFFAOYSA-N 2-[(3,4-dihydroxy-5-methoxyphenyl)methylidene]propanedinitrile Chemical compound COC1=CC(C=C(C#N)C#N)=CC(O)=C1O DWZFIFZYMHNRRV-UHFFFAOYSA-N 0.000 description 1
- VTJXFTPMFYAJJU-UHFFFAOYSA-N 2-[(3,4-dihydroxyphenyl)methylidene]propanedinitrile Chemical compound OC1=CC=C(C=C(C#N)C#N)C=C1O VTJXFTPMFYAJJU-UHFFFAOYSA-N 0.000 description 1
- PDMUGYOXRHVNMO-UHFFFAOYSA-N 2-[4-[3-(6-quinolinylmethyl)-5-triazolo[4,5-b]pyrazinyl]-1-pyrazolyl]ethanol Chemical compound C1=NN(CCO)C=C1C1=CN=C(N=NN2CC=3C=C4C=CC=NC4=CC=3)C2=N1 PDMUGYOXRHVNMO-UHFFFAOYSA-N 0.000 description 1
- FSPQCTGGIANIJZ-UHFFFAOYSA-N 2-[[(3,4-dimethoxyphenyl)-oxomethyl]amino]-4,5,6,7-tetrahydro-1-benzothiophene-3-carboxamide Chemical compound C1=C(OC)C(OC)=CC=C1C(=O)NC1=C(C(N)=O)C(CCCC2)=C2S1 FSPQCTGGIANIJZ-UHFFFAOYSA-N 0.000 description 1
- IGUBBWJDMLCRIK-UHFFFAOYSA-N 2-[[2-(2-methoxy-4-morpholin-4-ylanilino)-5-(trifluoromethyl)pyridin-4-yl]amino]-n-methylbenzamide Chemical compound CNC(=O)C1=CC=CC=C1NC1=CC(NC=2C(=CC(=CC=2)N2CCOCC2)OC)=NC=C1C(F)(F)F IGUBBWJDMLCRIK-UHFFFAOYSA-N 0.000 description 1
- HZTYDQRUAWIZRE-UHFFFAOYSA-N 2-[[2-[[1-[2-(dimethylamino)-1-oxoethyl]-5-methoxy-2,3-dihydroindol-6-yl]amino]-7H-pyrrolo[2,3-d]pyrimidin-4-yl]amino]-6-fluoro-N-methylbenzamide Chemical compound CNC(=O)C1=C(F)C=CC=C1NC1=C2C=CN=C2NC(NC=2C(=CC=3CCN(C=3C=2)C(=O)CN(C)C)OC)=N1 HZTYDQRUAWIZRE-UHFFFAOYSA-N 0.000 description 1
- DPLMXAYKJZOTKO-UHFFFAOYSA-N 2-methyl-2-[4-(2-oxo-9-quinolin-3-yl-4h-[1,3]oxazino[5,4-c]quinolin-1-yl)phenyl]propanenitrile Chemical compound C1=CC(C(C)(C#N)C)=CC=C1N1C2=C3C=C(C=4C=C5C=CC=CC5=NC=4)C=CC3=NC=C2COC1=O DPLMXAYKJZOTKO-UHFFFAOYSA-N 0.000 description 1
- QAZJUVDICQNITG-UHFFFAOYSA-N 3,4-dimethoxy-n-(5-phenyl-1h-pyrazol-3-yl)benzamide Chemical compound C1=C(OC)C(OC)=CC=C1C(=O)NC1=NNC(C=2C=CC=CC=2)=C1 QAZJUVDICQNITG-UHFFFAOYSA-N 0.000 description 1
- UHIZYQVRKSWIFO-UHFFFAOYSA-N 3-(4-chlorophenyl)-1-[3-[3-[(3-phenylphenyl)methyl]triazol-4-yl]phenyl]pyrazolo[3,4-d]pyrimidin-4-amine Chemical compound C1=2C(N)=NC=NC=2N(C=2C=C(C=CC=2)C=2N(N=NC=2)CC=2C=C(C=CC=2)C=2C=CC=CC=2)N=C1C1=CC=C(Cl)C=C1 UHIZYQVRKSWIFO-UHFFFAOYSA-N 0.000 description 1
- FGTCROZDHDSNIO-UHFFFAOYSA-N 3-(4-quinolinylmethylamino)-N-[4-(trifluoromethoxy)phenyl]-2-thiophenecarboxamide Chemical compound C1=CC(OC(F)(F)F)=CC=C1NC(=O)C1=C(NCC=2C3=CC=CC=C3N=CC=2)C=CS1 FGTCROZDHDSNIO-UHFFFAOYSA-N 0.000 description 1
- RCLQNICOARASSR-SECBINFHSA-N 3-[(2r)-2,3-dihydroxypropyl]-6-fluoro-5-(2-fluoro-4-iodoanilino)-8-methylpyrido[2,3-d]pyrimidine-4,7-dione Chemical compound FC=1C(=O)N(C)C=2N=CN(C[C@@H](O)CO)C(=O)C=2C=1NC1=CC=C(I)C=C1F RCLQNICOARASSR-SECBINFHSA-N 0.000 description 1
- HXHAJRMTJXHJJZ-UHFFFAOYSA-N 3-[(4-bromo-2,6-difluorophenyl)methoxy]-5-(4-pyrrolidin-1-ylbutylcarbamoylamino)-1,2-thiazole-4-carboxamide Chemical compound S1N=C(OCC=2C(=CC(Br)=CC=2F)F)C(C(=O)N)=C1NC(=O)NCCCCN1CCCC1 HXHAJRMTJXHJJZ-UHFFFAOYSA-N 0.000 description 1
- JUSFANSTBFGBAF-IRXDYDNUSA-N 3-[2,4-bis[(3s)-3-methylmorpholin-4-yl]pyrido[2,3-d]pyrimidin-7-yl]-n-methylbenzamide Chemical compound CNC(=O)C1=CC=CC(C=2N=C3N=C(N=C(C3=CC=2)N2[C@H](COCC2)C)N2[C@H](COCC2)C)=C1 JUSFANSTBFGBAF-IRXDYDNUSA-N 0.000 description 1
- OCUQMWSIGPQEMX-UHFFFAOYSA-N 3-[3-[N-[4-[(dimethylamino)methyl]phenyl]-C-phenylcarbonimidoyl]-2-hydroxy-1H-indol-6-yl]-N-ethylprop-2-ynamide Chemical compound CCNC(=O)C#CC1=CC2=C(C=C1)C(=C(N2)O)C(=NC3=CC=C(C=C3)CN(C)C)C4=CC=CC=C4 OCUQMWSIGPQEMX-UHFFFAOYSA-N 0.000 description 1
- CGJVMKJGKFEHTL-HSZRJFAPSA-N 3-[4-amino-1-[(3r)-1-prop-2-enoylpiperidin-3-yl]pyrazolo[3,4-d]pyrimidin-3-yl]-n-(3-methyl-4-propan-2-ylphenyl)benzamide Chemical compound C1=C(C)C(C(C)C)=CC=C1NC(=O)C1=CC=CC(C=2C3=C(N)N=CN=C3N([C@H]3CN(CCC3)C(=O)C=C)N=2)=C1 CGJVMKJGKFEHTL-HSZRJFAPSA-N 0.000 description 1
- WEVYNIUIFUYDGI-UHFFFAOYSA-N 3-[6-[4-(trifluoromethoxy)anilino]-4-pyrimidinyl]benzamide Chemical compound NC(=O)C1=CC=CC(C=2N=CN=C(NC=3C=CC(OC(F)(F)F)=CC=3)C=2)=C1 WEVYNIUIFUYDGI-UHFFFAOYSA-N 0.000 description 1
- ZGXOBLVQIVXKEB-UHFFFAOYSA-N 3-[N-[3-[(dimethylamino)methyl]phenyl]-C-phenylcarbonimidoyl]-2-hydroxy-N,N-dimethyl-1H-indole-6-carboxamide Chemical compound CN(C)CC1=CC(=CC=C1)N=C(C2=CC=CC=C2)C3=C(NC4=C3C=CC(=C4)C(=O)N(C)C)O ZGXOBLVQIVXKEB-UHFFFAOYSA-N 0.000 description 1
- DZQLVVLATXPWBK-UHFFFAOYSA-N 3-phenyl-1H-benzofuro[3,2-c]pyrazole Chemical compound C1=CC=CC=C1C1=NNC2=C1OC1=CC=CC=C12 DZQLVVLATXPWBK-UHFFFAOYSA-N 0.000 description 1
- AAALVYBICLMAMA-UHFFFAOYSA-N 4,5-dianilinophthalimide Chemical compound C=1C=CC=CC=1NC=1C=C2C(=O)NC(=O)C2=CC=1NC1=CC=CC=C1 AAALVYBICLMAMA-UHFFFAOYSA-N 0.000 description 1
- XXJWYDDUDKYVKI-UHFFFAOYSA-N 4-[(4-fluoro-2-methyl-1H-indol-5-yl)oxy]-6-methoxy-7-[3-(1-pyrrolidinyl)propoxy]quinazoline Chemical compound COC1=CC2=C(OC=3C(=C4C=C(C)NC4=CC=3)F)N=CN=C2C=C1OCCCN1CCCC1 XXJWYDDUDKYVKI-UHFFFAOYSA-N 0.000 description 1
- SKZQZGSPYYHTQG-UHFFFAOYSA-N 4-[2-[4-(3-phenylpyrazolo[1,5-a]pyrimidin-6-yl)phenoxy]ethyl]morpholine Chemical compound C=1C=C(C2=CN3N=CC(=C3N=C2)C=2C=CC=CC=2)C=CC=1OCCN1CCOCC1 SKZQZGSPYYHTQG-UHFFFAOYSA-N 0.000 description 1
- SYYMNUFXRFAELA-BTQNPOSSSA-N 4-[4-[[(1r)-1-phenylethyl]amino]-7h-pyrrolo[2,3-d]pyrimidin-6-yl]phenol;hydrobromide Chemical compound Br.N([C@H](C)C=1C=CC=CC=1)C(C=1C=2)=NC=NC=1NC=2C1=CC=C(O)C=C1 SYYMNUFXRFAELA-BTQNPOSSSA-N 0.000 description 1
- FPEIJQLXFHKLJV-UHFFFAOYSA-N 4-[6-(1h-indol-5-yl)-1-[1-(pyridin-3-ylmethyl)piperidin-4-yl]pyrazolo[3,4-d]pyrimidin-4-yl]morpholine Chemical compound C=1C=CN=CC=1CN(CC1)CCC1N(C1=NC(=N2)C=3C=C4C=CNC4=CC=3)N=CC1=C2N1CCOCC1 FPEIJQLXFHKLJV-UHFFFAOYSA-N 0.000 description 1
- IMXHGCRIEAKIBU-UHFFFAOYSA-N 4-[6-[4-(methoxycarbonylamino)phenyl]-4-(4-morpholinyl)-1-pyrazolo[3,4-d]pyrimidinyl]-1-piperidinecarboxylic acid methyl ester Chemical compound C1=CC(NC(=O)OC)=CC=C1C1=NC(N2CCOCC2)=C(C=NN2C3CCN(CC3)C(=O)OC)C2=N1 IMXHGCRIEAKIBU-UHFFFAOYSA-N 0.000 description 1
- KCODNOOPOPTZMO-UHFFFAOYSA-N 4-[[4-[[2-(4-amino-2,3,5,6-tetramethylanilino)acetyl]-methylamino]piperidin-1-yl]methyl]benzamide Chemical compound CC=1C(C)=C(N)C(C)=C(C)C=1NCC(=O)N(C)C(CC1)CCN1CC1=CC=C(C(N)=O)C=C1 KCODNOOPOPTZMO-UHFFFAOYSA-N 0.000 description 1
- ZCCPLJOKGAACRT-UHFFFAOYSA-N 4-methyl-3-[[1-methyl-6-(3-pyridinyl)-4-pyrazolo[3,4-d]pyrimidinyl]amino]-N-[3-(trifluoromethyl)phenyl]benzamide Chemical compound CC1=CC=C(C(=O)NC=2C=C(C=CC=2)C(F)(F)F)C=C1NC(C=1C=NN(C)C=1N=1)=NC=1C1=CC=CN=C1 ZCCPLJOKGAACRT-UHFFFAOYSA-N 0.000 description 1
- NODCQQSEMCESEC-UHFFFAOYSA-N 5-(1h-pyrrolo[2,3-b]pyridin-3-ylmethyl)-n-[[4-(trifluoromethyl)phenyl]methyl]pyridin-2-amine Chemical compound C1=CC(C(F)(F)F)=CC=C1CNC(N=C1)=CC=C1CC1=CNC2=NC=CC=C12 NODCQQSEMCESEC-UHFFFAOYSA-N 0.000 description 1
- XVECMUKVOMUNLE-UHFFFAOYSA-N 5-(2-phenyl-3-pyrazolo[1,5-a]pyridinyl)-2H-pyrazolo[3,4-c]pyridazin-3-amine Chemical compound C1=C2C(N)=NNC2=NN=C1C(=C1C=CC=CN1N=1)C=1C1=CC=CC=C1 XVECMUKVOMUNLE-UHFFFAOYSA-N 0.000 description 1
- LSFLAQVDISHMNB-UHFFFAOYSA-N 5-(3-phenylmethoxyphenyl)-7-[3-(pyrrolidin-1-ylmethyl)cyclobutyl]pyrrolo[2,3-d]pyrimidin-4-amine Chemical compound C1=2C(N)=NC=NC=2N(C2CC(CN3CCCC3)C2)C=C1C(C=1)=CC=CC=1OCC1=CC=CC=C1 LSFLAQVDISHMNB-UHFFFAOYSA-N 0.000 description 1
- GYLDXIAOMVERTK-UHFFFAOYSA-N 5-(4-amino-1-propan-2-yl-3-pyrazolo[3,4-d]pyrimidinyl)-1,3-benzoxazol-2-amine Chemical compound C12=C(N)N=CN=C2N(C(C)C)N=C1C1=CC=C(OC(N)=N2)C2=C1 GYLDXIAOMVERTK-UHFFFAOYSA-N 0.000 description 1
- JEGHXKRHKHPBJD-UHFFFAOYSA-N 5-(7-methylsulfonyl-2-morpholin-4-yl-5,6-dihydropyrrolo[2,3-d]pyrimidin-4-yl)pyrimidin-2-amine Chemical compound CS(=O)(=O)N1CCC2=C1N=C(N1CCOCC1)N=C2C1=CN=C(N)N=C1 JEGHXKRHKHPBJD-UHFFFAOYSA-N 0.000 description 1
- LULATDWLDJOKCX-UHFFFAOYSA-N 5-[(2,5-dihydroxyphenyl)methylamino]-2-hydroxybenzoic acid Chemical compound C1=C(O)C(C(=O)O)=CC(NCC=2C(=CC=C(O)C=2)O)=C1 LULATDWLDJOKCX-UHFFFAOYSA-N 0.000 description 1
- SRSGVKWWVXWSJT-ATVHPVEESA-N 5-[(z)-(5-fluoro-2-oxo-1h-indol-3-ylidene)methyl]-2,4-dimethyl-n-(2-pyrrolidin-1-ylethyl)-1h-pyrrole-3-carboxamide Chemical compound CC=1NC(\C=C/2C3=CC(F)=CC=C3NC\2=O)=C(C)C=1C(=O)NCCN1CCCC1 SRSGVKWWVXWSJT-ATVHPVEESA-N 0.000 description 1
- GYQRHHQPEMOLKH-UHFFFAOYSA-N 5-[3-(2-methoxyphenyl)-1h-pyrrolo[2,3-b]pyridin-5-yl]-n,n-dimethylpyridine-3-carboxamide Chemical compound COC1=CC=CC=C1C1=CNC2=NC=C(C=3C=C(C=NC=3)C(=O)N(C)C)C=C12 GYQRHHQPEMOLKH-UHFFFAOYSA-N 0.000 description 1
- MYQAUKPBNJWPIE-UHFFFAOYSA-N 5-[[3-methoxy-4-[(4-methoxyphenyl)methoxy]phenyl]methyl]pyrimidine-2,4-diamine Chemical compound C1=CC(OC)=CC=C1COC(C(=C1)OC)=CC=C1CC1=CN=C(N)N=C1N MYQAUKPBNJWPIE-UHFFFAOYSA-N 0.000 description 1
- XXSSGBYXSKOLAM-UHFFFAOYSA-N 5-bromo-n-(2,3-dihydroxypropoxy)-3,4-difluoro-2-(2-fluoro-4-iodoanilino)benzamide Chemical compound OCC(O)CONC(=O)C1=CC(Br)=C(F)C(F)=C1NC1=CC=C(I)C=C1F XXSSGBYXSKOLAM-UHFFFAOYSA-N 0.000 description 1
- NADLBPWBFGTESN-UHFFFAOYSA-N 6-(2,6-dichlorophenyl)-2-[4-[2-(diethylamino)ethoxy]anilino]-8-methylpyrido[2,3-d]pyrimidin-7-one;dihydrochloride Chemical compound Cl.Cl.C1=CC(OCCN(CC)CC)=CC=C1NC1=NC=C(C=C(C=2C(=CC=CC=2Cl)Cl)C(=O)N2C)C2=N1 NADLBPWBFGTESN-UHFFFAOYSA-N 0.000 description 1
- OONFNUWBHFSNBT-HXUWFJFHSA-N AEE788 Chemical compound C1CN(CC)CCN1CC1=CC=C(C=2NC3=NC=NC(N[C@H](C)C=4C=CC=CC=4)=C3C=2)C=C1 OONFNUWBHFSNBT-HXUWFJFHSA-N 0.000 description 1
- CMMDWEJTQUTCKG-WUXMJOGZSA-N AG-370 Chemical compound N#CC(C#N)=C(N)\C(C#N)=C\C1=CC=C2NC=CC2=C1 CMMDWEJTQUTCKG-WUXMJOGZSA-N 0.000 description 1
- TUSCYCAIGRVBMD-UHFFFAOYSA-N ANA-12 Chemical compound C=1C2=CC=CC=C2SC=1C(=O)NC1=CC=CC=C1C(=O)NC1CCCCNC1=O TUSCYCAIGRVBMD-UHFFFAOYSA-N 0.000 description 1
- BUROJSBIWGDYCN-GAUTUEMISA-N AP 23573 Chemical compound C1C[C@@H](OP(C)(C)=O)[C@H](OC)C[C@@H]1C[C@@H](C)[C@H]1OC(=O)[C@@H]2CCCCN2C(=O)C(=O)[C@](O)(O2)[C@H](C)CC[C@H]2C[C@H](OC)/C(C)=C/C=C/C=C/[C@@H](C)C[C@@H](C)C(=O)[C@H](OC)[C@H](O)/C(C)=C/[C@@H](C)C(=O)C1 BUROJSBIWGDYCN-GAUTUEMISA-N 0.000 description 1
- MGGBYMDAPCCKCT-UHFFFAOYSA-N ASP-3026 Chemical compound COC1=CC(N2CCC(CC2)N2CCN(C)CC2)=CC=C1NC(N=1)=NC=NC=1NC1=CC=CC=C1S(=O)(=O)C(C)C MGGBYMDAPCCKCT-UHFFFAOYSA-N 0.000 description 1
- KVLFRAWTRWDEDF-IRXDYDNUSA-N AZD-8055 Chemical compound C1=C(CO)C(OC)=CC=C1C1=CC=C(C(=NC(=N2)N3[C@H](COCC3)C)N3[C@H](COCC3)C)C2=N1 KVLFRAWTRWDEDF-IRXDYDNUSA-N 0.000 description 1
- OIRDTQYFTABQOQ-KQYNXXCUSA-N Adenosine Natural products C1=NC=2C(N)=NC=NC=2N1[C@@H]1O[C@H](CO)[C@@H](O)[C@H]1O OIRDTQYFTABQOQ-KQYNXXCUSA-N 0.000 description 1
- MLDQJTXFUGDVEO-UHFFFAOYSA-N BAY-43-9006 Chemical compound C1=NC(C(=O)NC)=CC(OC=2C=CC(NC(=O)NC=3C=C(C(Cl)=CC=3)C(F)(F)F)=CC=2)=C1 MLDQJTXFUGDVEO-UHFFFAOYSA-N 0.000 description 1
- YUXMAKUNSXIEKN-BTJKTKAUSA-N BGT226 Chemical compound OC(=O)\C=C/C(O)=O.C1=NC(OC)=CC=C1C1=CC=C(N=CC2=C3N(C=4C=C(C(N5CCNCC5)=CC=4)C(F)(F)F)C(=O)N2C)C3=C1 YUXMAKUNSXIEKN-BTJKTKAUSA-N 0.000 description 1
- 241000894006 Bacteria Species 0.000 description 1
- 241000283690 Bos taurus Species 0.000 description 1
- 239000002126 C01EB10 - Adenosine Substances 0.000 description 1
- 239000005461 Canertinib Substances 0.000 description 1
- 241000282465 Canis Species 0.000 description 1
- 206010057248 Cell death Diseases 0.000 description 1
- 241000699800 Cricetinae Species 0.000 description 1
- 102100038114 Cyclin-dependent kinase 13 Human genes 0.000 description 1
- 108090000695 Cytokines Proteins 0.000 description 1
- 102000004127 Cytokines Human genes 0.000 description 1
- 102000038566 DCAFs Human genes 0.000 description 1
- 108091007824 DCAFs Proteins 0.000 description 1
- AOZPVMOOEJAZGK-UHFFFAOYSA-N DDR1-IN-1 Chemical compound C1CN(CC)CCN1CC1=CC=C(C(=O)NC=2C=C(OC=3C=C4CC(=O)NC4=CC=3)C(C)=CC=2)C=C1C(F)(F)F AOZPVMOOEJAZGK-UHFFFAOYSA-N 0.000 description 1
- 108020004414 DNA Proteins 0.000 description 1
- 102100037799 DNA-binding protein Ikaros Human genes 0.000 description 1
- 206010011968 Decreased immune responsiveness Diseases 0.000 description 1
- 241000283073 Equus caballus Species 0.000 description 1
- HKVAMNSJSFKALM-GKUWKFKPSA-N Everolimus Chemical compound C1C[C@@H](OCCO)[C@H](OC)C[C@@H]1C[C@@H](C)[C@H]1OC(=O)[C@@H]2CCCCN2C(=O)C(=O)[C@](O)(O2)[C@H](C)CC[C@H]2C[C@H](OC)/C(C)=C/C=C/C=C/[C@@H](C)C[C@@H](C)C(=O)[C@H](OC)[C@H](O)/C(C)=C/[C@@H](C)C(=O)C1 HKVAMNSJSFKALM-GKUWKFKPSA-N 0.000 description 1
- 241000282324 Felis Species 0.000 description 1
- JRZJKWGQFNTSRN-UHFFFAOYSA-N Geldanamycin Natural products C1C(C)CC(OC)C(O)C(C)C=C(C)C(OC(N)=O)C(OC)CCC=C(C)C(=O)NC2=CC(=O)C(OC)=C1C2=O JRZJKWGQFNTSRN-UHFFFAOYSA-N 0.000 description 1
- MCAHMSDENAOJFZ-UHFFFAOYSA-N Herbimycin A Natural products N1C(=O)C(C)=CC=CC(OC)C(OC(N)=O)C(C)=CC(C)C(OC)C(OC)CC(C)C(OC)C2=CC(=O)C=C1C2=O MCAHMSDENAOJFZ-UHFFFAOYSA-N 0.000 description 1
- BYTORXDZJWWIKR-UHFFFAOYSA-N Hinokiol Natural products CC(C)c1cc2CCC3C(C)(CO)C(O)CCC3(C)c2cc1O BYTORXDZJWWIKR-UHFFFAOYSA-N 0.000 description 1
- 241000282412 Homo Species 0.000 description 1
- 101000884348 Homo sapiens Cyclin-dependent kinase 13 Proteins 0.000 description 1
- 101000599038 Homo sapiens DNA-binding protein Ikaros Proteins 0.000 description 1
- RFSMUFRPPYDYRD-CALCHBBNSA-N Ku-0063794 Chemical compound C1=C(CO)C(OC)=CC=C1C1=CC=C(C(=NC(=N2)N3C[C@@H](C)O[C@@H](C)C3)N3CCOCC3)C2=N1 RFSMUFRPPYDYRD-CALCHBBNSA-N 0.000 description 1
- 239000002147 L01XE04 - Sunitinib Substances 0.000 description 1
- 239000005511 L01XE05 - Sorafenib Substances 0.000 description 1
- 239000002136 L01XE07 - Lapatinib Substances 0.000 description 1
- 239000003798 L01XE11 - Pazopanib Substances 0.000 description 1
- 239000002118 L01XE12 - Vandetanib Substances 0.000 description 1
- 239000002146 L01XE16 - Crizotinib Substances 0.000 description 1
- 239000002144 L01XE18 - Ruxolitinib Substances 0.000 description 1
- 239000002138 L01XE21 - Regorafenib Substances 0.000 description 1
- 239000002139 L01XE22 - Masitinib Substances 0.000 description 1
- 239000002176 L01XE26 - Cabozantinib Substances 0.000 description 1
- 239000002177 L01XE27 - Ibrutinib Substances 0.000 description 1
- UCEQXRCJXIVODC-PMACEKPBSA-N LSM-1131 Chemical compound C1CCC2=CC=CC3=C2N1C=C3[C@@H]1C(=O)NC(=O)[C@H]1C1=CNC2=CC=CC=C12 UCEQXRCJXIVODC-PMACEKPBSA-N 0.000 description 1
- UIARLYUEJFELEN-LROUJFHJSA-N LSM-1231 Chemical compound C12=C3N4C5=CC=CC=C5C3=C3C(=O)NCC3=C2C2=CC=CC=C2N1[C@]1(C)[C@](CO)(O)C[C@H]4O1 UIARLYUEJFELEN-LROUJFHJSA-N 0.000 description 1
- 108060001084 Luciferase Proteins 0.000 description 1
- 239000005089 Luciferase Substances 0.000 description 1
- 108010047357 Luminescent Proteins Proteins 0.000 description 1
- 102000006830 Luminescent Proteins Human genes 0.000 description 1
- MZOPWQKISXCCTP-UHFFFAOYSA-N Malonoben Chemical compound CC(C)(C)C1=CC(C=C(C#N)C#N)=CC(C(C)(C)C)=C1O MZOPWQKISXCCTP-UHFFFAOYSA-N 0.000 description 1
- 239000005462 Mubritinib Substances 0.000 description 1
- 241001529936 Murinae Species 0.000 description 1
- GWOXVNOLEPNQGX-UHFFFAOYSA-N N-[(4-fluorophenyl)methyl]-6,7-dihydro-5H-pyrazolo[5,1-b][1,3]oxazine-3-carboxamide Chemical compound C1=CC(F)=CC=C1CNC(=O)C1=C2OCCCN2N=C1 GWOXVNOLEPNQGX-UHFFFAOYSA-N 0.000 description 1
- ZHXNIYGJAOPMSO-UHFFFAOYSA-N N-[5-[[5-[(4-acetyl-1-piperazinyl)-oxomethyl]-4-methoxy-2-methylphenyl]thio]-2-thiazolyl]-4-[(3,3-dimethylbutan-2-ylamino)methyl]benzamide Chemical compound C1=C(C(=O)N2CCN(CC2)C(C)=O)C(OC)=CC(C)=C1SC(S1)=CN=C1NC(=O)C1=CC=C(CNC(C)C(C)(C)C)C=C1 ZHXNIYGJAOPMSO-UHFFFAOYSA-N 0.000 description 1
- CXQHYVUVSFXTMY-UHFFFAOYSA-N N1'-[3-fluoro-4-[[6-methoxy-7-[3-(4-morpholinyl)propoxy]-4-quinolinyl]oxy]phenyl]-N1-(4-fluorophenyl)cyclopropane-1,1-dicarboxamide Chemical compound C1=CN=C2C=C(OCCCN3CCOCC3)C(OC)=CC2=C1OC(C(=C1)F)=CC=C1NC(=O)C1(C(=O)NC=2C=CC(F)=CC=2)CC1 CXQHYVUVSFXTMY-UHFFFAOYSA-N 0.000 description 1
- FTFRZXFNZVCRSK-UHFFFAOYSA-N N4-(3-chloro-4-fluorophenyl)-N6-(1-methyl-4-piperidinyl)pyrimido[5,4-d]pyrimidine-4,6-diamine Chemical compound C1CN(C)CCC1NC1=NC=C(N=CN=C2NC=3C=C(Cl)C(F)=CC=3)C2=N1 FTFRZXFNZVCRSK-UHFFFAOYSA-N 0.000 description 1
- SUDAHWBOROXANE-SECBINFHSA-N PD 0325901 Chemical compound OC[C@@H](O)CONC(=O)C1=CC=C(F)C(F)=C1NC1=CC=C(I)C=C1F SUDAHWBOROXANE-SECBINFHSA-N 0.000 description 1
- NHJSWORVNIOXIT-UHFFFAOYSA-N PD-166866 Chemical compound COC1=CC(OC)=CC(C=2C(=NC3=NC(N)=NC=C3C=2)NC(=O)NC(C)(C)C)=C1 NHJSWORVNIOXIT-UHFFFAOYSA-N 0.000 description 1
- KFHMLBXBRCITHF-UHFFFAOYSA-N PD158780 Chemical compound N1=CN=C2C=NC(NC)=CC2=C1NC1=CC=CC(Br)=C1 KFHMLBXBRCITHF-UHFFFAOYSA-N 0.000 description 1
- OYONTEXKYJZFHA-SSHUPFPWSA-N PHA-665752 Chemical compound CC=1C(C(=O)N2[C@H](CCC2)CN2CCCC2)=C(C)NC=1\C=C(C1=C2)/C(=O)NC1=CC=C2S(=O)(=O)CC1=C(Cl)C=CC=C1Cl OYONTEXKYJZFHA-SSHUPFPWSA-N 0.000 description 1
- TUVCWJQQGGETHL-UHFFFAOYSA-N PI-103 Chemical compound OC1=CC=CC(C=2N=C3C4=CC=CN=C4OC3=C(N3CCOCC3)N=2)=C1 TUVCWJQQGGETHL-UHFFFAOYSA-N 0.000 description 1
- NVRXTLZYXZNATH-UHFFFAOYSA-N PP121 Chemical compound N1=C(C=2C=C3C=CNC3=NC=2)C=2C(N)=NC=NC=2N1C1CCCC1 NVRXTLZYXZNATH-UHFFFAOYSA-N 0.000 description 1
- 241000288906 Primates Species 0.000 description 1
- 102000004245 Proteasome Endopeptidase Complex Human genes 0.000 description 1
- 108090000708 Proteasome Endopeptidase Complex Proteins 0.000 description 1
- 229940079156 Proteasome inhibitor Drugs 0.000 description 1
- 108090000412 Protein-Tyrosine Kinases Proteins 0.000 description 1
- 102000004022 Protein-Tyrosine Kinases Human genes 0.000 description 1
- FMETVQKSDIOGPX-UHFFFAOYSA-N RK-24466 Chemical compound C1=2C(N)=NC=NC=2N(C2CCCC2)C=C1C(C=C1)=CC=C1OC1=CC=CC=C1 FMETVQKSDIOGPX-UHFFFAOYSA-N 0.000 description 1
- 102000015097 RNA Splicing Factors Human genes 0.000 description 1
- 108010039259 RNA Splicing Factors Proteins 0.000 description 1
- 239000005464 Radotinib Substances 0.000 description 1
- ZESGNAJSBDILTB-UHFFFAOYSA-N Reveromycin A Natural products O1C(C=CC(C)=CC(O)=O)C(CCCC)(OC(=O)CCC(O)=O)CCC11OC(CC=C(C)C=CC(O)C(C)C=CC(O)=O)C(C)CC1 ZESGNAJSBDILTB-UHFFFAOYSA-N 0.000 description 1
- HDAJDNHIBCDLQF-RUZDIDTESA-N SCH772984 Chemical compound O=C([C@@H]1CCN(C1)CC(=O)N1CCN(CC1)C=1C=CC(=CC=1)C=1N=CC=CN=1)NC(C=C12)=CC=C1NN=C2C1=CC=NC=C1 HDAJDNHIBCDLQF-RUZDIDTESA-N 0.000 description 1
- XLBQNZICMYZIQT-GHXNOFRVSA-N SU5614 Chemical compound N1C(C)=CC(C)=C1\C=C/1C2=CC(Cl)=CC=C2NC\1=O XLBQNZICMYZIQT-GHXNOFRVSA-N 0.000 description 1
- GOUKLPYPPSXGLF-UHFFFAOYSA-N Sulochrin Natural products COC(=O)c1cc(O)cc(OC)c1C(=O)c2c(O)ccc(C)c2O GOUKLPYPPSXGLF-UHFFFAOYSA-N 0.000 description 1
- 239000005463 Tandutinib Substances 0.000 description 1
- CBPNZQVSJQDFBE-FUXHJELOSA-N Temsirolimus Chemical compound C1C[C@@H](OC(=O)C(C)(CO)CO)[C@H](OC)C[C@@H]1C[C@@H](C)[C@H]1OC(=O)[C@@H]2CCCCN2C(=O)C(=O)[C@](O)(O2)[C@H](C)CC[C@H]2C[C@H](OC)/C(C)=C/C=C/C=C/[C@@H](C)C[C@@H](C)C(=O)[C@H](OC)[C@H](O)/C(C)=C/[C@@H](C)C(=O)C1 CBPNZQVSJQDFBE-FUXHJELOSA-N 0.000 description 1
- 239000003819 Toceranib Substances 0.000 description 1
- 239000004012 Tofacitinib Substances 0.000 description 1
- GSNOZLZNQMLSKJ-UHFFFAOYSA-N Trapidil Chemical compound CCN(CC)C1=CC(C)=NC2=NC=NN12 GSNOZLZNQMLSKJ-UHFFFAOYSA-N 0.000 description 1
- DVEXZJFMOKTQEZ-JYFOCSDGSA-N U0126 Chemical compound C=1C=CC=C(N)C=1SC(\N)=C(/C#N)\C(\C#N)=C(/N)SC1=CC=CC=C1N DVEXZJFMOKTQEZ-JYFOCSDGSA-N 0.000 description 1
- 102000018478 Ubiquitin-Activating Enzymes Human genes 0.000 description 1
- 108010091546 Ubiquitin-Activating Enzymes Proteins 0.000 description 1
- 241000700605 Viruses Species 0.000 description 1
- QAPAJIZPZGWAND-UHFFFAOYSA-N XMD8-92 Chemical compound C=1C=C(NC=2N=C3N(C)C4=CC=CC=C4C(=O)N(C)C3=CN=2)C(OCC)=CC=1N1CCC(O)CC1 QAPAJIZPZGWAND-UHFFFAOYSA-N 0.000 description 1
- LUJZZYWHBDHDQX-QFIPXVFZSA-N [(3s)-morpholin-3-yl]methyl n-[4-[[1-[(3-fluorophenyl)methyl]indazol-5-yl]amino]-5-methylpyrrolo[2,1-f][1,2,4]triazin-6-yl]carbamate Chemical compound C=1N2N=CN=C(NC=3C=C4C=NN(CC=5C=C(F)C=CC=5)C4=CC=3)C2=C(C)C=1NC(=O)OC[C@@H]1COCCN1 LUJZZYWHBDHDQX-QFIPXVFZSA-N 0.000 description 1
- LNFBAYSBVQBKFR-UHFFFAOYSA-N [7-(6-aminopyridin-3-yl)-3,5-dihydro-2h-1,4-benzoxazepin-4-yl]-(3-fluoro-2-methyl-4-methylsulfonylphenyl)methanone Chemical compound CC1=C(F)C(S(C)(=O)=O)=CC=C1C(=O)N1CC2=CC(C=3C=NC(N)=CC=3)=CC=C2OCC1 LNFBAYSBVQBKFR-UHFFFAOYSA-N 0.000 description 1
- 230000005856 abnormality Effects 0.000 description 1
- 238000009825 accumulation Methods 0.000 description 1
- 230000003213 activating effect Effects 0.000 description 1
- 229960005305 adenosine Drugs 0.000 description 1
- 229960001686 afatinib Drugs 0.000 description 1
- ULXXDDBFHOBEHA-CWDCEQMOSA-N afatinib Chemical compound N1=CN=C2C=C(O[C@@H]3COCC3)C(NC(=O)/C=C/CN(C)C)=CC2=C1NC1=CC=C(F)C(Cl)=C1 ULXXDDBFHOBEHA-CWDCEQMOSA-N 0.000 description 1
- 238000004458 analytical method Methods 0.000 description 1
- 239000000427 antigen Substances 0.000 description 1
- 108091007433 antigens Proteins 0.000 description 1
- 102000036639 antigens Human genes 0.000 description 1
- 229940127079 antineoplastic immunimodulatory agent Drugs 0.000 description 1
- 238000013459 approach Methods 0.000 description 1
- 238000003491 array Methods 0.000 description 1
- 230000004900 autophagic degradation Effects 0.000 description 1
- 229960003005 axitinib Drugs 0.000 description 1
- RITAVMQDGBJQJZ-FMIVXFBMSA-N axitinib Chemical compound CNC(=O)C1=CC=CC=C1SC1=CC=C(C(\C=C\C=2N=CC=CC=2)=NN2)C2=C1 RITAVMQDGBJQJZ-FMIVXFBMSA-N 0.000 description 1
- 230000008901 benefit Effects 0.000 description 1
- 229950003054 binimetinib Drugs 0.000 description 1
- ACWZRVQXLIRSDF-UHFFFAOYSA-N binimetinib Chemical compound OCCONC(=O)C=1C=C2N(C)C=NC2=C(F)C=1NC1=CC=C(Br)C=C1F ACWZRVQXLIRSDF-UHFFFAOYSA-N 0.000 description 1
- 230000015572 biosynthetic process Effects 0.000 description 1
- 238000011138 biotechnological process Methods 0.000 description 1
- 229960001292 cabozantinib Drugs 0.000 description 1
- ONIQOQHATWINJY-UHFFFAOYSA-N cabozantinib Chemical compound C=12C=C(OC)C(OC)=CC2=NC=CC=1OC(C=C1)=CC=C1NC(=O)C1(C(=O)NC=2C=CC(F)=CC=2)CC1 ONIQOQHATWINJY-UHFFFAOYSA-N 0.000 description 1
- HFCFMRYTXDINDK-WNQIDUERSA-N cabozantinib malate Chemical compound OC(=O)[C@@H](O)CC(O)=O.C=12C=C(OC)C(OC)=CC2=NC=CC=1OC(C=C1)=CC=C1NC(=O)C1(C(=O)NC=2C=CC(F)=CC=2)CC1 HFCFMRYTXDINDK-WNQIDUERSA-N 0.000 description 1
- 229950002826 canertinib Drugs 0.000 description 1
- OMZCMEYTWSXEPZ-UHFFFAOYSA-N canertinib Chemical compound C1=C(Cl)C(F)=CC=C1NC1=NC=NC2=CC(OCCCN3CCOCC3)=C(NC(=O)C=C)C=C12 OMZCMEYTWSXEPZ-UHFFFAOYSA-N 0.000 description 1
- 229960002412 cediranib Drugs 0.000 description 1
- 230000022131 cell cycle Effects 0.000 description 1
- 230000025084 cell cycle arrest Effects 0.000 description 1
- 230000024245 cell differentiation Effects 0.000 description 1
- 239000002458 cell surface marker Substances 0.000 description 1
- 238000012054 celltiter-glo Methods 0.000 description 1
- 230000033077 cellular process Effects 0.000 description 1
- 230000036755 cellular response Effects 0.000 description 1
- 229960001602 ceritinib Drugs 0.000 description 1
- VERWOWGGCGHDQE-UHFFFAOYSA-N ceritinib Chemical compound CC=1C=C(NC=2N=C(NC=3C(=CC=CC=3)S(=O)(=O)C(C)C)C(Cl)=CN=2)C(OC(C)C)=CC=1C1CCNCC1 VERWOWGGCGHDQE-UHFFFAOYSA-N 0.000 description 1
- XDLYKKIQACFMJG-WKILWMFISA-N chembl1234354 Chemical compound C1=NC(OC)=CC=C1C(C1=O)=CC2=C(C)N=C(N)N=C2N1[C@@H]1CC[C@@H](OCCO)CC1 XDLYKKIQACFMJG-WKILWMFISA-N 0.000 description 1
- JROFGZPOBKIAEW-HAQNSBGRSA-N chembl3120215 Chemical compound N1C=2C(OC)=CC=CC=2C=C1C(=C1C(N)=NC=NN11)N=C1[C@H]1CC[C@H](C(O)=O)CC1 JROFGZPOBKIAEW-HAQNSBGRSA-N 0.000 description 1
- ZWVZORIKUNOTCS-OAQYLSRUSA-N chembl401930 Chemical compound C1([C@H](O)CNC2=C(C(NC=C2)=O)C=2NC=3C=C(C=C(C=3N=2)C)N2CCOCC2)=CC=CC(Cl)=C1 ZWVZORIKUNOTCS-OAQYLSRUSA-N 0.000 description 1
- XRZYELWZLNAXGE-KPKJPENVSA-N chembl539947 Chemical compound CC(C)(C)C1=CC(\C=C(/C#N)C(N)=S)=CC(C(C)(C)C)=C1O XRZYELWZLNAXGE-KPKJPENVSA-N 0.000 description 1
- 238000001311 chemical methods and process Methods 0.000 description 1
- 230000035605 chemotaxis Effects 0.000 description 1
- 238000002512 chemotherapy Methods 0.000 description 1
- 229960002271 cobimetinib Drugs 0.000 description 1
- BSMCAPRUBJMWDF-KRWDZBQOSA-N cobimetinib Chemical compound C1C(O)([C@H]2NCCCC2)CN1C(=O)C1=CC=C(F)C(F)=C1NC1=CC=C(I)C=C1F BSMCAPRUBJMWDF-KRWDZBQOSA-N 0.000 description 1
- RESIMIUSNACMNW-BXRWSSRYSA-N cobimetinib fumarate Chemical compound OC(=O)\C=C\C(O)=O.C1C(O)([C@H]2NCCCC2)CN1C(=O)C1=CC=C(F)C(F)=C1NC1=CC=C(I)C=C1F.C1C(O)([C@H]2NCCCC2)CN1C(=O)C1=CC=C(F)C(F)=C1NC1=CC=C(I)C=C1F RESIMIUSNACMNW-BXRWSSRYSA-N 0.000 description 1
- 230000005757 colony formation Effects 0.000 description 1
- 239000003086 colorant Substances 0.000 description 1
- 230000001268 conjugating effect Effects 0.000 description 1
- 229950009240 crenolanib Drugs 0.000 description 1
- DYNHJHQFHQTFTP-UHFFFAOYSA-N crenolanib Chemical compound C=1C=C2N(C=3N=C4C(N5CCC(N)CC5)=CC=CC4=CC=3)C=NC2=CC=1OCC1(C)COC1 DYNHJHQFHQTFTP-UHFFFAOYSA-N 0.000 description 1
- 229960005061 crizotinib Drugs 0.000 description 1
- KTEIFNKAUNYNJU-GFCCVEGCSA-N crizotinib Chemical compound O([C@H](C)C=1C(=C(F)C=CC=1Cl)Cl)C(C(=NC=1)N)=CC=1C(=C1)C=NN1C1CCNCC1 KTEIFNKAUNYNJU-GFCCVEGCSA-N 0.000 description 1
- 231100000433 cytotoxic Toxicity 0.000 description 1
- 230000001472 cytotoxic effect Effects 0.000 description 1
- LVXJQMNHJWSHET-AATRIKPKSA-N dacomitinib Chemical compound C=12C=C(NC(=O)\C=C\CN3CCCCC3)C(OC)=CC2=NC=NC=1NC1=CC=C(F)C(Cl)=C1 LVXJQMNHJWSHET-AATRIKPKSA-N 0.000 description 1
- 229950002205 dacomitinib Drugs 0.000 description 1
- 229950006418 dactolisib Drugs 0.000 description 1
- JOGKUKXHTYWRGZ-UHFFFAOYSA-N dactolisib Chemical compound O=C1N(C)C2=CN=C3C=CC(C=4C=C5C=CC=CC5=NC=4)=CC3=C2N1C1=CC=C(C(C)(C)C#N)C=C1 JOGKUKXHTYWRGZ-UHFFFAOYSA-N 0.000 description 1
- 230000034994 death Effects 0.000 description 1
- 238000001514 detection method Methods 0.000 description 1
- 230000004069 differentiation Effects 0.000 description 1
- 201000010099 disease Diseases 0.000 description 1
- 208000037265 diseases, disorders, signs and symptoms Diseases 0.000 description 1
- 231100000294 dose-dependent toxicity Toxicity 0.000 description 1
- 238000003182 dose-response assay Methods 0.000 description 1
- 230000002900 effect on cell Effects 0.000 description 1
- 229940125532 enzyme inhibitor Drugs 0.000 description 1
- YJGVMLPVUAXIQN-UHFFFAOYSA-N epipodophyllotoxin Natural products COC1=C(OC)C(OC)=CC(C2C3=CC=4OCOC=4C=C3C(O)C3C2C(OC3)=O)=C1 YJGVMLPVUAXIQN-UHFFFAOYSA-N 0.000 description 1
- 229960005167 everolimus Drugs 0.000 description 1
- 238000000799 fluorescence microscopy Methods 0.000 description 1
- 108091006047 fluorescent proteins Proteins 0.000 description 1
- 102000034287 fluorescent proteins Human genes 0.000 description 1
- 229950008692 foretinib Drugs 0.000 description 1
- 229950005309 fostamatinib Drugs 0.000 description 1
- GKDRMWXFWHEQQT-UHFFFAOYSA-N fostamatinib Chemical compound COC1=C(OC)C(OC)=CC(NC=2N=C(NC=3N=C4N(COP(O)(O)=O)C(=O)C(C)(C)OC4=CC=3)C(F)=CN=2)=C1 GKDRMWXFWHEQQT-UHFFFAOYSA-N 0.000 description 1
- QTQAWLPCGQOSGP-GBTDJJJQSA-N geldanamycin Chemical compound N1C(=O)\C(C)=C/C=C\[C@@H](OC)[C@H](OC(N)=O)\C(C)=C/[C@@H](C)[C@@H](O)[C@H](OC)C[C@@H](C)CC2=C(OC)C(=O)C=C1C2=O QTQAWLPCGQOSGP-GBTDJJJQSA-N 0.000 description 1
- 238000003633 gene expression assay Methods 0.000 description 1
- 230000002068 genetic effect Effects 0.000 description 1
- 229940045109 genistein Drugs 0.000 description 1
- TZBJGXHYKVUXJN-UHFFFAOYSA-N genistein Natural products C1=CC(O)=CC=C1C1=COC2=CC(O)=CC(O)=C2C1=O TZBJGXHYKVUXJN-UHFFFAOYSA-N 0.000 description 1
- 235000006539 genistein Nutrition 0.000 description 1
- ZCOLJUOHXJRHDI-CMWLGVBASA-N genistein 7-O-beta-D-glucoside Chemical compound O[C@@H]1[C@@H](O)[C@H](O)[C@@H](CO)O[C@H]1OC1=CC(O)=C2C(=O)C(C=3C=CC(O)=CC=3)=COC2=C1 ZCOLJUOHXJRHDI-CMWLGVBASA-N 0.000 description 1
- 230000013595 glycosylation Effects 0.000 description 1
- 238000006206 glycosylation reaction Methods 0.000 description 1
- 230000036541 health Effects 0.000 description 1
- 238000010438 heat treatment Methods 0.000 description 1
- MCAHMSDENAOJFZ-BVXDHVRPSA-N herbimycin Chemical compound N1C(=O)\C(C)=C\C=C/[C@H](OC)[C@@H](OC(N)=O)\C(C)=C\[C@H](C)[C@@H](OC)[C@@H](OC)C[C@H](C)[C@@H](OC)C2=CC(=O)C=C1C2=O MCAHMSDENAOJFZ-BVXDHVRPSA-N 0.000 description 1
- 231100000171 higher toxicity Toxicity 0.000 description 1
- FVYXIJYOAGAUQK-UHFFFAOYSA-N honokiol Chemical compound C1=C(CC=C)C(O)=CC=C1C1=CC(CC=C)=CC=C1O FVYXIJYOAGAUQK-UHFFFAOYSA-N 0.000 description 1
- VVOAZFWZEDHOOU-UHFFFAOYSA-N honokiol Natural products OC1=CC=C(CC=C)C=C1C1=CC(CC=C)=CC=C1O VVOAZFWZEDHOOU-UHFFFAOYSA-N 0.000 description 1
- 229960001507 ibrutinib Drugs 0.000 description 1
- XYFPWWZEPKGCCK-GOSISDBHSA-N ibrutinib Chemical compound C1=2C(N)=NC=NC=2N([C@H]2CN(CCC2)C(=O)C=C)N=C1C(C=C1)=CC=C1OC1=CC=CC=C1 XYFPWWZEPKGCCK-GOSISDBHSA-N 0.000 description 1
- 229950007440 icotinib Drugs 0.000 description 1
- QQLKULDARVNMAL-UHFFFAOYSA-N icotinib Chemical compound C#CC1=CC=CC(NC=2C3=CC=4OCCOCCOCCOC=4C=C3N=CN=2)=C1 QQLKULDARVNMAL-UHFFFAOYSA-N 0.000 description 1
- 230000002519 immonomodulatory effect Effects 0.000 description 1
- 230000001900 immune effect Effects 0.000 description 1
- 238000003364 immunohistochemistry Methods 0.000 description 1
- 238000000338 in vitro Methods 0.000 description 1
- 238000011503 in vivo imaging Methods 0.000 description 1
- 230000006698 induction Effects 0.000 description 1
- 208000015181 infectious disease Diseases 0.000 description 1
- 239000004615 ingredient Substances 0.000 description 1
- 230000003834 intracellular effect Effects 0.000 description 1
- 230000009545 invasion Effects 0.000 description 1
- 229960004891 lapatinib Drugs 0.000 description 1
- BCFGMOOMADDAQU-UHFFFAOYSA-N lapatinib Chemical compound O1C(CNCCS(=O)(=O)C)=CC=C1C1=CC=C(N=CN=C2NC=3C=C(Cl)C(OCC=4C=C(F)C=CC=4)=CC=3)C2=C1 BCFGMOOMADDAQU-UHFFFAOYSA-N 0.000 description 1
- 229960003784 lenvatinib Drugs 0.000 description 1
- WOSKHXYHFSIKNG-UHFFFAOYSA-N lenvatinib Chemical compound C=12C=C(C(N)=O)C(OC)=CC2=NC=CC=1OC(C=C1Cl)=CC=C1NC(=O)NC1CC1 WOSKHXYHFSIKNG-UHFFFAOYSA-N 0.000 description 1
- 229950001845 lestaurtinib Drugs 0.000 description 1
- 229950001762 linsitinib Drugs 0.000 description 1
- PKCDDUHJAFVJJB-VLZXCDOPSA-N linsitinib Chemical compound C1[C@](C)(O)C[C@@H]1C1=NC(C=2C=C3N=C(C=CC3=CC=2)C=2C=CC=CC=2)=C2N1C=CN=C2N PKCDDUHJAFVJJB-VLZXCDOPSA-N 0.000 description 1
- 230000004807 localization Effects 0.000 description 1
- 238000004020 luminiscence type Methods 0.000 description 1
- 229960004655 masitinib Drugs 0.000 description 1
- 239000000463 material Substances 0.000 description 1
- 230000007246 mechanism Effects 0.000 description 1
- 238000002483 medication Methods 0.000 description 1
- VDOCQQKGPJENHJ-UHFFFAOYSA-N methyl n-[4-[4-morpholin-4-yl-1-[1-(pyridin-3-ylmethyl)piperidin-4-yl]pyrazolo[3,4-d]pyrimidin-6-yl]phenyl]carbamate Chemical compound C1=CC(NC(=O)OC)=CC=C1C1=NC(N2CCOCC2)=C(C=NN2C3CCN(CC=4C=NC=CC=4)CC3)C2=N1 VDOCQQKGPJENHJ-UHFFFAOYSA-N 0.000 description 1
- BMGQWWVMWDBQGC-IIFHNQTCSA-N midostaurin Chemical compound CN([C@H]1[C@H]([C@]2(C)O[C@@H](N3C4=CC=CC=C4C4=C5C(=O)NCC5=C5C6=CC=CC=C6N2C5=C43)C1)OC)C(=O)C1=CC=CC=C1 BMGQWWVMWDBQGC-IIFHNQTCSA-N 0.000 description 1
- 229950010895 midostaurin Drugs 0.000 description 1
- 230000005012 migration Effects 0.000 description 1
- 238000013508 migration Methods 0.000 description 1
- 238000002156 mixing Methods 0.000 description 1
- 230000004048 modification Effects 0.000 description 1
- 238000012986 modification Methods 0.000 description 1
- 229950003968 motesanib Drugs 0.000 description 1
- RAHBGWKEPAQNFF-UHFFFAOYSA-N motesanib Chemical compound C=1C=C2C(C)(C)CNC2=CC=1NC(=O)C1=CC=CN=C1NCC1=CC=NC=C1 RAHBGWKEPAQNFF-UHFFFAOYSA-N 0.000 description 1
- 229950002212 mubritinib Drugs 0.000 description 1
- ZTFBIUXIQYRUNT-MDWZMJQESA-N mubritinib Chemical compound C1=CC(C(F)(F)F)=CC=C1\C=C\C1=NC(COC=2C=CC(CCCCN3N=NC=C3)=CC=2)=CO1 ZTFBIUXIQYRUNT-MDWZMJQESA-N 0.000 description 1
- WPOXAFXHRJYEIC-UHFFFAOYSA-N n-(2-chloro-5-methoxyphenyl)-6-methoxy-7-[(1-methylpiperidin-4-yl)methoxy]quinazolin-4-amine Chemical compound COC1=CC=C(Cl)C(NC=2C3=CC(OC)=C(OCC4CCN(C)CC4)C=C3N=CN=2)=C1 WPOXAFXHRJYEIC-UHFFFAOYSA-N 0.000 description 1
- ZNNWCQCQUKAMGL-UHFFFAOYSA-N n-(3-chloro-4-fluorophenyl)-6-[4-(diethylaminomethyl)piperidin-1-yl]pyrimido[5,4-d]pyrimidin-4-amine;dihydrochloride Chemical compound Cl.Cl.C1CC(CN(CC)CC)CCN1C1=NC=C(N=CN=C2NC=3C=C(Cl)C(F)=CC=3)C2=N1 ZNNWCQCQUKAMGL-UHFFFAOYSA-N 0.000 description 1
- ZDNURMVOKAERHZ-UHFFFAOYSA-N n-(3-fluorophenyl)-6,7-dimethoxy-1,4-dihydroindeno[1,2-c]pyrazol-3-amine Chemical compound N=1NC=2C=3C=C(OC)C(OC)=CC=3CC=2C=1NC1=CC=CC(F)=C1 ZDNURMVOKAERHZ-UHFFFAOYSA-N 0.000 description 1
- KSERXGMCDHOLSS-LJQANCHMSA-N n-[(1s)-1-(3-chlorophenyl)-2-hydroxyethyl]-4-[5-chloro-2-(propan-2-ylamino)pyridin-4-yl]-1h-pyrrole-2-carboxamide Chemical compound C1=NC(NC(C)C)=CC(C=2C=C(NC=2)C(=O)N[C@H](CO)C=2C=C(Cl)C=CC=2)=C1Cl KSERXGMCDHOLSS-LJQANCHMSA-N 0.000 description 1
- RDSACQWTXKSHJT-NSHDSACASA-N n-[3,4-difluoro-2-(2-fluoro-4-iodoanilino)-6-methoxyphenyl]-1-[(2s)-2,3-dihydroxypropyl]cyclopropane-1-sulfonamide Chemical compound C1CC1(C[C@H](O)CO)S(=O)(=O)NC=1C(OC)=CC(F)=C(F)C=1NC1=CC=C(I)C=C1F RDSACQWTXKSHJT-NSHDSACASA-N 0.000 description 1
- FDMQDKQUTRLUBU-UHFFFAOYSA-N n-[3-[2-[4-(4-methylpiperazin-1-yl)anilino]thieno[3,2-d]pyrimidin-4-yl]oxyphenyl]prop-2-enamide Chemical compound C1CN(C)CCN1C(C=C1)=CC=C1NC1=NC(OC=2C=C(NC(=O)C=C)C=CC=2)=C(SC=C2)C2=N1 FDMQDKQUTRLUBU-UHFFFAOYSA-N 0.000 description 1
- HUFOZJXAKZVRNJ-UHFFFAOYSA-N n-[3-[[2-[4-(4-acetylpiperazin-1-yl)-2-methoxyanilino]-5-(trifluoromethyl)pyrimidin-4-yl]amino]phenyl]prop-2-enamide Chemical compound COC1=CC(N2CCN(CC2)C(C)=O)=CC=C1NC(N=1)=NC=C(C(F)(F)F)C=1NC1=CC=CC(NC(=O)C=C)=C1 HUFOZJXAKZVRNJ-UHFFFAOYSA-N 0.000 description 1
- AZLXGJFHSLQCPV-UHFFFAOYSA-N n-[4-(3-chloro-4-fluoroanilino)-7-prop-2-ynoxyquinazolin-6-yl]prop-2-enamide Chemical compound C1=C(Cl)C(F)=CC=C1NC1=NC=NC2=CC(OCC#C)=C(NC(=O)C=C)C=C12 AZLXGJFHSLQCPV-UHFFFAOYSA-N 0.000 description 1
- DLPSDPPZXRJQOY-UHFFFAOYSA-N n-[4-(3-chloro-4-fluoroanilino)pyrido[3,4-d]pyrimidin-6-yl]but-2-ynamide Chemical compound N1=CN=C2C=NC(NC(=O)C#CC)=CC2=C1NC1=CC=C(F)C(Cl)=C1 DLPSDPPZXRJQOY-UHFFFAOYSA-N 0.000 description 1
- XNFHHOXCDUAYSR-SFHVURJKSA-N n-[[(2s)-oxolan-2-yl]methyl]-5,6-diphenylfuro[2,3-d]pyrimidin-4-amine Chemical compound C([C@H]1OCCC1)NC(C1=2)=NC=NC=2OC(C=2C=CC=CC=2)=C1C1=CC=CC=C1 XNFHHOXCDUAYSR-SFHVURJKSA-N 0.000 description 1
- 229950008835 neratinib Drugs 0.000 description 1
- ZNHPZUKZSNBOSQ-BQYQJAHWSA-N neratinib Chemical compound C=12C=C(NC\C=C\CN(C)C)C(OCC)=CC2=NC=C(C#N)C=1NC(C=C1Cl)=CC=C1OCC1=CC=CC=N1 ZNHPZUKZSNBOSQ-BQYQJAHWSA-N 0.000 description 1
- 230000001537 neural effect Effects 0.000 description 1
- 230000031972 neutrophil apoptotic process Effects 0.000 description 1
- 229960004378 nintedanib Drugs 0.000 description 1
- XZXHXSATPCNXJR-ZIADKAODSA-N nintedanib Chemical compound O=C1NC2=CC(C(=O)OC)=CC=C2\C1=C(C=1C=CC=CC=1)\NC(C=C1)=CC=C1N(C)C(=O)CN1CCN(C)CC1 XZXHXSATPCNXJR-ZIADKAODSA-N 0.000 description 1
- CGBJSGAELGCMKE-UHFFFAOYSA-N omipalisib Chemical compound COC1=NC=C(C=2C=C3C(C=4C=NN=CC=4)=CC=NC3=CC=2)C=C1NS(=O)(=O)C1=CC=C(F)C=C1F CGBJSGAELGCMKE-UHFFFAOYSA-N 0.000 description 1
- 229950008089 omipalisib Drugs 0.000 description 1
- 210000000056 organ Anatomy 0.000 description 1
- 229960003278 osimertinib Drugs 0.000 description 1
- DUYJMQONPNNFPI-UHFFFAOYSA-N osimertinib Chemical compound COC1=CC(N(C)CCN(C)C)=C(NC(=O)C=C)C=C1NC1=NC=CC(C=2C3=CC=CC=C3N(C)C=2)=N1 DUYJMQONPNNFPI-UHFFFAOYSA-N 0.000 description 1
- 229960000639 pazopanib Drugs 0.000 description 1
- CUIHSIWYWATEQL-UHFFFAOYSA-N pazopanib Chemical compound C1=CC2=C(C)N(C)N=C2C=C1N(C)C(N=1)=CC=NC=1NC1=CC=C(C)C(S(N)(=O)=O)=C1 CUIHSIWYWATEQL-UHFFFAOYSA-N 0.000 description 1
- WVUNYSQLFKLYNI-AATRIKPKSA-N pelitinib Chemical compound C=12C=C(NC(=O)\C=C\CN(C)C)C(OCC)=CC2=NC=C(C#N)C=1NC1=CC=C(F)C(Cl)=C1 WVUNYSQLFKLYNI-AATRIKPKSA-N 0.000 description 1
- 229950006299 pelitinib Drugs 0.000 description 1
- 230000035699 permeability Effects 0.000 description 1
- 229950010588 pevonedistat Drugs 0.000 description 1
- RYYNGWLOYLRZLK-RBUKOAKNSA-N pf03814735 Chemical compound C1([C@H]2CC[C@@H](C1=CC=1)N2C(=O)CNC(=O)C)=CC=1NC(N=1)=NC=C(C(F)(F)F)C=1NC1CCC1 RYYNGWLOYLRZLK-RBUKOAKNSA-N 0.000 description 1
- CCTIOCVIZPCTGO-BYPYZUCNSA-N phosphoarginine Chemical compound OC(=O)[C@@H](N)CCCNC(=N)NP(O)(O)=O CCTIOCVIZPCTGO-BYPYZUCNSA-N 0.000 description 1
- YJGVMLPVUAXIQN-HAEOHBJNSA-N picropodophyllotoxin Chemical compound COC1=C(OC)C(OC)=CC([C@@H]2C3=CC=4OCOC=4C=C3[C@H](O)[C@@H]3[C@H]2C(OC3)=O)=C1 YJGVMLPVUAXIQN-HAEOHBJNSA-N 0.000 description 1
- 230000004481 post-translational protein modification Effects 0.000 description 1
- 238000001556 precipitation Methods 0.000 description 1
- 239000002243 precursor Substances 0.000 description 1
- 229930010796 primary metabolite Natural products 0.000 description 1
- 230000000644 propagated effect Effects 0.000 description 1
- MDLUYYGRCGDKGL-UHFFFAOYSA-N propyl 4-[(1-hexyl-4-hydroxy-2-oxoquinoline-3-carbonyl)amino]benzoate Chemical compound O=C1N(CCCCCC)C2=CC=CC=C2C(O)=C1C(=O)NC1=CC=C(C(=O)OCCC)C=C1 MDLUYYGRCGDKGL-UHFFFAOYSA-N 0.000 description 1
- 239000003207 proteasome inhibitor Substances 0.000 description 1
- 238000003498 protein array Methods 0.000 description 1
- 238000000159 protein binding assay Methods 0.000 description 1
- 230000004850 protein–protein interaction Effects 0.000 description 1
- 238000011865 proteolysis targeting chimera technique Methods 0.000 description 1
- 229950001626 quizartinib Drugs 0.000 description 1
- CVWXJKQAOSCOAB-UHFFFAOYSA-N quizartinib Chemical compound O1C(C(C)(C)C)=CC(NC(=O)NC=2C=CC(=CC=2)C=2N=C3N(C4=CC=C(OCCN5CCOCC5)C=C4S3)C=2)=N1 CVWXJKQAOSCOAB-UHFFFAOYSA-N 0.000 description 1
- 229950004043 radotinib Drugs 0.000 description 1
- DUPWHXBITIZIKZ-UHFFFAOYSA-N radotinib Chemical compound C1=NC(C)=CN1C1=CC(NC(=O)C=2C=C(NC=3N=C(C=CN=3)C=3N=CC=NC=3)C(C)=CC=2)=CC(C(F)(F)F)=C1 DUPWHXBITIZIKZ-UHFFFAOYSA-N 0.000 description 1
- ZAHRKKWIAAJSAO-UHFFFAOYSA-N rapamycin Natural products COCC(O)C(=C/C(C)C(=O)CC(OC(=O)C1CCCCN1C(=O)C(=O)C2(O)OC(CC(OC)C(=CC=CC=CC(C)CC(C)C(=O)C)C)CCC2C)C(C)CC3CCC(O)C(C3)OC)C ZAHRKKWIAAJSAO-UHFFFAOYSA-N 0.000 description 1
- 238000011084 recovery Methods 0.000 description 1
- 229950008933 refametinib Drugs 0.000 description 1
- 229960004836 regorafenib Drugs 0.000 description 1
- FNHKPVJBJVTLMP-UHFFFAOYSA-N regorafenib Chemical compound C1=NC(C(=O)NC)=CC(OC=2C=C(F)C(NC(=O)NC=3C=C(C(Cl)=CC=3)C(F)(F)F)=CC=2)=C1 FNHKPVJBJVTLMP-UHFFFAOYSA-N 0.000 description 1
- 230000001105 regulatory effect Effects 0.000 description 1
- 229960001302 ridaforolimus Drugs 0.000 description 1
- 229950009855 rociletinib Drugs 0.000 description 1
- 229960000215 ruxolitinib Drugs 0.000 description 1
- HFNKQEVNSGCOJV-OAHLLOKOSA-N ruxolitinib Chemical compound C1([C@@H](CC#N)N2N=CC(=C2)C=2C=3C=CNC=3N=CN=2)CCCC1 HFNKQEVNSGCOJV-OAHLLOKOSA-N 0.000 description 1
- 229950009216 sapanisertib Drugs 0.000 description 1
- 238000007423 screening assay Methods 0.000 description 1
- 229930000044 secondary metabolite Natural products 0.000 description 1
- CYOHGALHFOKKQC-UHFFFAOYSA-N selumetinib Chemical compound OCCONC(=O)C=1C=C2N(C)C=NC2=C(F)C=1NC1=CC=C(Br)C=C1Cl CYOHGALHFOKKQC-UHFFFAOYSA-N 0.000 description 1
- 229950010746 selumetinib Drugs 0.000 description 1
- 230000035945 sensitivity Effects 0.000 description 1
- 229960002930 sirolimus Drugs 0.000 description 1
- QFJCIRLUMZQUOT-HPLJOQBZSA-N sirolimus Chemical compound C1C[C@@H](O)[C@H](OC)C[C@@H]1C[C@@H](C)[C@H]1OC(=O)[C@@H]2CCCCN2C(=O)C(=O)[C@](O)(O2)[C@H](C)CC[C@H]2C[C@H](OC)/C(C)=C/C=C/C=C/[C@@H](C)C[C@@H](C)C(=O)[C@H](OC)[C@H](O)/C(C)=C/[C@@H](C)C(=O)C1 QFJCIRLUMZQUOT-HPLJOQBZSA-N 0.000 description 1
- 108010026668 snake venom protein C activator Proteins 0.000 description 1
- 229960003787 sorafenib Drugs 0.000 description 1
- 230000004960 subcellular localization Effects 0.000 description 1
- 229960001796 sunitinib Drugs 0.000 description 1
- WINHZLLDWRZWRT-ATVHPVEESA-N sunitinib Chemical compound CCN(CC)CCNC(=O)C1=C(C)NC(\C=C/2C3=CC(F)=CC=C3NC\2=O)=C1C WINHZLLDWRZWRT-ATVHPVEESA-N 0.000 description 1
- 229950009893 tandutinib Drugs 0.000 description 1
- UXXQOJXBIDBUAC-UHFFFAOYSA-N tandutinib Chemical compound COC1=CC2=C(N3CCN(CC3)C(=O)NC=3C=CC(OC(C)C)=CC=3)N=CN=C2C=C1OCCCN1CCCCC1 UXXQOJXBIDBUAC-UHFFFAOYSA-N 0.000 description 1
- 229960000235 temsirolimus Drugs 0.000 description 1
- QFJCIRLUMZQUOT-UHFFFAOYSA-N temsirolimus Natural products C1CC(O)C(OC)CC1CC(C)C1OC(=O)C2CCCCN2C(=O)C(=O)C(O)(O2)C(C)CCC2CC(OC)C(C)=CC=CC=CC(C)CC(C)C(=O)C(OC)C(O)C(C)=CC(C)C(=O)C1 QFJCIRLUMZQUOT-UHFFFAOYSA-N 0.000 description 1
- ATFNSNUJZOYXFC-RQJHMYQMSA-N terreic acid Chemical compound O=C1C(C)=C(O)C(=O)[C@@H]2O[C@@H]21 ATFNSNUJZOYXFC-RQJHMYQMSA-N 0.000 description 1
- 229960003433 thalidomide Drugs 0.000 description 1
- 238000004448 titration Methods 0.000 description 1
- 229950005976 tivantinib Drugs 0.000 description 1
- 229960005048 toceranib Drugs 0.000 description 1
- MFAQYJIYDMLAIM-UHFFFAOYSA-N torkinib Chemical compound C12=C(N)N=CN=C2N(C(C)C)N=C1C1=CC2=CC(O)=CC=C2N1 MFAQYJIYDMLAIM-UHFFFAOYSA-N 0.000 description 1
- 229960004066 trametinib Drugs 0.000 description 1
- LIRYPHYGHXZJBZ-UHFFFAOYSA-N trametinib Chemical compound CC(=O)NC1=CC=CC(N2C(N(C3CC3)C(=O)C3=C(NC=4C(=CC(I)=CC=4)F)N(C)C(=O)C(C)=C32)=O)=C1 LIRYPHYGHXZJBZ-UHFFFAOYSA-N 0.000 description 1
- 238000013518 transcription Methods 0.000 description 1
- 230000035897 transcription Effects 0.000 description 1
- 230000002103 transcriptional effect Effects 0.000 description 1
- 229960000363 trapidil Drugs 0.000 description 1
- UOHFCPXBKJPCAD-UHFFFAOYSA-N tyrphostin 1 Chemical compound COC1=CC=C(C=C(C#N)C#N)C=C1 UOHFCPXBKJPCAD-UHFFFAOYSA-N 0.000 description 1
- GFNNBHLJANVSQV-UHFFFAOYSA-N tyrphostin AG 1478 Chemical compound C=12C=C(OC)C(OC)=CC2=NC=NC=1NC1=CC=CC(Cl)=C1 GFNNBHLJANVSQV-UHFFFAOYSA-N 0.000 description 1
- KXDONFLNGBQLTN-WUXMJOGZSA-N tyrphostin AG 825 Chemical compound COC1=CC(\C=C(/C#N)C(N)=O)=CC(CSC=2SC3=CC=CC=C3N=2)=C1O KXDONFLNGBQLTN-WUXMJOGZSA-N 0.000 description 1
- TUCIOBMMDDOEMM-RIYZIHGNSA-N tyrphostin B42 Chemical compound C1=C(O)C(O)=CC=C1\C=C(/C#N)C(=O)NCC1=CC=CC=C1 TUCIOBMMDDOEMM-RIYZIHGNSA-N 0.000 description 1
- 229950008878 ulixertinib Drugs 0.000 description 1
- 229960000241 vandetanib Drugs 0.000 description 1
- UHTHHESEBZOYNR-UHFFFAOYSA-N vandetanib Chemical compound COC1=CC(C(/N=CN2)=N/C=3C(=CC(Br)=CC=3)F)=C2C=C1OCC1CCN(C)CC1 UHTHHESEBZOYNR-UHFFFAOYSA-N 0.000 description 1
- 229950000578 vatalanib Drugs 0.000 description 1
- YCOYDOIWSSHVCK-UHFFFAOYSA-N vatalanib Chemical compound C1=CC(Cl)=CC=C1NC(C1=CC=CC=C11)=NN=C1CC1=CC=NC=C1 YCOYDOIWSSHVCK-UHFFFAOYSA-N 0.000 description 1
- GPXBXXGIAQBQNI-UHFFFAOYSA-N vemurafenib Chemical compound CCCS(=O)(=O)NC1=CC=C(F)C(C(=O)C=2C3=CC(=CN=C3NC=2)C=2C=CC(Cl)=CC=2)=C1F GPXBXXGIAQBQNI-UHFFFAOYSA-N 0.000 description 1
- 238000001262 western blot Methods 0.000 description 1
- 229950009819 zotarolimus Drugs 0.000 description 1
- CGTADGCBEXYWNE-JUKNQOCSSA-N zotarolimus Chemical compound N1([C@H]2CC[C@@H](C[C@@H](C)[C@H]3OC(=O)[C@@H]4CCCCN4C(=O)C(=O)[C@@]4(O)[C@H](C)CC[C@H](O4)C[C@@H](/C(C)=C/C=C/C=C/[C@@H](C)C[C@@H](C)C(=O)[C@H](OC)[C@H](O)/C(C)=C/[C@@H](C)C(=O)C3)OC)C[C@H]2OC)C=NN=N1 CGTADGCBEXYWNE-JUKNQOCSSA-N 0.000 description 1
Classifications
-
- C—CHEMISTRY; METALLURGY
- C12—BIOCHEMISTRY; BEER; SPIRITS; WINE; VINEGAR; MICROBIOLOGY; ENZYMOLOGY; MUTATION OR GENETIC ENGINEERING
- C12N—MICROORGANISMS OR ENZYMES; COMPOSITIONS THEREOF; PROPAGATING, PRESERVING, OR MAINTAINING MICROORGANISMS; MUTATION OR GENETIC ENGINEERING; CULTURE MEDIA
- C12N9/00—Enzymes; Proenzymes; Compositions thereof; Processes for preparing, activating, inhibiting, separating or purifying enzymes
- C12N9/10—Transferases (2.)
- C12N9/1025—Acyltransferases (2.3)
- C12N9/104—Aminoacyltransferases (2.3.2)
-
- C—CHEMISTRY; METALLURGY
- C12—BIOCHEMISTRY; BEER; SPIRITS; WINE; VINEGAR; MICROBIOLOGY; ENZYMOLOGY; MUTATION OR GENETIC ENGINEERING
- C12Y—ENZYMES
- C12Y203/00—Acyltransferases (2.3)
- C12Y203/02—Aminoacyltransferases (2.3.2)
-
- G—PHYSICS
- G01—MEASURING; TESTING
- G01N—INVESTIGATING OR ANALYSING MATERIALS BY DETERMINING THEIR CHEMICAL OR PHYSICAL PROPERTIES
- G01N33/00—Investigating or analysing materials by specific methods not covered by groups G01N1/00 - G01N31/00
- G01N33/48—Biological material, e.g. blood, urine; Haemocytometers
- G01N33/50—Chemical analysis of biological material, e.g. blood, urine; Testing involving biospecific ligand binding methods; Immunological testing
- G01N33/5005—Chemical analysis of biological material, e.g. blood, urine; Testing involving biospecific ligand binding methods; Immunological testing involving human or animal cells
- G01N33/5008—Chemical analysis of biological material, e.g. blood, urine; Testing involving biospecific ligand binding methods; Immunological testing involving human or animal cells for testing or evaluating the effect of chemical or biological compounds, e.g. drugs, cosmetics
- G01N33/5011—Chemical analysis of biological material, e.g. blood, urine; Testing involving biospecific ligand binding methods; Immunological testing involving human or animal cells for testing or evaluating the effect of chemical or biological compounds, e.g. drugs, cosmetics for testing antineoplastic activity
-
- G—PHYSICS
- G01—MEASURING; TESTING
- G01N—INVESTIGATING OR ANALYSING MATERIALS BY DETERMINING THEIR CHEMICAL OR PHYSICAL PROPERTIES
- G01N2440/00—Post-translational modifications [PTMs] in chemical analysis of biological material
- G01N2440/36—Post-translational modifications [PTMs] in chemical analysis of biological material addition of addition of other proteins or peptides, e.g. SUMOylation, ubiquitination
Definitions
- molecular glue compounds induce protein-protein interactions that, in the context of a ubiquitin ligase, lead to protein degradation (Stanton et al., Science 359 eaao5902 (2016)).
- proteolysis-targeting chimeric molecules PROTAC®s
- molecular glue compounds are small molecules (also known as small molecule degraders) that induce an interaction between a substrate receptor of an E3 ubiquitin ligase and a target protein leading to proteolysis of the target.
- IMiDs immune modulatory drugs
- thalidomide a substrate receptor
- CTL4 Cullin-RING ubiquitin ligase 4
- these molecular glue degraders act substoichiometrically to catalyze the rapid depletion of previously inaccessible targets (Chopra et al., Drug Discov. Today. Technol. 37:5-13 (2019)).
- the present invention is directed to a method for identifying a cullin dependent small molecule (e.g., monofunctional or multi-functional) degrader, comprising: incubating mammalian cells with a small molecule test compound in an amount sufficient to cause a change in phenotype in the cells, and a neddylation inhibitor at concentration nontoxic or of low toxicity to the cells; and measuring a difference in phenotype in the cells following incubation relative to a control that comprises the cells incubated with the test compound but not the neddylation inhibitor, wherein a difference in the phenotype in the cells relative to the control indicates that the test compound causes cullin dependent degradation of a target protein.
- a cullin dependent small molecule e.g., monofunctional or multi-functional
- the test compound is a natural product. In some embodiments, the test compound is a synthetic compound. In some embodiments, the test compound is a semi-synthetic compound.
- the test compound is a small molecule kinase inhibitor. In some embodiments, the test compound is a small molecule tyrosine kinase inhibitor.
- the test compound is a small molecule therapeutic agent.
- the test compound is a small molecule anticancer therapeutic agent.
- the small molecule inhibitor is CR8, CC-885, or indisulam.
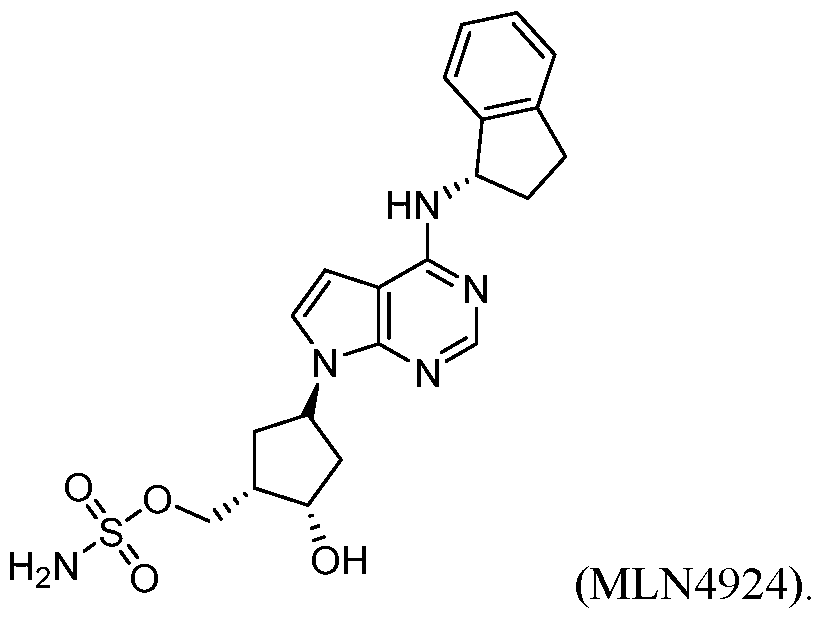
- the neddylation inhibitor is MLN4924.
- the concentration of the test compound is between about 0.001 pM to about 100 pM.
- the concentration of the neddylation inhibitor is less than 1 pM. [0013] In some embodiments, the concentration of the neddylation inhibitor is between about 10 nM to about 500 nM.
- the concentration of the neddylation inhibitor is about 100 nM.
- the cells are pretreated with the neddylation inhibitor prior to treatment with the test compound.
- the mammalian cells are human cells. In some embodiments, the cells are primary cells. In some embodiments, the primary cells are isolated from a tissue or a biopsy sample.
- the human cells are myeloid cells, lymphoid cells, neural cells, epithelial cells, endothelial cells, stem or progenitor cells, hepatocytes, myoblasts, osteoblasts, osteoclasts, lymphocytes, keratinocytes, melanocytes, mesothelial cells, germ cells, muscle cells, fibroblasts, transformed cells, non-transformed cells, or cancer cells.
- the transformed cells are Cas9 stable cells.
- the cells are SNGM, P31FU, OVK18, PFEIFFER, ES-2, OCIM1, K562, HEK293, or HEK293T cells.
- the difference in phenotype in the cells is increased protein abundance, decreased protein abundance, increased protein activity, decreased protein activity, increased gene expression, decreased gene expression, changed cellular proliferation, changed protein localization, increased or decreased mRNA transcript abundance of predetermined genes, or cell viability.
- the difference in phenotype in the cells is measured by flow cytometry, fluorescence-activated cell sorting (FACS), mass cytometry, and/or magnetic sorting, protein localization, cellular morphology, gene expression by RNA sequencing (RNAseq), Luminex® multiplex bead assay, quantitative polymerase chain reaction (qPCR), next generation sequencing (NGS), or immunoblotting.
- RNA sequencing RNA sequencing
- Luminex® multiplex bead assay is the L1000 assay.
- the invention is directed to high throughput screening (HTS) methods for identifying cullin dependent small molecule degraders.
- HTS high throughput screening
- the method of the present invention identifies small molecule (e.g, monofunctional and multiple-functional) degraders by observing a change phenotype in cells, e.g, cell viability or cell death, that is rescuable by co-treatment with the neddylation inhibitor.
- the rescue identifies compounds that cause protein degradation mediated by E3 ligase machinery which produces a toxic effect on the cell.
- the presence of the neddylation inhibitor prevents the degradation, thus rescuing the cell.
- This approach is generalizable to any phenotype, and presents a new way of identifying small molecule degrader compounds.
- FIG. 1A-FIG. IB are a set of graphs showing inhibition of neddylation as a readout for functional degradation (Slabicki et al., Nature DOI: 10.1038/s41586-020-2374-x (2020)).
- FIG. 1 A shows that neddylation inhibitor MLN4924 is not toxic to cells ⁇ 1 pM.
- FIG. IB shows that CR8 cell killing is rescued by low dose neddylation inhibitor MLN4924.
- FIG. 2A-FIG. 2H are a set of graphs showing that MLN4924 rescues CR8 (cyclin- dependent kinase (CDK) inhibitor) induced toxicity in SNGM (FIG. 2A), P31FUJ (FIG. 2B), OVK18 (FIG. 2C), PFEIFFER (FIG.2D), ES-2 (FIG. 2E), OCIM1 (FIG. 2F), K562 (FIG. 2G), and HEK293T (FIG. 2H) cell lines.
- CDK cyclin- dependent kinase
- FIG. 3 A-FIG. 3H are a set of graphs showing that MLN4924 rescues CC885 (cereblon (CRBN) modulator) induced toxicity in SNGM (FIG. 3A), P31FUJ (FIG. 3B), OVK18 (FIG. 3C), PFEIFFER (FIG. 3D), ES-2 (FIG. 3E), OCIM1 (FIG. 3F), K562 (FIG. 3G), and HEK293T (FIG. 3H) cell lines.
- CC885 cereblon (CRBN) modulator
- FIG. 4A-FIG. 4H are a set of graphs showing that MLN4924 rescues indisulam (CDK) induced toxicity in SNGM (FIG. 4A), P31FUJ (FIG. 4B), OVK18 (FIG. 4C), PFEIFFER (FIG. 4D), ES-2 (FIG. 4E), OCIM1 (FIG. 4F), K562 (FIG. 4G), and HEK293T (FIG. 4H) cell lines.
- CDK indisulam
- transitional term “comprising,” which is synonymous with “including,” “containing,” or “characterized by,” is inclusive or open-ended and does not exclude additional, unrecited elements or method steps.
- the transitional phrase “consisting of’ excludes any element, step, or ingredient not specified in the claim.
- the transitional phrase “consisting essentially of’ limits the scope of a claim to the specified materials or steps “and those that do not materially affect the basic and novel characteristic(s)” of the claimed invention.
- determining generally refer to any form of measurement, and include determining if an element is present or not. These terms include both quantitative and/or qualitative determinations. Assessing may be relative or absolute. “Assessing the presence of can include determining the amount of something present, as well as determining whether it is present or absent.
- small molecule refers to an organic molecule or compound that is monofunctional and that ranges in size from about 50 to about 10,000 daltons, usually from about 50 to about 5,000 daltons and more usually from about 100 to about 1000 daltons.
- small molecule degrader also known as “molecular glue,” refers to a small molecule that induces or stabilizes interactions between proteins.
- Known small molecule degraders e.g., thalidomide analogs, bind to the substrate receptors of E3 ubiquitin ligases and recruit target proteins for their ubiquitination and subsequent degradation by proteasomal degradation. Small molecule degraders also are referred to herein as cullin dependent degraders.
- concentration of low toxicity refers to a concentration that kills less than about 50% of untreated control cells.
- a low toxicity concentration of neddylation inhibitor MLN4924 is between about 10 nM to about 999 nM, or between about 10 nM to about 500 nM.
- a low toxicity concentration of neddylation inhibitor MLN4924 is about 200 nM.
- natural product refers to a small molecule organic compound produced by an organism, including a primary and secondary metabolite.
- synthetic compound refers to a small molecule organic compound made by chemical synthesis, especially to imitate a natural product.
- si-synthetic compound refers to a small molecule compound made by synthesis from a naturally occurring compound.
- the present invention is directed to a method for identifying a cullin dependent small molecule (e.g., monofunctional or multi-functional) degrader, comprising: incubating mammalian cells with a small molecule test compound in an amount sufficient to cause a change in phenotype in the cells, and a neddylation inhibitor at a concentration that is nontoxic or of low toxicity to the cells; and measuring a difference in phenotype in the cells following incubation relative to a control that comprises the cells incubated with the test compound but not the neddylation inhibitor, wherein a difference in the phenotype in the cells relative to the control indicates that the test compound causes cullin dependent degradation of a target protein.
- the control could be performed simultaneously or not, and with the same amount of the small molecule and for the same amount of incubation time.
- the test compound is a natural product. In some embodiments, the test compound is a synthetic compound. In some embodiments, the test compound is a semi-synthetic compound.
- the test compound is a small molecule kinase inhibitor. In some embodiments, the test compound is a small molecule tyrosine kinase inhibitor.
- Tyrosine kinase inhibitors are a class of chemotherapy medications that inhibit, or block, one or more of the enzyme tyrosine kinases. Over 25 TKIs have been approved by the U.S. Food and Drug Administration for use in humans (Roskoski, Pharmacol Res. 144'.19-50 (2019)).
- TKIs include Ruxolitinib, Tofacitinib, Lapatinib, Vandetanib, Sorafenib, Sunitinib, Axitinib, Nintedanib, Regorafenib, Pazopanib, Lenvatinib, Crizotinib, Ceritinib, Cabozantinib, DWF, Afatinib, Ibrutinib, B43, KU004, Foretinib, KRCA- 0008, PF-06439015, PF-06463922, Canertinib, GSA-10, GW2974, GW583340, WZ4002, CP- 380736, D2667, Mubritinib, PD153035, PD168393, Pelitinib, PF-06459988, PF-06672131, PF-6422899, PKI-166, Reveromycin A, Tyrphostin 1, Tyr
- the test compound is a member of a library or collection of compounds or agents of interest (e.g, TKIs).
- the library of compounds may be composed of a plurality of chemical compounds and agents that may be assembled from a single or multiple sources.
- a library may include chemical compounds and agents such as chemically synthesized substances, products of biotechnological processes, naturally occurring substances or products resulting from combinatorial chemistry techniques or combinations thereof.
- the library includes a plurality of clinical or preclinical small molecule drugs.
- the test compound is a small molecule therapeutic agent.
- the test compound is a small molecule anticancer therapeutic agent.
- test compound is CR8, CC-885, or indisulam.
- CR8 also known as (7?)-CR8 and (2R)-2-[[9-(Methylethyl)-6-[[[4-(2- pyridinyl)phenyl]methyl]amino]-9H-purin-2-yl]amino]-l-butanol, has the following structure:
- CC-885 also known as N-(3-chloro-4-methylphenyl)-N'-[[2-(2,6-dioxo-3- piperidinyl)-2,3-dihydro-l-oxo-lH-isoindol-5-yl]methyl]-urea, has the following structure:
- Indisulam also known as E7070 and N-(3-chloro-lH-indol-7-yl)benzene-l,4- disulfonamide, has the following structure: (Indisulam).
- the amount of the test compound is sufficient to cause a measurable change in phenotype in the cells.
- the concentration of the test compound is between about 0.001 pM to about 100 pM. In some embodiments, the concentration of the test compound is between about 0.001 pM to about 30 pM. In some embodiments, the concentration of the test compound is between about 0.03 pM to about 30 pM. In some embodiments, the concentration of the test compound is between about 0.1 pM to about 3 pM. In some embodiments, the concentration of the test compound is between about 0.001 pM to about 0.1 pM.
- Neddylation is a post-translational protein modification that conjugates ubiquitin-like protein neuronal precursor cell-expressed developmentally down-regulated protein 8 (NEDD8). Similar to ubiquitination, neddylation is mediated by a cascade of three NEDD8 specific enzymes, an El activating enzyme, an E2 conjugating enzyme and one of the several E3 ligases. By altering its substrates’ conformation, stability, subcellular localization or binding affinity to DNA or proteins, neddylation regulates diverse cellular processes including the ubiquitin-proteasome system-mediated protein degradation, protein transcription, cell signaling, etc. (Kandala et al., Am J Cardiovasc Dis. 4(4): 140-158 (2014)).
- the best- characterized substrates of neddylation are cullin family members that function as indispensable components of multiunit cullin-RING E3 ubiquitin ligases (CRLs), whose activity requires cullin neddylation (Zhou et al., Molecular cancer 75:77(2019)).
- CTLs cullin-RING E3 ubiquitin ligases
- the small molecule MLN4924 Panedistat
- NAE adenosine sulfamate analog
- Neddylation inhibitors effectively block cullin neddylation and inactivate CRLs. This in turn leads to accumulation of various CRL substrates, thus triggering multiple cellular responses, including cell cycle arrest, apoptosis, senescence and autophagy in a cell-type dependent manner (Zhou et al., Molecular cancer 18:77(2019)).
- the neddylation inhibitor is MLN4924, also known as ((lS,2S,4R)-4-(4-((lS)-2,3-Dihydro-lH-inden-l-ylamino)-7H-pyrrolo(2,3-d)pyrimidin-7-yl)- 2-hydroxycyclopentyl)methyl sulphamate, has the following structure:
- the neddylation inhibitor is a derivative or analog of MLN492.
- Analogs of MLN492 that may be useful as neddylation inhibitors are described in U.S. Patent 7,951,810 B2 and U.S. Patent Application Publication 2018/0086785 Al.
- TAS4464 Another exemplary NAE or neddylation inhibitor, which may be used in the present invention, is TAS4464, which is also known as 4-amino-7-[(2R,3R,4S,5R)-3,4-dihydroxy-5- [(sulfamoylamino)methyl]oxolan-2-yl]-5-[2-(2-ethoxy-6-fluorophenyl)ethynyl]pyrrolo[2,3- d] pyrimidine and has the following structure: [0056]
- the neddylation inhibitor is a derivative or analog of TAS4464. Analogs of TAS4464 that may be useful as neddylation inhibitors are described in U.S. Patent 9,963,456 B2.
- the concentration of the neddylation inhibitor is below IpM. In some embodiments, the concentration of the neddylation inhibitor is between about 10 nM to about 999 nM.
- the concentration of the neddylation inhibitor is between about 10 nM to about 500 nM.
- the concentration of the neddylation inhibitor is about 100 nM.
- the cells may be pretreated with different concentrations of the neddylation inhibitor, typically for about 10 minutes to about 24 hours prior to treatment with the test compound in dose response assays.
- the cells are incubated in the presence of the test compound and the neddylation inhibitor for an amount of time sufficient to allow for a measurable change in phenotype to occur.
- the cells are incubated with neddylation inhibitor and test compound for at least 3 to 5 days.
- the cells are incubated with neddylation inhibitor and test compound for at least 3 days.
- incubation time may be shortened to 10 minutes - 24 hours to investigate protein levels or other more general readout, e.g, cellular differentiation and/or gene expression by RNA sequencing (RNAseq) or quantitative polymerase chain reaction (qPCR).
- Mammalian cells suitable for use in the present methods are those that can be maintained or propagated in vitro and that naturally display, or that may be genetically modified to display, a phenotype (such as one described herein) the alteration of which is desired to be monitored for modulation.
- cells may display a phenotype whose inhibition within the assay is to be determined.
- Mammalian cells which display the appropriate phenotypic characteristics (phenotype) may be identified or obtained by any convenient technique or source, including those known to the person of ordinary skill in the art.
- Such cells may be stable cell lines, such as those obtainable from American Type Culture Collection (ATCC) or other cell repositories.
- ATCC American Type Culture Collection
- the mammalian cells employed may be primary cells derived from a tissue or organ of an individual organism.
- the mammalian cells may be transiently transfected with genetic constructs, such as those involved in the (desired) phenotype, for example a reporter gene.
- the mammalian cells may be infected with a virus or bacterium which leads to an alteration in the phenotype.
- human cells murine cells, hamster cells, rate cells, primate cells, and cells from domestic mammals (such as ovine, bovine, equine, canine or feline cells).
- domestic mammals such as ovine, bovine, equine, canine or feline cells.
- cells for employment in the present methods include mammalian cells derived from a cancer or tumor, or those associated with some other disease or abnormality of a mammal.
- the mammalian cells are primary cells.
- the primary cells are isolated from a tissue or a biopsy sample.
- the mammalian cells are human cells.
- the human cells are myeloid cells, lymphoid cells, neural cells, epithelial cells, endothelial cells, stem or progenitor cells, hepatocytes, myoblasts, osteoblasts, osteoclasts, lymphocytes, keratinocytes, melanocytes, mesothelial cells, germ cells, muscle cells, fibroblasts, transformed cells, non-transformed cells, or cancer cells.
- the transformed cells are Cas9 stable cells.
- the cells are SNGM, P31FU, OVK18, PFEIFFER, ES-2, 0CIM1, K562, HEK293, or HEK293T cells.
- the phenotype to be monitored for change may be one having been specifically engineered for a given cell type.
- a mammalian cell type may be genetically modified to result in a phenotype that is convenient or suitable for detection, such as a fluorescent protein/marker (such as GFP; eGFP), a luminescent protein/marker (such as luciferase) or a cell-surface marker than is detectable, e.g., with a labelled antibody, or intracellular proteins stained with labeled antibodies.
- the phenotype of the mammalian cell is any one of the following, namely: one associated with a cell signaling pathway, preferably an activated cell signaling pathway; and/or one selected from the list consisting of: luminescence, fluorescence, viability, senescence, differentiation, migration, invasion, chemotaxis, apoptosis, immunological anergy, surface marker expression, progress through the cell cycle, transcriptional activity, protein expression, glycosylation, resistance to infection, permeability and reporter-gene activity.
- the phenotype is not death and/or reduced (or decreased) growth, such as a phenotype that is not rescue of a cell from cytokine dependence, is not rescue from apoptosis (including neutrophil apoptosis/cell-death), is not induction of colony formation, or is not a rescue screen.
- a screen for such a phenotype may be designed to detect a trait or characteristic of the cell that is not a rescue from cell death or an increase in growth of the mammalian cell.
- the phenotypic difference in the cells is increased protein abundance, decreased protein abundance, increased protein activity, decreased protein activity, increased gene expression, decreased gene expression, changed cellular proliferation, changed protein localization, or cell viability.
- the phenotype difference is an increased or decreased mRNA transcript abundance of predetermined genes.
- immunoassays are binding assays involving binding between antibodies and antigen.
- Representative examples of immunoassays include enzyme linked immunosorbent assays (ELISAs), enzyme linked immunospot assay (ELISPOT), radioimmunoassays (RIA), radioimmune precipitation assays (RIP A), immunobead capture assays, Western blotting, dot blotting, gel-shift assays, Flow cytometry, immunohistochemistry, fluorescence microscopy, protein arrays, multiplexed bead arrays, magnetic capture, in vivo imaging, fluorescence resonance energy transfer (FRET), and fluorescence recovery/localization after photobleaching (FRAPZFLAP).
- ELISAs enzyme linked immunosorbent assays
- ELISPOT enzyme linked immunospot assay
- RIA radioimmunoassays
- RIP A radioimmune precipitation assays
- immunobead capture assays Western blotting, dot blotting, gel-
- the phenotypic difference in the cells is measured by flow cytometry, fluorescence-activated cell sorting (FACS), mass cytometry, and/or magnetic sorting, protein localization, cellular morphology, gene expression by RNA sequencing (RNAseq), Luminex® multiplex bead assay, quantitative polymerase chain reaction (qPCR), next generation sequencing (NGS), or immunoblotting.
- RNA sequencing RNA sequencing
- Luminex® multiplex bead assay e.g., the L1000 assay.
- L1000 is a high-throughput gene expression assay that measures the mRNA transcript abundance of 978 "landmark" genes from human cells. Landmark genes were chosen to be expressed across lineage which would enable the prediction of expression of other genes not directly measured.
- the "L” in LI 000 refers to the Landmark genes measured in the assay. Measurements are made using the 500 colors of Luminex® beads such that two transcripts are identified by a single bead color. Expression of 80 control transcripts, chosen for their invariant expression across cell states, as well as the 978 Landmark genes are also measured. LI 000 is highly reproducible, comparable to RNA sequencing, and suitable for computational inference of the expression levels of 81% of non-measured transcripts. Subramanian et al., Cell 777(6 1437-1452.el7 (2017).
- the disclosed methods entail high throughput screening (HTS) for identifying cullin dependent small molecule degraders.
- HTS high throughput screening
- the term “high-throughput” relates to an assay system for the rapid testing of a plurality of compounds in a short time. Thus, the assaying time per tested compound is minimized.
- the assay is preferably carried out in multiwell plates, e.g., 6-well plates, 12-well plates, 24-well plates, 96-well plates or 384-well plates, or 1536 plates, even more preferably in 96-well plates, 384-well or 1536 plates.
- the term “high-throughput” encompasses screening activity in which human intervention is minimized, and automation is maximized.
- high-throughput screening involves automated pipetting, mixing, and heating.
- a high-throughput method is one in which for example hundreds of compounds can be screened per 24-hour period by a single individual operating a single suitable apparatus.
- Automation refers to a method or any one or more steps thereof that is fully or partly controlled and/or carried out by one or more technical devices, preferably pipetting robots.
- Methods for conducting a variety of different HTS assays, in particular HTS screening assays are known in the art. The skilled person knows how to conduct HTS methods by the methods used in the context of the present invention.
- Example 1 Inhibition of neddylation as a readout for functional degradation.
- HEK293T cells were treated with various concentration of MG132 (proteasome inhibitor), MLN7243 (El ubiquitin-activating enzyme inhibitor) or MLN4924 (cullin neddylation inhibitor) for three days.
- Cell viability was assessed using CellTiter-Glo® Luminescent Cell Viability Assay (Promega®, G7572) on CLARIOstar® Plus Multi-mode Microplate Reader, MARS 3.4 (BMG LabTech).
- Cell viabilities were calculated relative to DMSO controls. Standard four-parameter log-logistic curves are fitted with the ‘dr4pl’ R package. The results are shown in FIG. 1A.
- FIG. 1A-FIG. IB show that titration of MLN4924 up to 1 pM did not have a significant effect on cell viability (FIG. 1 A), but a 100 nM dose of MLN4924 was able to rescue CR8 induced toxicity (FIG. IB). These results confirmed that addition of low dose of MLN4924 can achieve a functional effect of reduced toxicity, when the toxicity is caused by neddylation dependent mechanism, such as degradation of Cyclin K and CDK13 by CR8.
- Ubiquitin proteasome system (UPS) compounds including the neddylation inhibitor MLN4924, are known to be toxic at high to mid concentrations (>1 pM). It was believed that low doses would not inhibit neddylation. Applicant surprisingly found that low dose MLN4924 (e.g, -100 nM) was nontoxic to cells and it potently blocked neddylation generating a sufficient window to observe phenotypic difference in the cells.
- Example 2 MLN4924 rescue of CR8 (cyclin-dependent kinase (CDK) inhibitor) induced toxicity in multiple cell lines.
- CDK cyclin-dependent kinase
- HEK293T-Cas9 cells were resuspended at 0.15 x 10 6 per ml and plated on a 384-well plate with 50 pL per well and MLN4924, MLN7243 or MG132 with or without CR8 serially diluted with D300e Digital Dispenser (Tecan).
- a panel of SNGM, P31FUJ, OVK18, PFEIFFER, ES-2, 0CIM1 or HEK293T cells was resuspended at -0.15 x 10 6 per ml and plated on a 384-well plate with 50 pL per well. Cells were pretreated with MLN4924 at 0, 0.05, 0.1 or 0.2 pM concentration and treated with CR8 in dose response. [0085] After three days of drug exposure, cell viability was assessed using CellTiter-Glo® Luminescent Cell Viability Assay (Promega®, G7572) on CLARIOstar® Plus Multi-mode Microplate Reader, MARS 3.4 (BMG LabTech). Cell viabilities were calculated relative to DMSO controls. The half-maximum effective concentration (ECso) values were derived from standard four-parameter log-logistic curves fitted with the ‘dr4pl’ R package.
- Example 3 MLN4924 rescue of CC885 (cereblon (CRBN) modulator) induced toxicity in multiple cell lines.
- CC885 cereblon (CRBN) modulator
- HEK293T-Cas9 cells were resuspended at 0.15 x 10 6 per ml and plated on a 384-well plate with 50 pL per well and MLN4924, MLN7243 or MG132 with or without CC-885 serially diluted with D300e Digital Dispenser (Tecan).
- a panel of SNGM, P31FUJ, OVK18, PFEIFFER, ES-2, OCIM1 or HEK293T cells was resuspended at -0.15 x 10 6 per ml and plated on a 384-well plate with 50 pL per well. Cells were pretreated with MLN4924 at 0, 0.05, 0.1 or 0.2 pM concentration and treated with CC-885 in dose response.
- FIG.3A-FIG. 3H show that MLN4924 rescues CC-885 induced toxicity in multiple cell lines.
- Example 4 MLN4924 rescue of indisulam (CDK) induced toxicity in multiple cell lines.
- HEK293T-Cas9 cells were resuspended at 0.15 x 10 6 per ml and plated on a 384-well plate with 50 pL per well and MLN4924, MLN7243 or MG132 with or without indisulam serially diluted with D300e Digital Dispenser (Tecan).
- a panel of SNGM, P31FUJ, OVK18, PFEIFFER, ES-2, OCIM1 or HEK293T cells was resuspended at -0.15 x 10 6 per ml and plated on a 384-well plate with 50 pL per well. Cells were pretreated with MLN4924 at 0, 0.05, 0.1 or 0.2 pM concentration and treated with indisulam in dose response. [0095] After three days of drug exposure, cell viability was assessed using CellTiter-Glo® Luminescent Cell Viability Assay (Promega®, G7572) on CLARIOstar® Plus Multi-mode Microplate Reader, MARS 3.4 (BMG LabTech). Cell viabilities were calculated relative to DMSO controls. The half-maximum effective concentration (EC50) values were derived from standard four-parameter log-logistic curves fitted with the ‘dr4pl’ R package.
- CR8 induces the degradation of Cyclin K via direct binding of CR8-CDK12-Cyclin K to damage specific DNA binding protein 1 (DDB1).
- CC-885 is a CRBN based degrader of G1 to S phase transition 1 (GSPT1).
- Indisulam is a DDB1 and CUL4 associated factor 15 (DCAF15) based degrader of splicing factor RNA binding motif protein 39 (RBM39). All of the three compounds caused dose dependent cell killing via proteasomal degradation.
- FIG. 2A-FIG. 4H illustrate co-treatments of CC-885, CR8 and Indisulam with three doses of MLN4924 (50, 100, 200 nM).
- Increased doses of neddylation inhibitor decreased CellTiter-Glo® levels for different cell lines.
- Optimal concentration of MLN4924 may be characterized with increased toxicity.
Landscapes
- Health & Medical Sciences (AREA)
- Life Sciences & Earth Sciences (AREA)
- Chemical & Material Sciences (AREA)
- Engineering & Computer Science (AREA)
- Bioinformatics & Cheminformatics (AREA)
- Biomedical Technology (AREA)
- Organic Chemistry (AREA)
- Wood Science & Technology (AREA)
- Immunology (AREA)
- Molecular Biology (AREA)
- Zoology (AREA)
- General Health & Medical Sciences (AREA)
- Genetics & Genomics (AREA)
- Biochemistry (AREA)
- Biotechnology (AREA)
- Hematology (AREA)
- Microbiology (AREA)
- Medicinal Chemistry (AREA)
- Urology & Nephrology (AREA)
- General Engineering & Computer Science (AREA)
- Cell Biology (AREA)
- General Physics & Mathematics (AREA)
- Pathology (AREA)
- Analytical Chemistry (AREA)
- Physics & Mathematics (AREA)
- Tropical Medicine & Parasitology (AREA)
- Food Science & Technology (AREA)
- Toxicology (AREA)
- Medicines That Contain Protein Lipid Enzymes And Other Medicines (AREA)
Abstract
Disclosed is a method for identifying cullin dependent small molecule degraders via phenotypic screening.
Description
PHENOTYPIC ASSAY TO IDENTIFY PROTEIN DEGRADERS
RELATED APPLICATIONS
[0001] This application claims the benefit of priority under 35 U.S.C. § 119(e) to U.S. Provisional Application No: 63/140,605, filed on January 22, 2021, which is incorporated herein by reference in its entirety.
GOVERNMENT LICENSE RIGHTS
[0002] This invention was made with government support under grant number R35 CA253125 awarded by The National Institutes of Health. The government has certain rights in the invention.
BACKGROUND OF THE INVENTION
[0003] Molecular glue compounds induce protein-protein interactions that, in the context of a ubiquitin ligase, lead to protein degradation (Stanton et al., Science 359 eaao5902 (2018)). Unlike proteolysis-targeting chimeric molecules (PROTAC®s), molecular glue compounds are small molecules (also known as small molecule degraders) that induce an interaction between a substrate receptor of an E3 ubiquitin ligase and a target protein leading to proteolysis of the target. Examples of molecular glues that induce proteolysis of targets include IMiDs (immune modulatory drugs; e.g, thalidomide), which generate a novel interaction between a substrate (e.g., IKZF1/3) and cereblon, a substrate receptor (also known as DCAF) for Cullin-RING ubiquitin ligase 4 (CRL4) den Besten and Lipford, Nat. Chem. Biol. 16(11): 1157-1158 (2020). Unlike traditional enzyme inhibitors, these molecular glue degraders act substoichiometrically to catalyze the rapid depletion of previously inaccessible targets (Chopra et al., Drug Discov. Today. Technol. 37:5-13 (2019)). Although highly desirable, molecular glue degraders have only been found serendipitously. Strategies available for identifying or designing these compounds are limited (Slabicki et al., Nature DOI: 10.1038/s41586-020-2374-x (2020)).
SUMMARY OF THE INVENTION
[0004] The present invention is directed to a method for identifying a cullin dependent small molecule (e.g., monofunctional or multi-functional) degrader, comprising:
incubating mammalian cells with a small molecule test compound in an amount sufficient to cause a change in phenotype in the cells, and a neddylation inhibitor at concentration nontoxic or of low toxicity to the cells; and measuring a difference in phenotype in the cells following incubation relative to a control that comprises the cells incubated with the test compound but not the neddylation inhibitor, wherein a difference in the phenotype in the cells relative to the control indicates that the test compound causes cullin dependent degradation of a target protein.
[0005] In some embodiments, the test compound is a natural product. In some embodiments, the test compound is a synthetic compound. In some embodiments, the test compound is a semi-synthetic compound.
[0006] In some embodiments, the test compound is a small molecule kinase inhibitor. In some embodiments, the test compound is a small molecule tyrosine kinase inhibitor.
[0007] In some embodiments, the test compound is a small molecule therapeutic agent.
[0008] In some embodiments, the test compound is a small molecule anticancer therapeutic agent.
[0009] In some embodiments, the small molecule inhibitor is CR8, CC-885, or indisulam. [0010] In some embodiments, the neddylation inhibitor is MLN4924.
[0011] In some embodiments, the concentration of the test compound is between about 0.001 pM to about 100 pM.
[0012] In some embodiments, the concentration of the neddylation inhibitor is less than 1 pM. [0013] In some embodiments, the concentration of the neddylation inhibitor is between about 10 nM to about 500 nM.
[0014] In some embodiments, the concentration of the neddylation inhibitor is about 100 nM. [0015] In some embodiments, the cells are pretreated with the neddylation inhibitor prior to treatment with the test compound.
[0016] In some embodiments, the mammalian cells are human cells. In some embodiments, the cells are primary cells. In some embodiments, the primary cells are isolated from a tissue or a biopsy sample.
[0017] In some embodiments, the human cells are myeloid cells, lymphoid cells, neural cells, epithelial cells, endothelial cells, stem or progenitor cells, hepatocytes, myoblasts, osteoblasts, osteoclasts, lymphocytes, keratinocytes, melanocytes, mesothelial cells, germ cells, muscle cells, fibroblasts, transformed cells, non-transformed cells, or cancer cells.
[0018] In some embodiments, the transformed cells are Cas9 stable cells.
[0019] In some embodiments, the cells are SNGM, P31FU, OVK18, PFEIFFER, ES-2, OCIM1, K562, HEK293, or HEK293T cells.
[0020] In some embodiments, the difference in phenotype in the cells is increased protein abundance, decreased protein abundance, increased protein activity, decreased protein activity, increased gene expression, decreased gene expression, changed cellular proliferation, changed protein localization, increased or decreased mRNA transcript abundance of predetermined genes, or cell viability.
[0021] In some embodiments, the difference in phenotype in the cells is measured by flow cytometry, fluorescence-activated cell sorting (FACS), mass cytometry, and/or magnetic sorting, protein localization, cellular morphology, gene expression by RNA sequencing (RNAseq), Luminex® multiplex bead assay, quantitative polymerase chain reaction (qPCR), next generation sequencing (NGS), or immunoblotting. In some embodiments, the Luminex® multiplex bead assay is the L1000 assay.
[0022] In some aspects, the invention is directed to high throughput screening (HTS) methods for identifying cullin dependent small molecule degraders.
[0023] Without intending to be bound by any particular theory of operation, the method of the present invention identifies small molecule (e.g, monofunctional and multiple-functional) degraders by observing a change phenotype in cells, e.g, cell viability or cell death, that is rescuable by co-treatment with the neddylation inhibitor. The rescue identifies compounds that cause protein degradation mediated by E3 ligase machinery which produces a toxic effect on the cell. The presence of the neddylation inhibitor prevents the degradation, thus rescuing the cell. This approach is generalizable to any phenotype, and presents a new way of identifying small molecule degrader compounds.
BRIEF DESCRIPTION OF THE DRAWINGS
[0024] FIG. 1A-FIG. IB are a set of graphs showing inhibition of neddylation as a readout for functional degradation (Slabicki et al., Nature DOI: 10.1038/s41586-020-2374-x (2020)). FIG. 1 A shows that neddylation inhibitor MLN4924 is not toxic to cells <1 pM. FIG. IB shows that CR8 cell killing is rescued by low dose neddylation inhibitor MLN4924.
[0025] FIG. 2A-FIG. 2H are a set of graphs showing that MLN4924 rescues CR8 (cyclin- dependent kinase (CDK) inhibitor) induced toxicity in SNGM (FIG. 2A), P31FUJ (FIG. 2B),
OVK18 (FIG. 2C), PFEIFFER (FIG.2D), ES-2 (FIG. 2E), OCIM1 (FIG. 2F), K562 (FIG. 2G), and HEK293T (FIG. 2H) cell lines.
[0026] FIG. 3 A-FIG. 3H are a set of graphs showing that MLN4924 rescues CC885 (cereblon (CRBN) modulator) induced toxicity in SNGM (FIG. 3A), P31FUJ (FIG. 3B), OVK18 (FIG. 3C), PFEIFFER (FIG. 3D), ES-2 (FIG. 3E), OCIM1 (FIG. 3F), K562 (FIG. 3G), and HEK293T (FIG. 3H) cell lines.
[0027] FIG. 4A-FIG. 4H are a set of graphs showing that MLN4924 rescues indisulam (CDK) induced toxicity in SNGM (FIG. 4A), P31FUJ (FIG. 4B), OVK18 (FIG. 4C), PFEIFFER (FIG. 4D), ES-2 (FIG. 4E), OCIM1 (FIG. 4F), K562 (FIG. 4G), and HEK293T (FIG. 4H) cell lines.
DETAILED DESCRIPTION OF THE INVENTION
[0028] Unless defined otherwise, all technical and scientific terms used herein have the same meaning as is commonly understood by one of skill in the art to which the subject matter herein belongs. As used in the specification and the appended claims, unless specified to the contrary, the following terms have the meaning indicated in order to facilitate the understanding of the present invention.
[0029] As used in the description and the appended claims, the singular forms “a”, “an”, and “the” include plural referents unless the context clearly dictates otherwise. Thus, for example, reference to “a composition” includes mixtures of two or more such compositions, reference to “an inhibitor” includes mixtures of two or more such inhibitors, and the like.
[0030] Unless stated otherwise, the term “about” means within 10% (e.g., within 5%, 2% or 1%) of the particular value modified by the term “about.”
[0031] The transitional term “comprising,” which is synonymous with “including,” “containing,” or “characterized by,” is inclusive or open-ended and does not exclude additional, unrecited elements or method steps. By contrast, the transitional phrase “consisting of’ excludes any element, step, or ingredient not specified in the claim. The transitional phrase “consisting essentially of’ limits the scope of a claim to the specified materials or steps “and those that do not materially affect the basic and novel characteristic(s)” of the claimed invention.
[0032] The terms "determining", "measuring", "evaluating", "assessing" and "assaying" as used herein generally refer to any form of measurement, and include determining if an element
is present or not. These terms include both quantitative and/or qualitative determinations. Assessing may be relative or absolute. "Assessing the presence of can include determining the amount of something present, as well as determining whether it is present or absent.
[0033] The term “small molecule” as used herein refers to an organic molecule or compound that is monofunctional and that ranges in size from about 50 to about 10,000 daltons, usually from about 50 to about 5,000 daltons and more usually from about 100 to about 1000 daltons. [0034] The term “small molecule degrader,” also known as “molecular glue,” refers to a small molecule that induces or stabilizes interactions between proteins. Known small molecule degraders, e.g., thalidomide analogs, bind to the substrate receptors of E3 ubiquitin ligases and recruit target proteins for their ubiquitination and subsequent degradation by proteasomal degradation. Small molecule degraders also are referred to herein as cullin dependent degraders.
[0035] The term “concentration of low toxicity” as used herein refers to a concentration that kills less than about 50% of untreated control cells. For example, a low toxicity concentration of neddylation inhibitor MLN4924 is between about 10 nM to about 999 nM, or between about 10 nM to about 500 nM. For example, a low toxicity concentration of neddylation inhibitor MLN4924 is about 200 nM.
[0036] The term “natural product” as used herein refers to a small molecule organic compound produced by an organism, including a primary and secondary metabolite.
[0037] The term “synthetic compound” refers to a small molecule organic compound made by chemical synthesis, especially to imitate a natural product.
[0038] The term “semi-synthetic compound” refers to a small molecule compound made by synthesis from a naturally occurring compound.
[0039] The present invention is directed to a method for identifying a cullin dependent small molecule (e.g., monofunctional or multi-functional) degrader, comprising: incubating mammalian cells with a small molecule test compound in an amount sufficient to cause a change in phenotype in the cells, and a neddylation inhibitor at a concentration that is nontoxic or of low toxicity to the cells; and measuring a difference in phenotype in the cells following incubation relative to a control that comprises the cells incubated with the test compound but not the neddylation inhibitor, wherein a difference in the phenotype in the cells relative to the control indicates that the test compound causes cullin dependent degradation of a target protein. As persons skilled in the
art would appreciate, the control could be performed simultaneously or not, and with the same amount of the small molecule and for the same amount of incubation time.
[0040] In some embodiments, the test compound is a natural product. In some embodiments, the test compound is a synthetic compound. In some embodiments, the test compound is a semi-synthetic compound.
[0041] In some embodiments, the test compound is a small molecule kinase inhibitor. In some embodiments, the test compound is a small molecule tyrosine kinase inhibitor.
[0042] Tyrosine kinase inhibitors (TKIs) are a class of chemotherapy medications that inhibit, or block, one or more of the enzyme tyrosine kinases. Over 25 TKIs have been approved by the U.S. Food and Drug Administration for use in humans (Roskoski, Pharmacol Res. 144'.19-50 (2019)). Exemplary TKIs include Ruxolitinib, Tofacitinib, Lapatinib, Vandetanib, Sorafenib, Sunitinib, Axitinib, Nintedanib, Regorafenib, Pazopanib, Lenvatinib, Crizotinib, Ceritinib, Cabozantinib, DWF, Afatinib, Ibrutinib, B43, KU004, Foretinib, KRCA- 0008, PF-06439015, PF-06463922, Canertinib, GSA-10, GW2974, GW583340, WZ4002, CP- 380736, D2667, Mubritinib, PD153035, PD168393, Pelitinib, PF-06459988, PF-06672131, PF-6422899, PKI-166, Reveromycin A, Tyrphostin 1, Tyrphostin 23, Tyrphostin 51, Tyrphostin AG 528, Tyrphostin AG 658, Tyrphostin AG 825, Tyrphostin AG 835, Tyrphostin AG 1478, Tyrphostin RG 13022, Tyrphostin RG 14620, B178, GSK1838705A, PD-161570, PD 173074, SU-5402, Roslin 2, Picropodophyllotoxin, PQ401, I-OMe-Tyrphostin AG 538, GNF 5837, GW441756, Tyrphostin AG 879, DMPQ, JNJ-10198409, PLX647, Trapidil, Tyrphostin A9, Tyrphostin AG 370, Lestaurtinib, DMH4, Geldanamycin, Genistein, GW2580, Herbimycin A, Lavendustin C, Midostaurin, NVP-BHG712, PD158780, PD-166866, PF- 06273340, PP2, RPI, SU 11274, SU5614, Symadex, Tyrphostin AG 34, Tyrphostin AG 974, Tyrphostin AG 1007, UNC2881, Honokiol, SU1498, SKLB1002, CP-547632, JK-P3, KRN633, SC-1, ST638, SU 5416, Sulochrin, Tyrphostin SU 1498, S8567, rociletinib, Dacomitinib, Tivantinib, Neratinib, Masitinib, Vatalanib, Icotinib, XL-184, OSI-930, AB 1010, Quizartinib, AZD9291, Tandutinib, HM61713, Brigantinib, Vemurafenib (PLX-4032), Semaxanib, AZD2171, Crenolanib, Damnacanthal, Fostamatinib, Motesanib, Radotinib, OSI- 027, Linsitinib, BIX02189, PF-431396, PND-1186, PF-03814735, PF-431396, sirolimus, temsirolimus, everolimus, deforolimus, zotarolimus, BEZ235, INK128, Omipalisib, AZD8055, MHY1485, PI-103, KU-0063794, ETP-46464, GDC-0349, XL388, WYE-354, WYE-132, GSK1059615, WAY-600, PF-04691502, WYE-687, PP121, BGT226, AZD2014,
PP242, CH5132799, P529, GDC-0980, GDC-0994, XMD8-92, Ulixertinib, FR180204, SCH772984, Trametinib, PD184352, PD98059, Selumetinib, PD325901, U0126, Pimasertinib, TAK-733, AZD8330, Binimetinib, PD318088, SL-327, Refametinib, GDC- 0623, Cobimetinib, BI-847325, Adaphostin, GNF 2, PPY A, AIM-100, ASP 3026, LFM A13, PF 06465469, (-)-Terreic acid, AG-490, BIBU 1361, BIBX 1382, BMS 599626, CGP 52411, GW 583340, HDS 029, HKI 357, JNJ 28871063, WHI-P 154, PF 431396, PF 573228, FUN 1, PD 166285, SUN 11602, SR 140333, TCS 359, BMS 536924, NVP ADW 742, PQ 401, BMS 509744, CP 690550, NSC 33994, WHI-P 154, KB SRC 4, DDR1-IN-1, PF 04217903, PHA 665752, SU 16f, A 419259, AZM 475271, PP 1, PP 2, 1-Naphthyl PPI, Src II, ANA 12, PD 90780, Ki 8751, Ki 20227, ZM 306416, ZM 323881, AEE 788, GTP 14564, PD 180970, R 1530, SU 6668, and Toceranib.
[0043] In some embodiments, the test compound is a member of a library or collection of compounds or agents of interest (e.g, TKIs). The library of compounds may be composed of a plurality of chemical compounds and agents that may be assembled from a single or multiple sources. A library may include chemical compounds and agents such as chemically synthesized substances, products of biotechnological processes, naturally occurring substances or products resulting from combinatorial chemistry techniques or combinations thereof. In some embodiments, the library includes a plurality of clinical or preclinical small molecule drugs. [0044] In some embodiments, the test compound is a small molecule therapeutic agent.
[0045] In some embodiments, the test compound is a small molecule anticancer therapeutic agent.
[0046] In some embodiments, the test compound is CR8, CC-885, or indisulam.
[0047] CR8, also known as (7?)-CR8 and (2R)-2-[[9-(Methylethyl)-6-[[[4-(2- pyridinyl)phenyl]methyl]amino]-9H-purin-2-yl]amino]-l-butanol, has the following structure:
[0048] CC-885, also known as N-(3-chloro-4-methylphenyl)-N'-[[2-(2,6-dioxo-3- piperidinyl)-2,3-dihydro-l-oxo-lH-isoindol-5-yl]methyl]-urea, has the following structure:
[0049] Indisulam, also known as E7070 and N-(3-chloro-lH-indol-7-yl)benzene-l,4- disulfonamide, has the following structure:
(Indisulam).
[0050] The amount of the test compound is sufficient to cause a measurable change in phenotype in the cells. In some embodiments, the concentration of the test compound is between about 0.001 pM to about 100 pM. In some embodiments, the concentration of the test compound is between about 0.001 pM to about 30 pM. In some embodiments, the concentration of the test compound is between about 0.03 pM to about 30 pM. In some embodiments, the concentration of the test compound is between about 0.1 pM to about 3 pM. In some embodiments, the concentration of the test compound is between about 0.001 pM to about 0.1 pM.
Neddylation inhibitors
[0051] Neddylation is a post-translational protein modification that conjugates ubiquitin-like protein neuronal precursor cell-expressed developmentally down-regulated protein 8 (NEDD8). Similar to ubiquitination, neddylation is mediated by a cascade of three NEDD8 specific enzymes, an El activating enzyme, an E2 conjugating enzyme and one of the several E3 ligases. By altering its substrates’ conformation, stability, subcellular localization or binding affinity to DNA or proteins, neddylation regulates diverse cellular processes including the ubiquitin-proteasome system-mediated protein degradation, protein transcription, cell signaling, etc. (Kandala et al., Am J Cardiovasc Dis. 4(4): 140-158 (2014)). The best- characterized substrates of neddylation are cullin family members that function as indispensable components of multiunit cullin-RING E3 ubiquitin ligases (CRLs), whose activity requires cullin neddylation (Zhou et al., Molecular cancer 75:77(2019)). The small molecule MLN4924 (Pevonedistat), an adenosine sulfamate analog, potently and selectively
inhibits NEDD8 activating enzyme (NAE), the El enzyme (Soucy et al., Nature 45<S': 732-736 (2009).
[0052] Neddylation inhibitors effectively block cullin neddylation and inactivate CRLs. This in turn leads to accumulation of various CRL substrates, thus triggering multiple cellular responses, including cell cycle arrest, apoptosis, senescence and autophagy in a cell-type dependent manner (Zhou et al., Molecular cancer 18:77(2019)).
[0053] In some embodiments, the neddylation inhibitor is MLN4924, also known as ((lS,2S,4R)-4-(4-((lS)-2,3-Dihydro-lH-inden-l-ylamino)-7H-pyrrolo(2,3-d)pyrimidin-7-yl)- 2-hydroxycyclopentyl)methyl sulphamate, has the following structure:
[0054] In some embodiments, the neddylation inhibitor is a derivative or analog of MLN492. Analogs of MLN492 that may be useful as neddylation inhibitors are described in U.S. Patent 7,951,810 B2 and U.S. Patent Application Publication 2018/0086785 Al.
[0055] Another exemplary NAE or neddylation inhibitor, which may be used in the present invention, is TAS4464, which is also known as 4-amino-7-[(2R,3R,4S,5R)-3,4-dihydroxy-5- [(sulfamoylamino)methyl]oxolan-2-yl]-5-[2-(2-ethoxy-6-fluorophenyl)ethynyl]pyrrolo[2,3- d] pyrimidine and has the following structure:
[0056] In some embodiments, the neddylation inhibitor is a derivative or analog of TAS4464. Analogs of TAS4464 that may be useful as neddylation inhibitors are described in U.S. Patent 9,963,456 B2.
[0057] In some embodiments, the concentration of the neddylation inhibitor is below IpM. In some embodiments, the concentration of the neddylation inhibitor is between about 10 nM to about 999 nM.
[0058] In some embodiments, the concentration of the neddylation inhibitor is between about 10 nM to about 500 nM.
[0059] In some embodiments, the concentration of the neddylation inhibitor is about 100 nM. [0060] The cells may be pretreated with different concentrations of the neddylation inhibitor, typically for about 10 minutes to about 24 hours prior to treatment with the test compound in dose response assays.
[0061] The cells are incubated in the presence of the test compound and the neddylation inhibitor for an amount of time sufficient to allow for a measurable change in phenotype to occur. In some embodiments, the cells are incubated with neddylation inhibitor and test compound for at least 3 to 5 days. In some embodiments, the cells are incubated with neddylation inhibitor and test compound for at least 3 days. In some embodiments, incubation time may be shortened to 10 minutes - 24 hours to investigate protein levels or other more general readout, e.g, cellular differentiation and/or gene expression by RNA sequencing (RNAseq) or quantitative polymerase chain reaction (qPCR).
Cells
[0062] Mammalian cells suitable for use in the present methods are those that can be maintained or propagated in vitro and that naturally display, or that may be genetically modified to display, a phenotype (such as one described herein) the alteration of which is desired to be monitored for modulation. For example, cells may display a phenotype whose inhibition within the assay is to be determined. Mammalian cells which display the appropriate phenotypic characteristics (phenotype) may be identified or obtained by any convenient technique or source, including those known to the person of ordinary skill in the art.
[0063] Such cells may be stable cell lines, such as those obtainable from American Type Culture Collection (ATCC) or other cell repositories. Alternatively, the mammalian cells employed may be primary cells derived from a tissue or organ of an individual organism. In certain embodiments, the mammalian cells may be transiently transfected with genetic
constructs, such as those involved in the (desired) phenotype, for example a reporter gene. In other embodiments, the mammalian cells may be infected with a virus or bacterium which leads to an alteration in the phenotype. Of particular utility are human cells, murine cells, hamster cells, rate cells, primate cells, and cells from domestic mammals (such as ovine, bovine, equine, canine or feline cells). Yet other cells for employment in the present methods include mammalian cells derived from a cancer or tumor, or those associated with some other disease or abnormality of a mammal.
[0064] In some embodiments, the mammalian cells are primary cells. In some embodiments, the primary cells are isolated from a tissue or a biopsy sample.
[0065] In some embodiments, the mammalian cells are human cells.
[0066] In some embodiments, the human cells are myeloid cells, lymphoid cells, neural cells, epithelial cells, endothelial cells, stem or progenitor cells, hepatocytes, myoblasts, osteoblasts, osteoclasts, lymphocytes, keratinocytes, melanocytes, mesothelial cells, germ cells, muscle cells, fibroblasts, transformed cells, non-transformed cells, or cancer cells.
[0067] In some embodiments, the transformed cells are Cas9 stable cells.
[0068] In some embodiments, the cells are SNGM, P31FU, OVK18, PFEIFFER, ES-2, 0CIM1, K562, HEK293, or HEK293T cells.
Difference in phenotype in cells
[0069] In some embodiments, the phenotype to be monitored for change may be one having been specifically engineered for a given cell type. For example, a mammalian cell type may be genetically modified to result in a phenotype that is convenient or suitable for detection, such as a fluorescent protein/marker (such as GFP; eGFP), a luminescent protein/marker (such as luciferase) or a cell-surface marker than is detectable, e.g., with a labelled antibody, or intracellular proteins stained with labeled antibodies.
[0070] In some embodiments, the phenotype of the mammalian cell is any one of the following, namely: one associated with a cell signaling pathway, preferably an activated cell signaling pathway; and/or one selected from the list consisting of: luminescence, fluorescence, viability, senescence, differentiation, migration, invasion, chemotaxis, apoptosis, immunological anergy, surface marker expression, progress through the cell cycle, transcriptional activity, protein expression, glycosylation, resistance to infection, permeability and reporter-gene activity. In some embodiments, the phenotype is not death and/or reduced (or decreased) growth, such as a phenotype that is not rescue of a cell from cytokine
dependence, is not rescue from apoptosis (including neutrophil apoptosis/cell-death), is not induction of colony formation, or is not a rescue screen. For example, a screen for such a phenotype may be designed to detect a trait or characteristic of the cell that is not a rescue from cell death or an increase in growth of the mammalian cell.
[0071] In some embodiments, the phenotypic difference in the cells is increased protein abundance, decreased protein abundance, increased protein activity, decreased protein activity, increased gene expression, decreased gene expression, changed cellular proliferation, changed protein localization, or cell viability. In some embodiments, the phenotype difference is an increased or decreased mRNA transcript abundance of predetermined genes.
Analytical methods
[0072] Methods to assess phenotypic difference in cells are known in the art. One exemplary method is an immunoassay. In their simplest and most direct sense, immunoassays are binding assays involving binding between antibodies and antigen. Representative examples of immunoassays include enzyme linked immunosorbent assays (ELISAs), enzyme linked immunospot assay (ELISPOT), radioimmunoassays (RIA), radioimmune precipitation assays (RIP A), immunobead capture assays, Western blotting, dot blotting, gel-shift assays, Flow cytometry, immunohistochemistry, fluorescence microscopy, protein arrays, multiplexed bead arrays, magnetic capture, in vivo imaging, fluorescence resonance energy transfer (FRET), and fluorescence recovery/localization after photobleaching (FRAPZFLAP).The steps of various useful immunodetection methods have been described in the scientific literature, such as, e.g, Maggio et al., ed. (1980), Enzyme Immunoassay, CRC, Boca Raton, FL and Nakamura, et al., Enzyme Immunoassays: Heterogeneous and Homogeneous Systems in Handbook of Experimental Immunology 4th Edition, Vol. 1: Immunochemistry, Chapter 27, edited by D. M. Weir, Blackwell Scientific Publications, Oxford, (1986), each of which is incorporated herein by reference in its entirety and specifically for its teaching regarding immunodetection methods.
[0073] In some embodiments, the phenotypic difference in the cells is measured by flow cytometry, fluorescence-activated cell sorting (FACS), mass cytometry, and/or magnetic sorting, protein localization, cellular morphology, gene expression by RNA sequencing (RNAseq), Luminex® multiplex bead assay, quantitative polymerase chain reaction (qPCR), next generation sequencing (NGS), or immunoblotting.
[0074] In some embodiments, an increase in abundance of mRNA transcripts of predetermined genes is determined via a Luminex® multiplex bead assay, e.g., the L1000 assay. L1000 is a high-throughput gene expression assay that measures the mRNA transcript abundance of 978 "landmark" genes from human cells. Landmark genes were chosen to be expressed across lineage which would enable the prediction of expression of other genes not directly measured. The "L" in LI 000 refers to the Landmark genes measured in the assay. Measurements are made using the 500 colors of Luminex® beads such that two transcripts are identified by a single bead color. Expression of 80 control transcripts, chosen for their invariant expression across cell states, as well as the 978 Landmark genes are also measured. LI 000 is highly reproducible, comparable to RNA sequencing, and suitable for computational inference of the expression levels of 81% of non-measured transcripts. Subramanian et al., Cell 777(6 1437-1452.el7 (2017).
High throughput screenings
[0075] In some embodiments, the disclosed methods entail high throughput screening (HTS) for identifying cullin dependent small molecule degraders. The term “high-throughput” relates to an assay system for the rapid testing of a plurality of compounds in a short time. Thus, the assaying time per tested compound is minimized. The assay is preferably carried out in multiwell plates, e.g., 6-well plates, 12-well plates, 24-well plates, 96-well plates or 384-well plates, or 1536 plates, even more preferably in 96-well plates, 384-well or 1536 plates. Thus, the term “high-throughput” encompasses screening activity in which human intervention is minimized, and automation is maximized. For example, high-throughput screening involves automated pipetting, mixing, and heating. Alternatively, a high-throughput method is one in which for example hundreds of compounds can be screened per 24-hour period by a single individual operating a single suitable apparatus. “Automation” refers to a method or any one or more steps thereof that is fully or partly controlled and/or carried out by one or more technical devices, preferably pipetting robots. Methods for conducting a variety of different HTS assays, in particular HTS screening assays are known in the art. The skilled person knows how to conduct HTS methods by the methods used in the context of the present invention.
[0076] These and other aspects of the present invention will be further appreciated upon consideration of the following Examples, which are intended to illustrate certain particular embodiments of the invention but are not intended to limit its scope, as defined by the claims.
EXAMPLES
[0077] Example 1 : Inhibition of neddylation as a readout for functional degradation.
[0078] HEK293T cells were treated with various concentration of MG132 (proteasome inhibitor), MLN7243 (El ubiquitin-activating enzyme inhibitor) or MLN4924 (cullin neddylation inhibitor) for three days. Cell viability was assessed using CellTiter-Glo® Luminescent Cell Viability Assay (Promega®, G7572) on CLARIOstar® Plus Multi-mode Microplate Reader, MARS 3.4 (BMG LabTech). Cell viabilities were calculated relative to DMSO controls. Standard four-parameter log-logistic curves are fitted with the ‘dr4pl’ R package. The results are shown in FIG. 1A.
[0079] HEK293T-Cas9 cells that were also treated with 100 nM MLN4924 or DMSO in combination with compound CR8 for three days (n = 3). The results are shown in FIG. 2B.
[0080] The results illustrated in FIG. 1A-FIG. IB show that titration of MLN4924 up to 1 pM did not have a significant effect on cell viability (FIG. 1 A), but a 100 nM dose of MLN4924 was able to rescue CR8 induced toxicity (FIG. IB). These results confirmed that addition of low dose of MLN4924 can achieve a functional effect of reduced toxicity, when the toxicity is caused by neddylation dependent mechanism, such as degradation of Cyclin K and CDK13 by CR8.
[0081] Ubiquitin proteasome system (UPS) compounds, including the neddylation inhibitor MLN4924, are known to be toxic at high to mid concentrations (>1 pM). It was believed that low doses would not inhibit neddylation. Applicant surprisingly found that low dose MLN4924 (e.g, -100 nM) was nontoxic to cells and it potently blocked neddylation generating a sufficient window to observe phenotypic difference in the cells.
[0082] Example 2: MLN4924 rescue of CR8 (cyclin-dependent kinase (CDK) inhibitor) induced toxicity in multiple cell lines.
[0083] HEK293T-Cas9 cells were resuspended at 0.15 x 106 per ml and plated on a 384-well plate with 50 pL per well and MLN4924, MLN7243 or MG132 with or without CR8 serially diluted with D300e Digital Dispenser (Tecan).
[0084] A panel of SNGM, P31FUJ, OVK18, PFEIFFER, ES-2, 0CIM1 or HEK293T cells was resuspended at -0.15 x 106 per ml and plated on a 384-well plate with 50 pL per well. Cells were pretreated with MLN4924 at 0, 0.05, 0.1 or 0.2 pM concentration and treated with CR8 in dose response.
[0085] After three days of drug exposure, cell viability was assessed using CellTiter-Glo® Luminescent Cell Viability Assay (Promega®, G7572) on CLARIOstar® Plus Multi-mode Microplate Reader, MARS 3.4 (BMG LabTech). Cell viabilities were calculated relative to DMSO controls. The half-maximum effective concentration (ECso) values were derived from standard four-parameter log-logistic curves fitted with the ‘dr4pl’ R package.
[0086] The results illustrated in FIG.2A-FIG. 2H show that MLN4924 rescued CR8 induced toxicity in tested cell lines.
[0087] Example 3: MLN4924 rescue of CC885 (cereblon (CRBN) modulator) induced toxicity in multiple cell lines.
[0088] HEK293T-Cas9 cells were resuspended at 0.15 x 106 per ml and plated on a 384-well plate with 50 pL per well and MLN4924, MLN7243 or MG132 with or without CC-885 serially diluted with D300e Digital Dispenser (Tecan).
[0089] A panel of SNGM, P31FUJ, OVK18, PFEIFFER, ES-2, OCIM1 or HEK293T cells was resuspended at -0.15 x 106 per ml and plated on a 384-well plate with 50 pL per well. Cells were pretreated with MLN4924 at 0, 0.05, 0.1 or 0.2 pM concentration and treated with CC-885 in dose response.
[0090] After three days of drug exposure, cell viability was assessed using CellTiter-Glo® Luminescent Cell Viability Assay (Promega®, G7572) on CLARIOstar® Plus Multi-mode Microplate Reader, MARS 3.4 (BMG LabTech). Cell viabilities were calculated relative to DMSO controls. The half-maximum effective concentration (EC50) values were derived from standard four-parameter log-logistic curves fitted with the ‘dr4pl’ R package.
[0091] The results illustrated in FIG.3A-FIG. 3H show that MLN4924 rescues CC-885 induced toxicity in multiple cell lines.
[0092] Example 4: MLN4924 rescue of indisulam (CDK) induced toxicity in multiple cell lines.
[0093] HEK293T-Cas9 cells were resuspended at 0.15 x 106 per ml and plated on a 384-well plate with 50 pL per well and MLN4924, MLN7243 or MG132 with or without indisulam serially diluted with D300e Digital Dispenser (Tecan).
[0094] A panel of SNGM, P31FUJ, OVK18, PFEIFFER, ES-2, OCIM1 or HEK293T cells was resuspended at -0.15 x 106 per ml and plated on a 384-well plate with 50 pL per well. Cells were pretreated with MLN4924 at 0, 0.05, 0.1 or 0.2 pM concentration and treated with indisulam in dose response.
[0095] After three days of drug exposure, cell viability was assessed using CellTiter-Glo® Luminescent Cell Viability Assay (Promega®, G7572) on CLARIOstar® Plus Multi-mode Microplate Reader, MARS 3.4 (BMG LabTech). Cell viabilities were calculated relative to DMSO controls. The half-maximum effective concentration (EC50) values were derived from standard four-parameter log-logistic curves fitted with the ‘dr4pl’ R package.
[0096] The results illustrated in FIG.4A-FIG. 4H show that MLN4924 reduced indisulam induced toxicity in limited number of cell lines.
[0097] Established molecules CR8, CC-885, and indisulam, were confirmed as cullin dependent small molecule degraders using the method described herein. CR8 induces the degradation of Cyclin K via direct binding of CR8-CDK12-Cyclin K to damage specific DNA binding protein 1 (DDB1). CC-885 is a CRBN based degrader of G1 to S phase transition 1 (GSPT1). Indisulam is a DDB1 and CUL4 associated factor 15 (DCAF15) based degrader of splicing factor RNA binding motif protein 39 (RBM39). All of the three compounds caused dose dependent cell killing via proteasomal degradation. Treatment of CR8, CC-885 and indisulam induced dose dependent toxicity in a panel of cell lines, which was rescued by cotreatment with neddylation inhibitor MLN4924. The rescue of CR8 and CC-885 induced toxicity was near universal. However, co-treatment of neddylation inhibitor MLN4924 and indisulam of ES-2 or HEK293T cells failed to reduce the cytotoxic effect. This may have been due to differential expression of the DCAF15 E3 ligase or reduced sensitivity to RBM39 degradation.
[0098] FIG. 2A-FIG. 4H illustrate co-treatments of CC-885, CR8 and Indisulam with three doses of MLN4924 (50, 100, 200 nM). Increased doses of neddylation inhibitor decreased CellTiter-Glo® levels for different cell lines. Optimal concentration of MLN4924 may be characterized with increased toxicity.
[0099] All patent publications and non-patent publications are indicative of the level of skill of those skilled in the art to which this invention pertains. All these publications are herein incorporated by reference to the same extent as if each individual publication were specifically and individually indicated as being incorporated by reference.
[00100] Although the invention herein has been described with reference to particular embodiments, it is to be understood that these embodiments are merely illustrative of the principles and applications of the present invention. It is therefore to be understood that numerous modifications may be made to the illustrative embodiments and that other
arrangements may be devised without departing from the spirit and scope of the present invention as defined by the appended claims.
Claims
1. A method for identifying a cullin dependent small molecule degrader of a target protein in a cellular screen, comprising: incubating mammalian cells with a small molecule test compound in an amount sufficient to cause a change in phenotype in the cells, and a neddylation inhibitor at concentration that is nontoxic or of low toxicity to the cells; and measuring a difference in phenotype in the cells following incubation relative to a control that comprises the cells incubated with the test compound but not the neddylation inhibitor, wherein a difference in the phenotype in the cells relative to the control indicates that the test compound causes cullin dependent degradation of the target protein.
2. The method of claim 1, wherein the test compound is a natural product, a synthetic compound, or semi-synthetic compound.
3. The method of any one of claims 1-2, wherein the test compound is a small molecule kinase inhibitor.
4. The method of any one of claims 1-3, wherein the test compound is a small molecule tyrosine kinase inhibitor.
5. The method of any one of claims 1-2, wherein the test compound is a clinical or preclinical small molecule drug.
6. The method of any one of claims 1-2, wherein the test compound is a small molecule therapeutic agent.
7. The method of claim 6, wherein the test compound is a small molecule anti cancer therapeutic agent.
8. The method of claim 1, wherein the test compound is CR8, CC-885, or indisulam.
9. The method of any one of claims 1-8, wherein the concentration of the test compound is between about 0.001 pM to about 100 pM.
10. The method of any one of claims 1-9, wherein the neddylation inhibitor is MLN4924.
11. The method of any one of claims 1 -9, wherein the neddylation inhibitor is a MLN4924 analog.
12. The method of any one of claims 1-9, wherein the neddylation inhibitor is TAS4464.
13. The method of any one of claims 1-9, wherein the neddylation inhibitor is a TAS4464 analog.
14. The method of any one of claims 1-13, wherein the concentration of the neddylation inhibitor is below IpM.
15. The method of any one of claims 1-14, wherein the concentration of the neddylation inhibitor is between about 10 nM to about 500 nM.
16. The method of any one of claims 1-15, wherein the concentration of the neddylation inhibitor is about 100 nM.
17. The method of any one of claims 1-16, wherein the cells are pretreated with the neddylation inhibitor prior to incubation with the test compound.
18. The method of any one of claims 1-17, wherein the mammalian cells are human cells.
19. The method of any one of claims 1-18, wherein the cells are myeloid cells, lymphoid cells, neural cells, epithelial cells, endothelial cells, stem or progenitor cells, hepatocytes, myoblasts, osteoblasts, osteoclasts, lymphocytes, keratinocytes, melanocytes, mesothelial cells, germ cells, muscle cells, fibroblasts, transformed cells, non-transformed cells, or cancer cells.
20. The method of any one of claims 1-19, wherein the cells are primary cells.
21. The method of claim 20, wherein the primary cells are isolated from a tissue or a biopsy sample.
22. The method of any one of claims 1-19, wherein the cells are transformed cells.
23. The method of claim 22, wherein the transformed cells are Cas9 stable cells.
24. The method of any one of claims 1-19, wherein the cells are SNGM, P31FU, OVK18, PFEIFFER, ES-2, OCIM1, K562, HEK293, or HEK293T cells.
25. The method of claim 1-24, wherein the difference in the phenotype in the cells is cell viability.
26. The method of any one of claims 1-24, wherein the difference in the phenotype in the cells is increased protein abundance, decreased protein abundance, increased protein activity, decreased protein activity, increased gene expression, decreased gene expression, changed cellular proliferation, increased or decreased abundance of mRNA transcripts of landmark genes, or changed protein localization.
27. The method of any one of claims 1-26, wherein the difference in the phenotype in the cells is measured by flow cytometry, fluorescence-activated cell sorting (FACS), mass cytometry, and/or magnetic sorting, protein localization, cellular morphology, gene expression by RNA sequencing (RNAseq), Luminex multiplex bead assay, quantitative polymerase chain reaction (qPCR), next generation sequencing (NGS), or immunoblotting.
28. The method of claim 27, wherein the Luminex multiplex bead assay is L1000 assay.
Applications Claiming Priority (2)
| Application Number | Priority Date | Filing Date | Title |
|---|---|---|---|
| US202163140605P | 2021-01-22 | 2021-01-22 | |
| US63/140,605 | 2021-01-22 |
Publications (1)
| Publication Number | Publication Date |
|---|---|
| WO2022159687A1 true WO2022159687A1 (en) | 2022-07-28 |
Family
ID=82549805
Family Applications (1)
| Application Number | Title | Priority Date | Filing Date |
|---|---|---|---|
| PCT/US2022/013294 WO2022159687A1 (en) | 2021-01-22 | 2022-01-21 | Phenotypic assay to identify protein degraders |
Country Status (1)
| Country | Link |
|---|---|
| WO (1) | WO2022159687A1 (en) |
Citations (3)
| Publication number | Priority date | Publication date | Assignee | Title |
|---|---|---|---|---|
| US20180140578A1 (en) * | 2016-11-23 | 2018-05-24 | Board Of Regents Of The University Of Texas System | Methods and compositions for the diagnosis and selective treatment of cancer |
| US20190374657A1 (en) * | 2017-02-08 | 2019-12-12 | Dana-Farber Cancer Institute, Inc. | Tunable endogenous protein degradation with heterobifunctional compounds |
| US20200348285A1 (en) * | 2017-11-09 | 2020-11-05 | Dana-Farber Cancer Institute, Inc. | Methods to prevent teratogenicity of imid like molecules and imid based degraders/protacs |
-
2022
- 2022-01-21 WO PCT/US2022/013294 patent/WO2022159687A1/en active Application Filing
Patent Citations (3)
| Publication number | Priority date | Publication date | Assignee | Title |
|---|---|---|---|---|
| US20180140578A1 (en) * | 2016-11-23 | 2018-05-24 | Board Of Regents Of The University Of Texas System | Methods and compositions for the diagnosis and selective treatment of cancer |
| US20190374657A1 (en) * | 2017-02-08 | 2019-12-12 | Dana-Farber Cancer Institute, Inc. | Tunable endogenous protein degradation with heterobifunctional compounds |
| US20200348285A1 (en) * | 2017-11-09 | 2020-11-05 | Dana-Farber Cancer Institute, Inc. | Methods to prevent teratogenicity of imid like molecules and imid based degraders/protacs |
Non-Patent Citations (1)
| Title |
|---|
| SłABICKI MIKOłAJ; KOZICKA ZUZANNA; PETZOLD GEORG; LI YEN-DER; MANOJKUMAR MANISHA; BUNKER RICHARD D.; DONOVAN KATHERINE A: "The CDK inhibitor CR8 acts as a molecular glue degrader that depletes cyclin K", NATURE, NATURE PUBLISHING GROUP UK, LONDON, vol. 585, no. 7824, 3 June 2020 (2020-06-03), London, pages 293 - 297, XP037241512, ISSN: 0028-0836, DOI: 10.1038/s41586-020-2374-x * |
Similar Documents
| Publication | Publication Date | Title |
|---|---|---|
| Cardenas et al. | Rationally designed BCL6 inhibitors target activated B cell diffuse large B cell lymphoma | |
| EP2454598B1 (en) | Drug selection for gastric cancer therapy using antibody-based arrays | |
| Botbol et al. | Common γ-chain cytokine signaling is required for macroautophagy induction during CD4+ T-cell activation | |
| Argüello et al. | SunRiSE–measuring translation elongation at single-cell resolution by means of flow cytometry | |
| JP2016515383A (en) | Dendritic cell response gene expression, composition and method of use thereof | |
| JP2010533842A (en) | Drug selection for the treatment of lung cancer using antibody arrays | |
| US8765713B2 (en) | Method for determination of sensitivity to anti-cancer agent | |
| EP2656077A1 (en) | Drug selection for malignant cancer therapy using antibody-based arrays | |
| TW201726169A (en) | Pd-1 signal inhibitor combination therapy | |
| JP2014532184A (en) | Multiplexed kinase inhibitor beads and uses thereof | |
| JP7369524B2 (en) | Method for evaluating the immunogenicity of a test substance | |
| EP3906942A1 (en) | Prediction and/or determination marker for effectiveness of treatment with drug containing pd-1 signal inhibitor | |
| KR20190103154A (en) | Combination of BRD4 Inhibitors and Antifolates for the Treatment of Cancer | |
| Barret et al. | Inhibition of nucleotide excision repair and sensitisation of cells to DNA cross-linking anticancer drugs by F 11782, a novel fluorinated epipodophylloid | |
| US20150352086A1 (en) | Heat shock protein (hsp) inhibition and monitoring effectiveness thereof | |
| Ippolito et al. | Aneuploidy-driven genome instability triggers resistance to chemotherapy | |
| Vladychenskaya et al. | Rat lymphocytes express NMDA receptors that take part in regulation of cytokine production | |
| WO2022159687A1 (en) | Phenotypic assay to identify protein degraders | |
| US9459254B2 (en) | Method for determining sensitivity to an anticancer agent | |
| KR20220111319A (en) | Interleukin-4-induced gene 1 (IL4I1) and respective metabolites as biomarkers for cancer | |
| Heuser et al. | Cytotoxicity determination without photochemical artifacts | |
| EP3421994B1 (en) | Method for predicting and monitoring clinical response to immunomodulatory therapy | |
| Wang et al. | Addressing soluble target interference in the development of a functional assay for the detection of neutralizing antibodies against a BCMA-CD3 bispecific antibody | |
| Chen et al. | Identification of mediators of T-cell receptor signaling via the screening of chemical inhibitor libraries | |
| US20240241128A1 (en) | Perivascular accumulation of immune cells in the diagnosis and treatment of cancer |
Legal Events
| Date | Code | Title | Description |
|---|---|---|---|
| 121 | Ep: the epo has been informed by wipo that ep was designated in this application |
Ref document number: 22743236 Country of ref document: EP Kind code of ref document: A1 |
|
| NENP | Non-entry into the national phase |
Ref country code: DE |
|
| 122 | Ep: pct application non-entry in european phase |
Ref document number: 22743236 Country of ref document: EP Kind code of ref document: A1 |