NOVEL PROTEINS AND NUCLEIC ACIDS ENCODING SAME
BACKGROUND OF THE INVENTION
The invention generally relates to nucleic acids and polypeptides. More particularly, the invention relates to nucleic acids encoding novel G-protein coupled receptor (GPCR) polypeptides, as well as vectors, host cells, antibodies, and recombinant methods for producing these nucleic acids and polypeptides.
SUMMARY OF THE INVENTION
The invention is based in part upon the discovery of nucleic acid sequences encoding novel polypeptides. The novel nucleic acids and polypeptides are referred to herein as GPCRX, or GPCR1, GPCR2, GPCR3, GPCR4, GPCR5, GPCR6, GPCR7 and GPCR8 nucleic acids and polypeptides. These nucleic acids and polypeptides, as well as derivatives,, homologs, analogs and fragments thereof, will hereinafter be collectively designated as "GPCRX" nucleic acid or polypeptide sequences.
In one aspect, the invention provides an isolated GPCRX nucleic acid molecule encoding a GPCRX polypeptide that includes a nucleic acid sequence that has identity to the nucleic acids disclosed in SEQ ID NOS:l, 3, 5, 7, 9, 11, 13, 15, 17, 19, 21, 23, 25, 27, 29, 31, 33, 35, 37, 39, 41 and 43. hi some embodiments, the GPCRX nucleic acid molecule will hybridize under stringent conditions to a nucleic acid sequence complementary to a nucleic acid molecule that includes a protein-coding sequence of a GPCRX nucleic acid sequence. The invention also includes an isolated nucleic acid that encodes a GPCRX polypeptide, or a fragment, homolog, analog or derivative thereof. For example, the nucleic acid can encode a polypeptide at least 80% identical to a polypeptide comprising the amino acid sequences of SEQ ID NOS:2, 4, 6, 8, 10, 12, 14, 16, 18, 20, 22, 24, 26, 28, 30, 32, 34, 36, 38, 40, 42 and 44. The nucleic acid can be, for example, a genomic DNA fragment or a cDNA molecule that includes the nucleic acid sequence of any of SEQ ID NOS: 1, 3, 5, 7, 9, 11, 13, 15, 17, 19, 21, 23, 25, 27, 29, 31, 33, 35, 37, 39, 41 and 43.
Also included in the invention is an oligonucleotide, e.g., an oligonucleotid which includes at least 6 contiguous nucleotides of a GPCRX nucleic acid (e.g., SEQ ID NOS: 1, 3, 5,
7, 9, 11, 13, 15, 17, 19, 21, 23, 25, 27, 29, 31, 33, 35, 37, 39, 41 and 43) or a complement of said oligonucleotide.
Also included in the invention are substantially purified GPCRX polypeptides (SEQ ID NOS: 2, 4, 6, 8, 10, 12, 14, 16, 18, 20, 22, 24, 26, 28, 30, 32, 34, 36, 38, 40, 42 and 44). In certain embodiments, the GPCRX polypeptides include an amino acid sequence that is substantially identical to the amino acid sequence of a human GPCRX polypeptide.
The invention also features antibodies that immunoselectively bind to GPCRX polypeptides, or fragments, homologs, analogs or derivatives thereof.
In another aspect, the invention includes pharmaceutical compositions that include therapeutically- or prophylactically-effective amounts of a therapeutic and a pharmaceutically- acceptable carrier. The therapeutic can be, e.g., a GPCRX nucleic acid, a GPCRX polypeptide, or an antibody specific for a GPCRX polypeptide. In a further aspect, the invention includes, in one or more containers, a therapeutically- or prophylactically-effective amount of this pharmaceutical composition. In a further aspect, the invention includes a method of producing a polypeptide by culturing a cell that includes a GPCRX nucleic acid, under conditions allowing for expression of the GPCRX polypeptide encoded by the DNA. If desired, the GPCRX polypeptide can then be recovered.
In another aspect, the invention includes a method of detecting the presence of a GPCRX polypeptide in a sample. In the method, a sample is contacted with a compound that selectively binds to the polypeptide under conditions allowing for formation of a complex between the polypeptide and the compound. The complex is detected, if present, thereby identifying the GPCRX polypeptide within the sample.
The invention also includes methods to identify specific cell or tissue types based on their expression of a GPCRX.
Also included in the invention is a method of detecting the presence of a GPCRX nucleic acid molecule in a sample by contacting the sample with a GPCRX nucleic acid probe or primer, and detecting whether the nucleic acid probe or primer bound to a GPCRX nucleic acid molecule in the sample. In a further aspect, the invention provides a method for modulating the activity of a
GPCRX polypeptide by contacting a cell sample that includes the GPCRX polypeptide with a compound that binds to the GPCRX polypeptide in an amount sufficient to modulate the activity of said polypeptide. The compound can be, e.g., a small molecule, such as a nucleic
acid, peptide, polypeptide, peptidomimetic, carbohydrate, lipid or other organic (carbon containing) or inorganic molecule, as further described herein.
Also within the scope of the invention is the use of a therapeutic in the manufacture of a medicament for treating or preventing disorders or syndromes including, e.g., diabetes, metabolic disturbances associated with obesity, the metabolic syndrome X, anorexia, wasting disorders associated with chronic diseases, metabolic disorders, diabetes, obesity, infectious disease, anorexia, cancer-associated cachexia, cancer, neurodegenerative disorders, Alzheimer's Disease, Parkinson's Disorder, immune disorders, and hematopoietic disorders, or other disorders related to cell signal processing and metabolic pathway modulation. The therapeutic can be, e.g., a GPCRX nucleic acid, a GPCRX polypeptide, or a GPCRX-specific antibody, or biologically-active derivatives or fragments thereof.
For example, the compositions of the present invention will have efficacy for treatment of patients suffering from: developmental diseases, MHCII and III diseases (immune diseases), taste and scent detectability Disorders, Burkitt's lymphoma, corticoneurogenic disease, signal transduction pathway disorders, Retinal diseases including those involving photoreception, Cell growth rate disorders; cell shape disorders, feeding disorders; control of feeding; potential obesity due to over-eating; potential disorders due to starvation (lack of appetite), noninsulin- dependent diabetes mellitus (NTDDM1), bacterial, fungal, protozoal and viral infections (particularly infections caused by HIV-1 or HTV-2), pain, cancer (including but not limited to neoplasm; adenocarcinoma; lymphoma; prostate cancer; uterus cancer), anorexia, bulimia, asthma, Parkinson's disease, acute heart failure, hypotension, hypertension, urinary retention, osteoporosis, Crohn's disease; multiple sclerosis; Albright Hereditary Ostoeodystrophy, angina pectoris, myocardial infarction, ulcers, allergies, benign prostatic hypertrophy, and psychotic and neurological disorders, including anxiety, schizophrenia, manic depression, delirium, dementia, severe mental retardation. Dentatorubro-pallidoluysian atrophy (DRPLA)
Hypophosphatemic rickets, autosomal dominant (2) Acrocallosal syndrome and dyskinesias, such as Huntington's disease or Gilles de la Tourette syndrome and/or other pathologies and disorders of the like.
The polypeptides can be used as immunogens to produce antibodies specific for the invention, and as vaccines. They can also be used to screen for potential agonist and antagonist compounds. For example, a cDNA encoding GPCRX may be useful in gene therapy, and GPCRX may be useful when administered to a subject in need thereof. By way of nonlimiting example, the compositions of the present invention will have efficacy for treatment of patients suffering from bacterial, fungal, protozoal and viral infections (particularly infections caused by
HIV-1 or HIV-2), pain, cancer (including but not limited to Neoplasm; adenocarcinoma; lymphoma; prostate cancer; uterus cancer), anorexia, bulimia, asthma, Parkinson's disease, acute heart failure, hypotension, hypertension, urinary retention, osteoporosis, Crohn's disease; multiple sclerosis; and Treatment of Albright Hereditary Ostoeodystrophy, angina pectoris, myocardial infarction, ulcers, allergies, benign prostatic hypertrophy, and psychotic and neurological disorders, including anxiety, schizophrenia, manic depression, delirium, dementia, severe mental retardation and dyskinesias, such as Huntington's disease or Gilles de la Tourette syndrome and/or other pathologies and disorders.
The invention further includes a method for screening for a modulator of disorders or syndromes including, e.g., diabetes, metabolic disturbances associated with obesity, the metabolic syndrome X, anorexia, wasting disorders associated with chronic diseases, metabolic disorders, diabetes, obesity, infectious disease, anorexia, cancer-associated cachexia, cancer, neurodegenerative disorders, Alzheimer's Disease, Parkinson's Disorder, immune disorders, and hematopoietic disorders or other disorders related to cell signal processing and metabolic pathway modulation. The method includes contacting a test compound with a GPCRX polypeptide and determining if the test compound binds to said GPCRX polypeptide. Binding of the test compound to the GPCRX polypeptide indicates the test compound is a modulator of activity, or of latency or predisposition to the aforementioned disorders or syndromes.
Also within the scope of the invention is a method for screening for a modulator of activity, or of latency or predisposition to an disorders or syndromes including, e.g., diabetes, metabolic disturbances associated with obesity, the metabolic syndrome X, anorexia, wasting disorders associated with chronic diseases, metabolic disorders, diabetes, obesity, infectious disease, anorexia, cancer-associated cachexia, cancer, neurodegenerative disorders, Alzheimer's Disease, Parkinson's Disorder, immune disorders, and hematopoietic disorders or other disorders related to cell signal processing and metabolic pathway modulation by administering a test compound to a test animal at increased risk for the aforementioned disorders or syndromes. The test animal expresses a recombinant polypeptide encoded by a GPCRX nucleic acid. Expression or activity of GPCRX polypeptide is then measured in the test animal, as is expression or activity of the protein in a control animal which recombinantly-expresses GPCRX polypeptide and is not at increased risk for the disorder or syndrome. Next, the expression of GPCRX polypeptide in both the test animal and the control animal is compared. A change in the activity of GPCRX polypeptide in the test animal relative to the control animal indicates the test compound is a modulator of latency of the disorder or syndrome.
In yet another aspect, the invention includes a method for determining the presence of or predisposition to a disease associated with altered levels of a GPCRX polypeptide, a GPCRX nucleic acid, or both, in a subject (e.g., a human subject). The method includes measuring the amount of the GPCRX polypeptide in a test sample from the subject and comparing the amount of the polypeptide in the test sample to the amount of the GPCRX polypeptide present in a control sample. An alteration in the level of the GPCRX polypeptide in the test sample as compared to the control sample indicates the presence of or predisposition to a disease in the subject. Preferably, the predisposition includes, e.g., diabetes, metabolic disturbances associated with obesity, the metabolic syndrome X, anorexia, wasting disorders associated with chronic diseases, metabolic disorders, diabetes, obesity, infectious disease, anorexia, cancer- associated cachexia, cancer, neurodegenerative disorders, Alzheimer's Disease, Parkinson's Disorder, immune disorders, and hematopoietic disorders. Also, the expression levels of the new polypeptides of the invention can be used in a method to screen for various cancers as well as to determine the stage of cancers. In a further aspect, the invention includes a method of treating or preventing a pathological condition associated with a disorder in a mammal by administering to the subject a GPCRX polypeptide, a GPCRX nucleic acid, or a GPCRX-specific antibody to a subject (e.g., a human subject), in an amount sufficient to alleviate or prevent the pathological condition. In preferred embodiments, the disorder, includes, e.g., diabetes, metabolic disturbances associated with obesity, the metabolic syndrome X, anorexia, wasting disorders associated with chronic diseases, metabolic disorders, diabetes, obesity, infectious disease, anorexia, cancer-associated cachexia, cancer, neurodegenerative disorders, Alzheimer's Disease, Parkinson's Disorder, immune disorders, and hematopoietic disorders.
In yet another aspect, the invention can be used in a method to identity the cellular receptors and downstream effectors of the invention by any one of a number of techniques commonly employed in the art. These include but are not limited to the two-hybrid system, affinity purification, co-precipitation with antibodies or other specific-interacting molecules. Unless otherwise defined, all technical and scientific terms used herein have the same meaning as commonly understood by one of ordinary skill in the art to which this invention belongs. Although methods and materials similar or equivalent to those described herein can be used in the practice or testing of the present invention, suitable methods and materials are described below. All publications, patent applications, patents, and other references mentioned herein are incorporated by reference in their entirety. In the case of conflict, the present
specification, including definitions, will control. In addition, the materials, methods, and examples are illustrative only and not intended to be limiting.
Other features and advantages of the invention will be apparent from the following detailed description and claims.
DETAILED DESCRIPTION OF THE INVENTION
The invention is based, in part, upon the discovery of novel nucleic acid sequences that encode novel polypeptides. The novel nucleic acids and their encoded polypeptides are referred to individually as GPCR1, GPCR2, GPCR3, GPCR4, GPCR5, GPCR6, GPCR7 and GPCR8. The nucleic acids, and their encoded polypeptides, are collectively designated herein as "GPCRX".
The novel GPCRX nucleic acids of the invention include the nucleic acids whose sequences are provided in Tables 1 A, ID, 2A, 2D, 2G, 3A, 3D, 4A, 5A, 5E, 5H, 5L, 6A, 6D, 6G, 6K, 7A, 8A, 8D, 8H, 8L and 8P, inclusive ("Tables 1A- 8P"), or a fragment, derivative, analog or homolog thereof. The novel GPCRX proteins of the invention include the protein fragments whose sequences are provided in Tables IB, IE, 2B, 2E, 2H, 3B, 3E, 4B, 5B, 5F, 51, 5M, 6B, 6E, 6H, 6L, 7B, 8B, 8E, 81, 8M and 8Q, inclusive ("Tables IB - 8Q"). The individual GPCRX nucleic acids and proteins are described below. Within the scope of this invention is a method of using these nucleic acids and peptides in the treatment or prevention of a disorder related to cell signaling or metabolic pathway modulation. G-Protein Coupled Receptor proteins (GPCRs) have been identified as a large family of
G protein-coupled receptors in a number of species. These receptors share a seven transmembrane domain structure with many neurotransmitter and hormone receptors, and are likely to underlie the recognition and G-protein-mediated transduction of various signals. Human GPCR generally do not contain introns and belong to four different gene subfamilies, displaying great sequence variability. These genes are dominantly expressed in olfactory epithelium. See, e.g., Ben-Arie et al., Hum. Mol. Genet. 1994 3:229-235; and, Online Mendelian Inheritance in Man (OMIM) entry # 164342 (http://www.ncbi.nlm.nih.gov/entrez/ dispomim.cgi?).
The olfactory receptor (OR) gene family constitutes one of the largest GPCR multigene families and is distributed among many chromosomal sites in the human genome. See
Rouquier et al., Hum. Mol. Genet. 7(9): 1337-45 (1998); Malnic et al., Cell 96:713-23 (1999). Olfactory receptors constitute the largest family among G protein-coupled receptors, with up to
1000 members expected. See Vanderhaeghen et al, Genomics 39(3):239-46 (1997); Xie et al., Mamm. Genome l l(12):1070-78 (2000); Issel-Tarver et al., Proc. Natl. Acad. Sci. USA 93(20):10897-902 (1996). The recognition of odorants by olfactory receptors is the first stage in odor discrimination. See Krautwurst et al, Cell 95(7):917-26 (1998); Buck et al., Cell 65(l):175-87 (1991). Many ORs share some characteristic sequence motifs and have a central variable region corresponding to a putative ligand binding site. See Issel-Tarver et al., Proc. Natl. Acad. Sci. USA 93:10897-902 (1996).
Other examples of seven membrane spanning proteins that are related to GPCRs are chemoreceptors. See Thomas et al., Gene 178(1-2): 1-5 (1996). Chemoreceptors have been identified in taste, olfactory, and male reproductive tissues. See id.; Walensky et al., J. Biol.
Chem. 273(16):9378-87 (1998); Parmentier et al., Nature 355(6359):453-55 (1992); Asai et al., Biochem. Biophys. Res. Commun. 221(2):240-47 (1996).
GPCR1
GPCR1 includes two novel G-protein coupled receptor ("GPCR") proteins disclosed below. The disclosed proteins have been named GPCRla, GPCRlb, and are related to olfactory receptors.
GPCRla
A disclosed GPCRla nucleic acid of 964 nucleotides (also referred to as AP001884_A) is shown in Table 1A. The disclosed GPCRla open reading frame ("ORF") begins at the ATG initiation codon at nucleotides 39-41, shown in bold in Table 1 A. The disclosed GPCRla ORF terminates at a TGA codon at nucleotides 933-935. As shown in Table 1 A, putative untranslated regions 5' to the start codon and 3' to the stop codon are underlined, and the start and stop codons are in bold letters.
Table 1A. GPCRla nucleotide sequence (SEQ ID NO:l).
CTTTTCACA6ACACCCAAGAGTTGATTCCTCCCCAG6aATGA6ΑΑATCACACAATG6TGACTGAATTCATCCTTCTG GGAATCCCTGAGACAGAGGGCCTAGAGACAGCCCTTTTATTCCTGTTCTCCTCATTTTATTTATGCACCCTCTTGGG AAACGTGCTTATCCTTACAGCTATCATCTCCTCCACTCGACTTCACACTCCTATGTATTTTTTCTTGGGAAACCTCT CCATCTTTGACCTGGGTTTCTCTTCAACGACTGTTCCCAAGATGTTGTTCTACCTTTCGGGGAACAGCCATGCTATC TCGTATGCAGGCTGCGTGTCCCAGCTTTTCTTCTACCATTTCCTAGGCTGTACTGAGTGTTTCCTCTACACAGTGAT GGCCTGTGACCGCTTTGTTGCCATATGTTTTCCTTTGAGATACACGGTCATCATGAACCACAGGGTGTGCTTTATGT TGGCCACGGGGACCTGGATGATTGGCTGTGTCCATGCCATGATCCTAACTCCCCTCACCTTCCAGTTACCTTACTGT GGCCCTAACAAGGTGGGCTATTACTTCTGTGATATTCCTGCAGTGTTACCTCTAGCCTGTAAGGACACATCCTTAGC CCAGAGGGTAGGTTTTACAAATGTTGGTCTTTTGTCTCTCATTTGCTTTTTTCTCATCCTTGTTTCCTATACTTGCA TTGGGATTTCCATATCAAAAATCCGCTCAGCAGAGGGCAGGCAGCGGGCCTTCTCCACCTGCAGCGCTCACCTCACT GCAATCCTTTGTGCTTATGGGCCAGTCATCGTTATCTATCTACAACCCAATCCCAGTGCCTTGCTTGGTTCCATAAT TCAGATATTGAATAATCTGGTAACCCCAATGTTGAATCCACTAATCTATAGCCTTAGGAATAAGGATGTAAAATCAG ATCAGCCCTGAGGAATGTATTTCCCAAGAAAAGCTTTGCT
A disclosed encoded GPCRla protein has 298 amino acid residues, referred to as the GPCRla protein and is presented in Table IB using the one-letter amino acid code. The Psort profile for GPCRla predicts that this sequence has a signal peptide and is likely to be localized at the plasma membrane with a certainty of 0.6000. The most likely cleavage site for a GPCRla peptide is between amino acids 46 and 47, at: LTA-IL, based on the SignalP result. GPCRla has a molecular weight of 33106.8.
Table IB. Encoded GPCRla protein sequence (SEQ ID NO:2).
MRNHTMVTEFILLGIPETEGLETALLFLFSSFY CTLLGNV ILTAIISSTRLHTPMYFF GNLSIFDLGFSSTTVP KMLFYLSGNSHAISYAGCVSQLFFYHFLGCTECFLYTVMACDRFVAICFPLRYTVIMNHRVCFMLATGT MIGCVHA MILTPLTFQLPYCGPNKVGYYFCDIPAVLPLACKDTSLAQRVGFTNVGL SLICFFLILVSYTCIGISISKIRSAEG RQRAFSTCSAHLTAILCAYGPVIVIYLQPNPSAL GSIIQILNNLVTPM NPLIYSLRNKDVKSDQP
The amino acid sequence of GPCRla had high homology to other proteins as shown in Table IC.
Table IC. BLASTX results for GPCRla
Smallest
Sum
Reading High Prob
Sequences producing High-scoring Segment Pairs: Frame Score P(N) N patp:AAY90873 Human G protein-coupled receptor GTAR14-. .. +3 751 1.3e-73 1 patp:AAY90872 Human G protein-coupled receptor GTAR14-. .. +3 737 3.8e-72 1 patp:AAY90874 Human G protein-coupled receptor GTAR14-. .. +3 732 1.3e-71 1 patp:AAB43266 Human ORFX ORF3030 polypeptide sequence . .. +3 658 9.0e-64 1 patp:AAR27876 Odorant receptor clone 115 - Rattus ratt. .. +3 635 2.5e-61 1 patp:AAY92364 G protein-coupled receptor protein 4 - H. .. +3 632 5.1e-61 1 patp:AAR27874 Odorant receptor clone 19 - Rattus rattu. .. +3 628 1.4e-60 1
GPCRlb
In the present invention, the target sequence identified previously, Accession Number AP001884_A, was subjected to the exon linking process to confirm the sequence. PCR primers were designed by starting at the most upstream sequence available, for the forward primer, and at the most downstream sequence available for the reverse primer. In each case, the sequence was examined, walking inward from the respective termini toward the coding sequence, until a suitable sequence that is either unique or highly selective was encountered, or, in the case of the reverse primer, until the stop codon was reached. Such primers were designed based on in silico predictions for the full length cDNA, part (one or more exons) of the DNA or protein sequence of the target sequence, or by translated homology of the predicted exons to closely related human sequences sequences from other species. These primers were then employed in
PCR amplification based on the following pool of human cDNAs: adrenal gland, bone marrow, brain - amygdala, brain - cerebellum, brain - hippocampus, brain - substantia nigra, brain - thalamus, brain -whole, fetal brain, fetal kidney, fetal liver, fetal lung, heart, kidney, lymphoma - Raji, mammary gland, pancreas, pituitary gland, placenta, prostate, salivary gland, skeletal muscle, small intestine, spinal cord, spleen, stomach, testis, thyroid, trachea, uterus. Usually the resulting amplicons were gel purified, cloned and sequenced to high redundancy. The resulting sequences from all clones were assembled with themselves, with other fragments in CuraGen Coφoration' s database and with public ESTs. Fragments and ESTs were included as components for an assembly when the extent of their identity with another component of the assembly was at least 95% over 50 bp.. In addition, sequence traces were evaluated manually and edited for corrections if appropriate. These procedures provide the sequence reported below, which is designated Accession Number AP001884_A_dal.
A disclosed GPCRlb (also referred to as AP001884_A_dal) nucleic acid of 906 nucleotides is shown in Table ID. An open reading frame was identified beginning with an ATG initiation codon at nucleotides 5-7 and ending with a TGA codon at nucleotides 899-901. A putative untranslated region upstream from the initiation codon and downstream from the termination codon is underlined in Table ID, and the start and stop codons are in bold letters.
Table ID. GPCRlb Nucleic acid sequence (SEQ ID NO:3).
AGGAATGAGAAATCACACAATGGTGACTGAATTCATCCTTCTGGGAATCCCTGAGACAGAGGGCCTAGAGACAGCCCT TTTATTCCTGTTCTCCTCATTTTATTTATGCACCCTCTTGGGAAACGTGCTTATCCTTACAGCTATCATCTCCTCCAG TCGACTTCACACTCCTATGTATTTTTTCTTGGGAAACCTCTCCATCTTTGACCTGGGTTTCTCTTCAACGACTGCTCC CAAGATGTTGTTCTACCTTTCGGGGAACAGCCATGCTATCTCGTATGCAGGCTGCGTGTCCCAGCTTTTCTTCTACCA TTTCCTAGGCTGTACTGAGTGTTTCCTCTACACAGTGATGGCCTGTGACCGCTTTGTTGCCATATGTTTTCCTTTGAG ATACACGGTCATCATGAACCACAGGGTGTGCTTTATGTTGGCCACGGGGACCTGGATGATTGGCTGTGTCCATGCCAT GATCCTAACTCCCCTCACCTTCCAGTTACCTTACTGTGGCCCTAACAAGGTGGGCTATTACTTCTGTGATATTCCTGC AGTGTTACCTCTAGCCTGTAAGGACACATCCTTAGCCCAGAGGGTAGGTTTTACAAATGTTGGTCTTTTGTCTCTCAT TTGCTTTTTTCTCATCCTTGTTTCCTATACTTGCATTGGGATTTCCATATCAAAAATCCGCTCAGCAGAGGGCAGGCA GCGGGCCTTCTCCACCTGCAGCGCTCACCTCACTGCAATCCTTTGTGCTTATGGGCCAGTCATCGTTATCTATCTACA ACCCAATCCCAGTGCCTTGCTTGGTTCCATAATTCAGATATTGAATAATCTGGTAACCCCAATGTTGAATCCACTAAT CTATAGCCTTAGGAATAAGGATGTAAAATCAGATCAGTCCTGAGGAAT
The disclosed encoded GPCRlb protein has 298 amino acid residues, referred to as the
GPCRlb protein, is presented in Table IE using the one-letter amino acid code. The Psort profile for GPCRlb predicts that this sequence has a signal peptide and is likely to be localized at the plasma membrane with a certainty of 0.6000. The most likely cleavage site for a GPCRlb peptide is between amino acids 46 and 47, at: LTA-IL, based on the SignalP result. GPCRlb has a molecular weight of 33068.7.
Table IE. Encoded GPCRlb protein sequence (SEQ ID NO:4).
MRNHTMVTEFILLGIPETEGLETALLFLFSSFYLCTLLGNVLILTAIISSTRLHTPMYFFLGNLSIFDLGFSSTTAP KMLFYLSGNSHAISYAGCVSQLFFYHFLGCTECFLYTVMACDRFVAICFPLRYTVIM HRVCFMLATGTWMIGCVHA MILTPLTFQLPYCGPNKVGYYFCDIPAVLPLACKDTSLAQRVGFTNVGLLSLICFFLILVSYTCIGISISKIRSAEG RQRAFSTCSAHLTAILCAYGPVIVIYLQPNPSALLGSIIQILNNLVTPMLNPLIYSLRNKDVKSDQS
BLASTP (Non-Redundant Composite database) analysis of the best hits for alignments with GPCRlb are listed in Table IF.
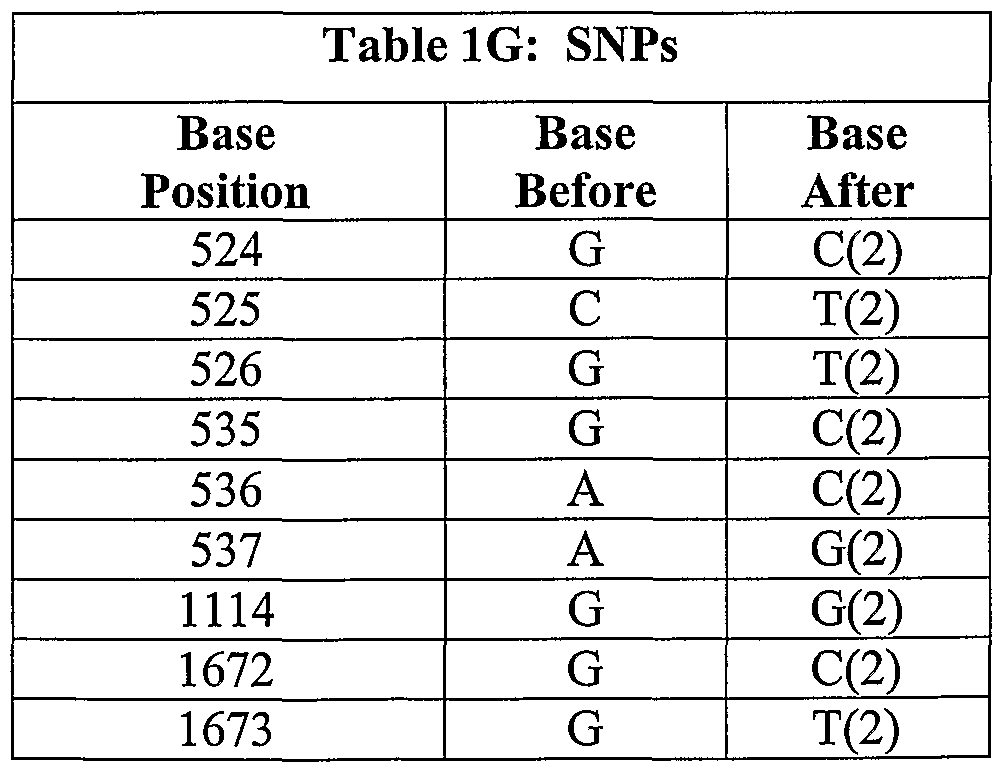
Possible SNPs found for GPCRlb are listed in Table 1G.
GPCRla and lb are related to each other as shown in the alignment listed in Table
1H.
Table 1H: Clustal W of GPCR1 Variants
10 20 30 40 50 60
I.. • I. .1. ■ I . .1 GPCR 1A Protein Sequence (AP00 RNHTMVTEFILLGIPETEGLETAL FLFSSFYLCTLLGNVLILTAIISSTRLHTPMYF GPCR IB Protein Sequence (AP00 [MRNHTMVTEFIL GIPETEGLETA F FSSFYLCTL GNV ILTAIISSTRLHTPMYFF
70 80 90 100 110 120
. . I . . . I GPCR 1A Protein Sequence (AP00 LGNLSIFDLGFSSTTVPKMLFYLSGNSHAISYAGCVSQLFFYHFLGCTECFLYTVMACDR
GPCR IB Protein Sequence (AP00 jGNLSIFDLGFSSTTWPKMLFYLSGNSHAISYAGCV
130 140 150 160 170 180 |.-..|....|....|....]....|....[....!.... I ....I GPCR 1A Protein Sequence (AP00 eVAICFPLRYTVIMNHRVCFM ATGTW IGCVHAM uiruj.- u_r ∞UΛIU I ij. ωiJ GPCR IB Protein Sequence (AP00 FVAICFPLRYTVIMNHRVCFM ATGTW IGCVHAMILTPLTFQ PYCGPNKVGYYFCDI]
190 200 210 220 230 240
GPCR 1A Protein Sequence (AP00
GPCR IB Protein Sequence (AP00 VLPLACKDTSLAORVGFTNVGLLSLICFFLILVSYTCIGISISKIRSAEGRORAFSTC
250 260 270 280 290 ■■■■l....|....l....|....|....|....|....|....l....l....|.
GPCR 1A Protein Sequence (AP00 IHLTAILCAYGPVIVIYLQPNPSAL GSIIQILNNLVTPMLNPLIYSLRNKDVKSDQ GPCR IB Protein Sequence (AP0O IHLTAI CAYGPVIVIYLQPNPSA LGSIIQILNNLVTPMLNPLIYS RNKDVKSDQ!
It was also found that GPCRla had homology to the amino acid sequences shown in the BLASTP data listed in Table II.
The homology of these sequences is shown graphically in the ClustalW analysis shown in Table 1 J.
Table U. ClustalW Analysis of GPCRla
1) Novel GPCRla (SEQ ID NO:2)
2) gi|11692587|gb|AAG39890.1|AF282305_l odorant receptor M37 [Mus musculus] (SEQ ID NO:45)
3). gi|11692585|gb|AAG39889.1|AF282304_l odorant receptor M36 [Mus musculus] (SEQ ID
NO:46)
4) gi|11692583|gb|AAG39888.1|AF282303_l odorant receptor M34 [Mus musculus] (SEQ ID NO:47)
5) gi|11692581|gb|AAG39887.1|AF282302_l odorant receptor M32 [Mus musculus] (SEQ ID NO:48)
6) gi|11692579|gb|AAG39886.1|AF282301_l odorant receptor M30 [Mus musculus] (SEQ ID NO:49)
310
I
GPCR 1A gilll692587| FLKRCLS EVNENS gi|11692585] FL RCLSLBVNENI gi 111692583 I LR PAFAPBQ gi|11692581| LQQGPI SKK gi|11692579| FQNITFHGQK
The homologies shown above are shared by GPCRlb insofar as GPCRla and lb are homologous as shown in Table 1H.
The presence of identifiable domains in GPCRla, as well as all other GPCRX proteins, was determined by searches using software algorithms such as PROSITE, DOMAIN, Blocks, Pfam, ProDomain, and Prints, and then determining the Interpro number by crossing the domain match (or numbers) using the Interpro website (http://www.ebi.ac.uk/interproy DOMAIN results, e.g., for GPCRla as disclosed in Table IK, were collected from the
Conserved Domain Database (CDD) with Reverse Position Specific BLAST analyses. This BLAST analysis software samples domains found in the Smart and Pfam collections. For Table 1 J and all successive DOMAIN sequence alignments, fully conserved single residues are indicated by black shading and "strong" semi-conserved residues are indicated by grey shading.
The "strong" group of conserved amino acid residues may be any one of the following groups of amino acids: STA, NEQK, NHQK, NDEQ, QHRK, MILV, MILF, HY, FYW.
Table IK lists the domain description from DOMAIN analysis results against GPCRla. This indicates that the GPCRla sequence has properties similar to those of other proteins known to contain this domain as well as to the 377 amino acid 7tm domain itself.
Table IK. Domain Analysis of GPCRla gnl I famlpfamOOOOl, 7tm_l, 7 transmembrane receptor (rhodopsin family) (SEQ ID NO: 65) Length: 254 Score = 103 bits (256) , Expect = 2e-23
10 20 30 40 50 60
I I GPCR 1A SVHUJTAΠISST ';RfflH-Bg YFpSlGraSIFg ΠHlGBSSΠTVHKIV ISBNSHAI S YΘGHVS
7 transmembrane receptor (rhod ataVILVfli TKK R-0@TN IHJLS VAM: LBLLSLPBWS IVHGD VFGDI HKL
70 80 90 100 110 120 I. I. I I I . . . . I GPCR 1A Q FFYHFLgCTECFfflYnVMACn [TVπMNHgvCFMfflATGTfeΪGCVHAMI
7 transmembrane receptor (rhod VGAL VVNBYASILHLHAISIIJ IRRBRTPSR KVΪILLVIVIIAIΛLSIIP-
130 140 150 160 170 180 GPCR 1A TrajTgQLPYCGPNKVG Y YFCDI PAVLPLA CKDTSfflAQ: iTNVGffl Sl
7 transmembrane receptor (rhod -fflliBsWLRTVEEGNT TVC IDFPEESVK RSYVLHSTLI LPLpivi
190 200 210 220 230 240 GPCR 1A
7 transmembrane receptor (rhod
250 260 270 280 290 300 GPCR 1A -SI
7 transmembrane receptor (rhod -RS
310 320 330 340 350 360 GPCR 1A aFSTCSAHIiTAIfi IrYfflQPNPSA IiG- -SIIQI NNli-
7 transmembrane receptor (rhod aKML VVWVFvB tliHDSLCLLSΪW- -RVLPTAL IT
370 GPCR 1A
7 transmembrane receptor (rhod L LΆY'NSCIBBI"
The 7 transmembrane receptor family includes a number of different proteins, including, for example, serotonin receptors, dopamine receptors, histamine receptors, andrenergic receptors, cannabinoid receptors, angiotensin II receptors, chemokine receptors, opioid receptors, G-protein coupled receptor (GPCR) proteins, olfactory receptors (OR), and the like. Some proteins and the Protein Data Base Ids/gene indexes include, for example: rhodopsin (129209); 5-hydroxytryptamine receptors; (112821, 8488960, 112805, 231454, 1168221, 398971, 112806); G protein-coupled receptors (119130, 543823, 1730143, 132206, 137159, 6136153, 416926, 1169881, 136882, 134079); gustatory receptors (544463, 462208); c-x-c
chemokine receptors (416718, 128999, 416802, 548703, 1352335); opsins (129193, 129197, 129203); and olfactory receptor-like proteins (129091, 1171893, 400672, 548417).
The nucleic acids and proteins of GPCR1 are useful in potential therapeutic applications implicated in various GPCR- or olfactory receptor (OR)-related pathologies and/or disorders. For example, a cDNA encoding the G-protein coupled receptor-like protein may be useful in gene therapy, and the G-protein coupled receptor-like protein may be useful when administered to a subject in need thereof. The novel nucleic acid encoding GPCR1 protein, or fragments thereof, may further be useful in diagnostic applications, wherein the presence or amount of the nucleic acid or the protein are to be assessed. These materials are further useful in the generation of antibodies that bind immunospecifically to the novel substances of the invention for use in therapeutic or diagnostic methods. The GPCRX nucleic acids and proteins are useful in potential diagnostic and therapeutic applications implicated in various diseases and disorders described below and/or other pathologies. For example, the compositions of the present invention will have efficacy for treatment of patients suffering from: developmental diseases, MHCπ and III diseases (immune diseases), Taste and scent detectability Disorders, Burkitt's lymphoma, Corticoneurogenic disease, Signal Transduction pathway disorders, Retinal diseases including those involving photoreception, Cell Growth rate disorders; Cell Shape disorders, Feeding disorders;control of feeding; potential obesity due to over-eating; potential disorders due to starvation (lack of apetite), noninsulin-dependent diabetes mellitus (NIDDMl), bacterial, fungal, protozoal and viral infections (particularly infections caused by HIV-1 or HTV-2), pain, cancer (including but not limited to Neoplasm; adenocarcinoma; lymphoma; prostate cancer; uterus cancer), anorexia, bulimia, asthma, Parkinson's disease, acute heart failure, hypotension, hypertension, urinary retention, osteoporosis, Crohn's disease; multiple sclerosis; and Treatment of Albright Hereditary Ostoeodystrophy, angina pectoris, myocardial infarction, ulcers, asthma, allergies, benign prostatic hypertrophy, and psychotic and neurological disorders, including anxiety, schizophrenia, manic depression, delirium, dementia, severe mental retardation. Dentatorubro-pallidoluysian atrophy(DRPLA) Hypophosphatemic rickets, autosomal dominant (2) Acrocallosal syndrome and dyskinesias, such as Huntington's disease or Gilles de la Tourette syndrome and/or other pathologies and disorders of the like. By way of nonlimiting example, the compositions of the present invention will have efficacy for treatment of patients suffering from neoplasm, adenocarcinoma, lymphoma, prostate cancer, uterus cancer, immune response, AIDS, asthma, Crohn's disease, multiple sclerosis, and
Albright Hereditary Ostoeodystrophy. Additional GPCR-related diseases and disorders are mentioned throughout the Specification.
The novel nucleic acid encoding the GPCR-like protein of the invention, or fragments thereof, may further be useful in diagnostic applications, wherein the presence or amount of the nucleic acid or the protein are to be assessed. These materials are further useful in the generation of antibodies that bind immunospecifically to the novel substances of the invention for use in therapeutic or diagnostic methods. These antibodies may be generated according to methods known in the art, using prediction from hydrophobicity charts, as described in the "Anti-GPCRX Antibodies" section below. The disclosed GPCRla protein has multiple hydrophilic regions, each of which can be used as an immunogen. In one embodiment, a contemplated GPCRla epitope is from about amino acids 160 to 170. In another embodiment, a GPCRla epitope is from about amino acids 220 to 240. In an additional embodiment, GPCRla epitopes are from about amino acids 280 to 290.
GPCR2
GPCR2 includes three novel G-protein coupled receptor-like proteins disclosed below. The disclosed proteins have been named GPCR2a, GPCR2b and GPCR2c.
GPCR2a
A novel nucleic acid of 1051 nucleotides (AP001884_B) encoding a novel G-protein coupled receptor-like protein, referred to herein as GPCR2a, is shown in Table 2A.
Table 2A. GPCR2a Nucleotide Sequence (SEQ ID NO:5)
TATCAATTAATTGGTAAATGCTGGGTGCTCCTTATATCCCCAGAGGGAGAGAGACCAAGGGTGAGAAGAAATGTCCA AGACCAGCCTCGTGACAGCGTTCATCCTCACGGGCCTTCCCCATGCCCCAGGGCTGGACGCCCCACTCTTTGGAATC TTCCTGGTGGTTTACGTGCTCACTGTGCTGGGGAACCTCCTCATCCTGCTGGTGATCAGGGTGGATTCTCACCTCCA CACCCCCATGTACTACTTCCTCACCAACCTGTCCTTCATTGACATGTGGTTCTCCACTGTCACGGTGCCCAAAATGC TGATGACCTTGGTGTCCCCAAGCGGCAGGGCTATCTCCTTCCACAGCTGCGTGGCTCAGCTCTATTTTTTCCACTTC CTGGGGAGCACCGAGTGTTTCCTCTACACAGTCATGTCCTATGATCGCTACTTGGCCATCAGTTACCCGCTCAGGTA CACCAGCATGATGAGTGGGAGCAGATGTGCCCTCCTGGCCACCAGCACTTGGCTCAGTGGCTCTCTGCACTCTGCTG TCCAGACCATATTGACTTTCCATTTGCCCTACTGTGGACCCAACCAGATCCAGCACTATTTGTGTGATGCACCGCCC ATCCTGAAACTGGCCTGTGCAGACACCTCAGCCAACGAGATGGTCATCTTTGTGGACATTGGGCTAGTGGCCTCGGG CTGCTTTCTCCTGATAGTGCTGTCTTATGTGTCCATCGTCTGTTCCATCCTGCGGATCCACACCTCAGAGGGGAGGC ACAGAGCCTTTCAGACCTGTGCCTCCCACTGCATCGTGGTCCTTTGCTTTTTTGTNNCCTGTGTTTTCATTTACCTG AGACCAGGCTCCAGGGACGTCGTGGATGGAGTTGTGGCCATTTTCTACACTGTGCTGACACCCCTTCTCAACCCTGT TGTGTACACCCTGAGAAACAAGGAGGTGAAGAAAGCTGTGTTGAAACTGAGAGACAAAGTAGCACATTCTCAGGGAG AATAAATACTAGGAAGTAGATACACTAGTTTGTTTAAAAATAGTAATATA
An open reading frame was identified beginning with an ATG initiation codon at nucleotides 71-73 and ending with a TAA codon at nucleotides 1004-1006. A putative untranslated region upstream from the initiation codon and downstream from the termination codon is underlined in Table 2A, and the start and stop codons are in bold letters. The disclosed
GPCR2a polypeptide (SEQ ID NO:6) encoded by SEQ ID NO:5 is 311 amino acid residues and is presented using the one-letter code in Table 2B. The Psort profile for GPCR2a predicts that this sequence has a signal peptide and is likely to be localized at the plasma membrane with a certainty of 0.6400. The most likely cleavage site for a GPCR2a peptide is between amino acids 51 and 52, at: VDS-HL., based on the SignalP result. GPCR2a has a molecular weiglit of 34476.4.
Table 2B. Encoded GPCR2a protein sequence (SEQ ID NO:6).
MSKTSLVTAFILTGLPHAPGLDAPLFGIFLVVYVLTVLGNLLILLVIRVDSHLHTPMYYFLTNLSFIDMWFSTVTVP KMLMTLVSPSGRAISFHSCVAQLYFFHFLGSTECFLYTVMSYDRYLAISYPLRYTSMMSGSRCALLATSTWLSGSLH SAVQTILTFHLPYCGPNQIQHYLCDAPPILKLACADTSANE VIFVDIGLVASGCFLLIVLSYVSIVCSILRIHTSE GRHRAFQTCASHCIVVLCFFVXCVFIYLRPGSRDVVDGWAIFYTVLTPLLNPVVYTLRNKEVK AVLKLRDKVAHS QGE
The amino acid sequence of GPCR2a had high homology to other proteins as shown in Table 2C.
Table 2C. BLASTX results for GPCR2a
Smallest
Sum
Reading High Prob
Sequences producing High-scoring Segment Pairs: Frame Score P(N) N patp:AAY90874 Human G protein-coupled receptor GTAR14-.. . +2 940 1.2e-93 1 patp:AAY90873 Human G protein-coupled receptor GTAR14-.. . +2 883 1.3e-87 1 patp:AAY90872 Human G protein-coupled receptor GTAR14-.. . +2 736 4.9e-72 1 patp:AAR27868 Odorant receptor clone F5 - Rattus rattu.. . +2 690 3.7e-67 1 patp:AAY90877 Human G protein-coupled receptor GTAR11-.. . +2 686 9.7e-67 1 patp:AAR27876 Odorant receptor clone 115 - Rattus ratt .. . +2 684 1.6e-66 1 patp:AAR27874 Odorant receptor clone 9 - Rattus rattu.. . +2 673 2.3e-65 1 patp:AAY90876 Human G protein-coupled receptor GTAR11-.. . +2 673 2.3e-65 1 patp:AAY83387 Olfactory receptor protein OLF-2 - Homo .. . +2 672 3.0e-65 1
GPCR2b
A novel nucleic acid of 1101 nucleotides (AP001884_C) encoding a novel G-protein coupled receptor-like protein is shown in Table 2D. An open reading frame was identified beginning with an ATG initiation codon at nucleotides 76-78 and ending with a TAA codon at nucleotides 1009-1011. Putative untranslated regions upstream from the initiation codon and downstream from the termination codon are underlined in Table 2D, and the start and stop codons are in bold letters.
Table 2D. GPCR2b Nucleotide Sequence (SEQ ID NO:7)
TCATTTCTCAATTAAGTGCTAAATGCTGGGTGCTCNTTATATCCCCAGAGGGAGΆGAGACCAAGGGTGAGAAGAA"
ATGTCCAACGCCAGCCTCGTGACAGCATTCATCCTCACAGGCCTTCCCCATGCCCCAGGGCTGGACGCCCTCCTC TTTGGAATCTTCCTGGTGGTTTACGTGCTCACTGTGCTGGGGAACCTCCTCATCCTGCTGGTGATCAGGGTGGAT TCTCACCTCCACACCCCCATGTACTACTTCCTCACCAACCTGTCCTTCATTGACATGTGGTTCTCCACTGTCACG GTGCCCAAAATGCTGATGACCTTGGTGTCCCCAAGCGGCAGGGCTATCTCCTTCCACAGCTGCGTGGCTCAGCTC TATTTTTTCCACTTCCTGGGGAGCACCGAGTGTTTCCTCTACACAGTCATGTCCTATGATCGCTACTTGGCCATC AGTTACCCGCTCAGGTACACCAGCATGATGAGTGGGAGCAGGTGTGCCCTCCTGGCCACCGGCACTTGGCTCAGT GGCTCTCTGCACTCTGCTGTCCAGACCATATTGACTTTCCATTTGCCCTACTGTGGACCCAACCAGATCCAGCAC TACTTCTGTGACGCACCGCCCATCCTGAAACTGGCCTGTGCAGACACCTCAGCCAACGTGATGGTCATCTTTGTG GACATTGGGATAGTGGCCTCAGGCTGCTTTGTCCTGATAGTGCTGTCCTATGTGTCCATCGTCTGTTCCATCCTG CGGATCCGCACCTCAGATGGGAGGCGCAGAGCCTTTCAGACCTGTGCCTCCCACTGTATTGTGGTCCTTTGCTTC TTTGTTCCCTGTGTTGTCATTTATCTGAGGCCAGGCTCCATGGATGCCATGGATGGAGTTGTGGCCATTTTCTAC ACTGTGCTGACGCCCCTTCTCAACCCTGTTGTGTACACCCTGAGAAACAAGGAGGTGAAGAAAGCTGTGTTGAAA CTTAGAGACAAAGTAGCACATCCTCAGAGGAAATAAATACTAGGAAGTAAATACACTAGTTTGTTTAAAAATAGT AATCTAATTTAGTTATTCATGTGAAATTGATTATATGTATAGTTCTCAGTG
The disclosed GPCR2b polypeptide (SEQ ID NO:8) encoded by SEQ ID NO:7 is 311 amino acid residues, and is presented using the one-letter code in Table 2E. The Psort profile for GPCR2b predicts that this sequence has a signal peptide and is likely to be localized at the plasma membrane with a certainty of0.6400. The most likely cleavage site for a GPCR2b peptide is between amino acids 51 and 52, at: VDS-HL., based on the SignalP result. GPCR2b has a molecular weight of34568.7.
Table 2E. Encoded GPCR2b protein sequence (SEQ ID NO:8).
MSNASLVTAFILTGLPHAPGLDALLFGIFLVVYVLTVLGNLLILLVIRVDSHLHTPMYYFLTNLSFIDM FSTVT VPKMLMTLVSPSGRAISFHSCVAQLYFFHFLGSTECFLYTVMSYDRYLAISYPLRYTS MSGSRCALLATGTWLS GSLHSAVQTILTFHLPYCGPNQIQHYFCDAPPILKLACADTSANVMVIFVDIGIVASGCFVLIVLSYVSIVCSIL RIRTSDGRRRAFQTCASHCIVVLCFFVPCVVIYLRPGSMDAMDGWAIFYTVLTPLLNPWYTLRNKEVKKAVLK LRDKVAHPQRK
The amino acid sequence of GPCR2b had high homology to other proteins as shown in
Table 2F.
Table 2F. BLASTX results for GPCR2b
Smallest
Sum
Reading High Prob
Sequences producing High-scoring Segment Pairs : Frame Score P(N) N patp:AAY90874 Human G protein-coupled receptor GTAR14-. .. +1 959 l.le-95 1 patp:AAY90873 Human G protein-coupled receptor GTAR14-. .. +1 906 4.7e-90 1 patp:AAY90872 Human G protein-coupled receptor GTAR14-. .. +1 740 1.8e-72 1 patp:AAR27868 Odorant receptor clone F5 - Rattus rattu. .. +1 708 4.5e-69 1 patp:AAR27872 Odorant receptor clone 17 - Rattus rattu. .. +1 675 1.4e-65 1 patp:AAR27876 Odorant receptor clone 115 - Rattus ratt. .. +1 675 1.4e-65 1 patp:AAY83387 Olfactory receptor protein OLF-2 - Homo . .. +1 672 3.0e-65 1 patp:AAY83390 Olfactory receptor protein OLF-5 - Homo . .. +1 667 1.0e-64 1 patp:AAR27874 Odorant receptor clone 19 - Rattus rattu. .. +1 665 1.6e-64 1 patp:AAY83387 Olfactory receptor protein OLF-2 - Homo . .. +2 672 3.0e-65 1
GPCR2c
In the present invention, the target sequence identified previously, Accession Number AP001884_C, was subjected to the exon linking process to confirm the sequence. PCR primers were designed by starting at the most upstream sequence available, for the forward primer, and at the most downstream sequence available for the reverse primer. In each case, the sequence was examined, walking inward from the respective termini toward the coding sequence, until a suitable sequence that is either unique or highly selective was encountered, or, in the case of the reverse primer, until the stop codon was reached. Such primers were designed based on in silico predictions for the full length cDNA, part (one or more exons) of the DNA or protein sequence of the target sequence, or by translated homology of the predicted exons to closely related human sequences sequences from other species. These primers were then employed in PCR amplification based on the following pool of human cDNAs: adrenal gland, bone marrow, brain - amygdala, brain - cerebellum, brain - hippocampus, brain - substantia nigra, brain - thalamus, brain -whole, fetal brain, fetal kidney, fetal liver, fetal lung, heart, kidney, lymphoma - Raji, mammary gland, pancreas, pituitary gland, placenta, prostate, salivary gland, skeletal muscle, small intestine, spinal cord, spleen, stomach, testis, thyroid, trachea, uterus. Usually the resulting amplicons were gel purified, cloned and sequenced to high redundancy. The resulting sequences from all clones were assembled with themselves, with other fragments in CuraGen Corporation's database and with public ESTs. Fragments and ESTs were included as components for an assembly when the extent of their identity with another component of the assembly was at least 95% over 50 bp.. In addition, sequence traces were evaluated manually and edited for corrections if appropriate. These procedures provide the sequence reported below, which is designated Accession Number CG54395-02.
A disclosed GPCR2c (also referred to as CG54395-02) nucleic acid of 1401 nucleotides is shown in Table 2G. An open reading frame was identified beginning with an ATG initiation codon at nucleotides 175-177 and ending with a TAA codon at nucleotides 1108-1110. A putative untranslated region upstream from the initiation codon and downstream from the termination codon is underlined in Table 2G, and the start and stop codons are in bold letters.
Table 2G. GPCR2c Nucleic acid sequence (SEQ ID NO:9).
TCATCATCTCTGTGAGGGAAGCTTTGTAACAAGCGAAGTGCAGGATAACTCCAGAATTATCTACCTGGTTGATGCAGT TTCCACATAGAGAATGGATTCTCATTTCTCAATTAAGTGCTAAATGCTGGGTGCTCTTTATATCCCCAGAGGGAGAGA GACCAAGGGTGAGAAGAAATGTCCAACGCCAGCCTCGTGACAGCGTTCATCCTCACGGGCCTTCCCCATGCCCCAGGG CTGGACGCCCTCCTCTTTGGAATCTTCCTGGTGGTTTACGTGCTCACTGTGCTGGGGAACCTCCTCATCCTGCTGGTG ATCAGGGTGGATTCTCACCTCCACACCCCCATGTACTACTTCCTCACCAACCTGTCCTTCATTGACATGTGGTTCTCC ACTGTCACGGTGCCCAAAATGCTGATGACCTTGGTGTCCCCAAGCGGCAGGGCTATCTCCTTCCACAGCTGCGTGGCT CAGCTCTATTTTTTCCACTTCCTGGGGAGCACCGAGTGTTTCCTCTACACAGTCATGTCCTATGATCGCTACTTGGCC ATCAGTTACCCGCTCAGGTACACCAGCATGATGAGTGGGAGCAGGTGTGCCCTCCTGGCCACCGGCACTTGGCTCAGT
GGCTCTCTGCACTCTGCTGTCCAGACCATATTGACTTTCCATTTGCCCTACTGTGGACCCAACCAGATCCAGCACTAC TTCTGTGACGCACCGCCCATCCTGAAACTGGCCTGTGCAGACACCTCAGCCAACGTGATGGTCATCTTTGTGGACAAT GGGATAGTGGCCTCAGGCTGCTTTGTCCTGATAGTGCTGTCCTATGTGTCCATCGTCTGTTCCATCCTGCGGATCCGC ACCTCAGATGGGAGGCGCAGAGCCTTTCAGACCTGTGCCTCCCACTGTATTGTGGTCCTTTGCTTCTTTGTTCCCTGT GTTGTCATTTATCTGAGGCCAGGCTCCATGGATGCCATGGATGGAGTTGTGGCCATTTTCTACACTGTGCTGACGCCC CTTCTCAACCCTGTTGTGTACACCCTGAGAAACAAGGAGGTGAAGAAAGCTGTGTTGAAACTTAGAGACAAAGTAGCA CATCCTCAGAGGAAATAAATACTAGGAAGTAAATACACTAGTTTGTTTAAAAATAGTAATCTAATTTAGTTATTCATG TGAAATTGATTATATGTATAGTTCTCAGTGTTAAACATTATTCCAAAACACCTGCACAGTTATAATTCTTCCACAGAT TGTCTAAGACAGTTTTAACCTCACAGCTAGACTTATATTTATGATGAACATGATTATATTCTGAATTATTGACTCATT TCTCATCAATAGGTTTATATTAAGTTTAAAACATATTTTAATCAAATCTCAGGGATAGATGATTAATTCATGTTT
The disclosed nucleic acid sequence has 595 of 949 bases (62%) identical to a Homo sapiens T-cell receptor alpha mRNA (gb:GENBANK-ID:HUAE000658|acc:AE000658.1).
The GPCR2c protein endcoded by SEQ ID NO:9 has 311 amino acid residues, referred to as the GPCR2c protein, is presented in Table 2H using the one-letter amino acid code. The Psort profile for GPCR2c predicts that this sequence has a signal peptide and is likely to be localized at the plasma membrane with a certainty of 0.6400. The most likely cleavage site for a GPCR2c peptide is between amino acids 51 and 52, at: VDS-HL, based on the SignalP result.
Table 2H. Encoded GPCR2c protein sequence (SEQ ID NO:10).
MSNASLVTAFILTGLPHAPGLDALLFGIFLVVYVLTVLGNLLILLVIRVDSHLHTPMYYFLTNLSFIDMWFSTVTVP K LMTLVSPSGRAISFHSCVAQLYFFHFLGSTECFLYTVMSYDRYLAISYPLRYTSMMSGSRCALLATGTWLSGSLH SAVQTILTFHLPYCGPNQIQHYFCDAPPILKLACADTSANVMVIFVDNGIVASGCFVLIVLSYVSIVCSILRIRTSD GRRRAFQTCASHCIWLCFFVPCVVIYLRPGSMDAMDGVVAIFYTVLTPLLNPVVYTLRNKEVKKAVLKLRDKVAHP QRK
BLASTP analysis of the best hits for alignments with GPCR2c are listed in Table 21.
GPCR2a, 2b and 2c are related to each other as shown in the alignment listed in Table 2J.
Table 2J: Clustal W of GPCR2 Variants
GPCR 2A Protein Sequence (AP00 GPCR 2B Protein Sequence (AP00 GPCR 2C Protein Sequence (CG54
70 80 90 100 110 120 .1 •■ ..|..
GPCR 2A Protein Sequence (AP00 iTNLSFIDM FSTV VPKML TLVSPSGRAISFHSCVAQLYFFHFLGSTECF YTVMSY GPCR 2B Protein Sequence (AP00 iTNLSFIDM FSTVTVPKMLMTLVSPSGRAISFHSCVAQLYFFHFLGSTECFLYTVMSYI
GPCR 2C Protein Sequence (CG54
130 140 150 160 170 180
■ ■■■ I ....1^.. I .... I .... I .... I .... I .... I .... I ■■■■I ....I ....| GPCR 2A Protein Sequence (APOO YLAISYPLRYTSMMSGSRCAL ATgTWLSGSLHSAVQTILTFHLPYCGPNQIQHY GPCR 2B Protein Sequence (APOO RYLAISYPLRYTSMMSGSRCALLATGTWLSGSLHSAVQTILTFHLPYCGPNQIQHYFCD2 GPCR 2C Protein Sequence (CG54 RYLAISYPLRYTSMMΞGSRCALLATGT SGSLHSAVQTILTFHLPYCGPHQIQHYFCP;
190 200 210 220 230 240 1 ----1 I — — 1 I 1 — — 1 1 - — -I — . I ... -I ....I
GPCR 2A Protein Sequence (APOO [53CTS^«kM^Ji]F& ι BlB iffϊf3-^ GPCR 2B Protein Sequence (APOO 'PI KLACADTSANV VIFVDIGIVASGCFV IVLSYVSIVCSILRIRTSDGRRRAFQTI GPCR 2C Protein Sequence (CG54 »PILK ACADTSANVMVIFVDEGIVASGCFVLIV SYVSIVCSILRIRTSDGRRRAFQTI
250 260 270 280 290 300
■ I ....1.... i ....i .... I .... I .... I .... I .... I
GPCR 2A Protein Sequence (APOO fflffτtt. ff?i-xfflF ^ΨtiRHwR GPCR 2B Protein Sequence (APOO WIY RPGSMDAMDGVVAIFYTVLTPLLNPVVYTLRNKEVKKAVL GPCR 2C Protein Sequence (CG54 SHCIVVLCFFVPCVVIYLRPGSMDAMDGVVAIFYTVLTPLLNPVVYTLRNKEVKKAV
310 • ■ ■ ■ 1 - ■ ■ - 1 ■
GPCR 2A Protein Sequence (APOO SsSSSSIIsfflGE GPCR 2B Protein Sequence (APOO LRDKVAHPQRK GPCR 2C Protein Sequence (CG54 LRDKVAHPQRK
It was also found that GPCR2a had homology to the amino acid sequences shown in the BLASTP data listed in Table 2K.
The homology of these sequences is shown graphically in the ClustalW analysis shown in Table 2L.
Table 2L. ClustalW Analysis of GPCR2a
1) Novel GPCR2a (SEQ ID NO:5)
2) gi|2143658|pir||JC5202chemoreceptorTB641 [Rat] (SEQ ID NO: 50)
3) gi|11692583|gb|AAG39888.1|AF282303_l odorant receptor M34 [Mus musculus] (SEQ ID
NO:47)
4) gi|l 1692581|gb|AAG39887.1|AF282302_l odorant receptor M32 [Mus musculus] (SEQ ID
NO:48)
5) gi|l 1692585|gb|AAG39889.1|AF282304_l odorant receptor M36 [Mus musculus] (SEQ ID NO:46)
6) gi|11692587|gb|AAG39890.1|AF282305_l odorant receptor M37 [Mus musculus] (SEQ ID
NO:45)
310 320 I....|....I....|..
GPCR 2A gκ LKIιRDι3vAHSQGE gi|2143658| ~JKRJ_.RAGRGNVGGDK gi|11692583| iSHWKV RIjJPAFAPUQIj— gi|11692581| gCSfflKKMLQQGPILSKK gi|116925851 RHVFLgRC LEVNENI gi| 11692587 I RHVFLHRCLSLEVNENS
The homologies shown above are shared by GPCR2b and 2c insofar as they are themselves homologous to GPCR2a as shown in Table 2J.
Table 2M lists the domain description from DOMAIN analysis results against GPCR2a. This indicates that the GPCR2a sequence has properties similar to those of other proteins known to contain this domain as well as to the 377 amino acid 7tm domain itself.
Table 2M. Domain Analysis of GPCR2a gnl I Pfaml famOOOOl, 7tm_l, 7 transmembrane receptor (rhodopsin family) (SEQ ID NO: 65) Length: 254 Score = 81.6 bits (200), Expect = 5e-17
70 80 90 100 110 120
GPCR 2A QLYFraHF gΞTECFfflYraVMgY SYEgjgTSMMSGsBc llj TSTirfSGSJΪHl VQ
7 transmembrane receptor (rhod VGALHVVNHYAS ILBLBAISI VHF3SRRXRT?PR@AK HILLV| LAL3I |LPP
130 140 150 160 170 180
GPCR 2A TlLTFHLPYCGP Q IQHYIiCDAPgiLKIA CADTSANEMMI|
7 transmembrane receptor (rhod LJIFSWLRTVEEGHT TVCLID—FHEESVK RSYVLLSTj-fflGB
190 200 210 220 230 240 ..|..
GPCR 2A t—PSSYVSIVC-
7 transmembrane receptor (rhod RT3RKRARSQ-
250 260 270 280 290 300
GPCR 2A -SI BRIHTHEGSH [FQTCASHCtV
7 transmembrane receptor (rhod -RSEKRRSSSEIK KMLLVVVWF
310 320 330 340 350
. . . I . . . . | . . . . | . . . | . . . . | . . . . | . . . . I
GPCR 2A FFVXCVFIYrfRPGSRDVVD- R VAIFYTVIiTPLl avv
7 transmembrane receptor (rhod ""LPYHIVILJP DSLC LSIW- BLPTALLITIW AYVNSC an
The GPCR2c gene maps to chromosome 11 and is expressed in at least the following tissues: Apical microvilli of the retinal pigment epithelium, arterial (aortic), basal forebrain, brain, Burkitt lymphoma cell lines, corpus callosum, cardiac (atria and ventricle), caudate nucleus, CNS and peripheral tissue, cerebellum, cerebral cortex, colon, cortical neurogenic cells, endothelial (coronary artery and umbilical vein) cells, palate epithelia, eye, neonatal eye, frontal cortex, fetal hematopoietic cells, heart, hippocampus, hypothalamus, leukocytes, liver, fetal liver, lung, lung lymphoma cell lines, fetal lymphoid tissue, adult lymphoid tissue, Those that express MHC II and III nervous, medulla, subthalamic nucleus, ovary, pancreas, pituitary, placenta, pons, prostate, putamen, serum, skeletal muscle, small intestine, smooth muscle (coronary artery in aortic) spinal cord, spleen, stomach, taste receptor cells of the tongue, testis, thalamus, and thymus tissue. This information was derived by determining the tissue sources of
the sequences that were included in the invention including but not limited to SeqCalling sources, Public EST sources, Literature sources, and/or RACE sources.
GPCR2a-c include the nucleic acids whose sequences are provided in Table 2A, 2D and 2G, or a fragment thereof. The invention also includes a mutant or variant nucleic acid any of whose bases may be changed from the corresponding base shown in Table 2A, 2D and 2G while still encoding a protein that maintains its G-Protein Coupled Receptor-like activities and physiological functions, or a fragment of such a nucleic acid. GPCR2a-c further includes nucleic acids whose sequences are complementary to those just described, including nucleic acid fragments that are complementary to any of the nucleic acids just described. GPCR2a-c additionally includes nucleic acids or nucleic acid fragments, or complements thereto, whose structures include chemical modifications. Such modifications include, by way of nonlimiting example, modified bases, and nucleic acids whose sugar phosphate backbones are modified or derivatized. These modifications are carried out at least in part to enhance the chemical stability of the modified nucleic acid, such that they may be used, for example, as antisense binding nucleic acids in therapeutic applications in a subject, hi the mutant or variant nucleic acids, and their complements, up to about 38 percent of the bases may be so changed.
The protein similarity information, expression pattern, cellular localization, and map location for the protein and nucleic acid disclosed herein suggest that this Olfactory Receptorlike protein may have important structural and/or physiological functions characteristic of the Olfactory Receptor family. Therefore, the nucleic acids and proteins of the invention are useful in potential diagnostic and therapeutic applications and as a research tool. These include serving as a specific or selective nucleic acid or protein diagnostic and/or prognostic marker, wherein the presence or amount of the nucleic acid or the protein are to be assessed. These also include potential therapeutic applications such as the following: (i) a protein therapeutic, (ii) a small molecule drug target, (iii) an antibody target (therapeutic, diagnostic, drug targeting/cytotoxic antibody), (iv) a nucleic acid useful in gene therapy (gene delivery/gene ablation), (v) an agent promoting tissue regeneration in vitro and in vivo, and (vi) a biological defense weapon.
The nucleic acids and proteins of the invention are useful in potential diagnostic and therapeutic applications implicated in various diseases and disorders described below and/or other pathologies. For example, the compositions of the present invention will have efficacy for treatment of patients suffering from: developmental diseases, MHCII and III diseases (immune diseases), Taste and scent detectability Disorders, Burkitt's lymphoma, Corticoneurogenic disease, Signal Transduction pathway disorders, Retinal diseases including those involving photoreception, Cell Growth rate disorders; Cell Shape disorders, Feeding disorders;control of
feeding; potential obesity due to over-eating; potential disorders due to starvation (lack of apetite), noninsulin-dependent diabetes mellitus (NTDDM1), bacterial, fungal, protozoal and viral infections (particularly infections caused by HTV-l or HTV-2), pain, cancer (including but not limited to Neoplasm; adenocarcinoma; lymphoma; prostate cancer; uterus cancer), anorexia, bulimia, asthma, Parkinson's disease, acute heart failure, hypotension, hypertension, urinary retention, osteoporosis, Crohn's disease; multiple sclerosis; and Treatment of Albright Hereditary Ostoeodystrophy, angina pectoris, myocardial infarction, ulcers, asthma, allergies, benign prostatic hypertrophy, and psychotic and neurological disorders, including anxiety, schizophrenia, manic depression, delirium, dementia, severe mental retardation. Dentatorubro- pallidoluysian atrophy(DRPLA) Hypophosphatemic rickets, autosomal dominant (2)
Acrocallosal syndrome and dyskinesias, such as Huntington's disease or Gilles de la Tourette syndrome and/or other pathologies and disorders of the like. The polypeptides can be used as immunogens to produce antibodies specific for the invention, and as vaccines. They can also be used to screen for potential agonist and antagonist compounds. For example, a cDNA encoding the OR -like protein may be useful in gene therapy, and the OR-like protein may be useful when administered to a subject in need thereof. By way of nonlimiting example, the compositions of the present invention will have efficacy for treatment of patients suffering from bacterial, fungal, protozoal and viral infections (particularly infections caused by HTV-l or HΓV-2), pain, cancer (including but not limited to Neoplasm; adenocarcinoma; lymphoma; prostate cancer; uterus cancer), anorexia, bulimia, asthma, Parkinson's disease, acute heart failure, hypotension, hypertension, urinary retention, osteoporosis, Crohn's disease; multiple sclerosis; and Treatment of Albright Hereditary Ostoeodystrophy, angina pectoris, myocardial infarction, ulcers, asthma, allergies, benign prostatic hypertrophy, and psychotic and neurological disorders, including anxiety, schizophrenia, manic depression, delirium, dementia, severe mental retardation and dyskinesias, such as Huntington's disease or Gilles de la Tourette syndrome and/or other pathologies and disorders.
The novel nucleic acid encoding the GPCR-like protein of the invention, or fragments thereof, may further be useful in diagnostic applications, wherein the presence or amount of the nucleic acid or the protein are to be assessed. These materials are further useful in the generation of antibodies that bind immunospecifically to the novel substances of the invention for use in therapeutic or diagnostic methods. These antibodies may be generated according to methods known in the art, using prediction from hydrophobicity charts, as described in the
"Anti-GPCRX Antibodies" section below. The disclosed GPCR2a-c proteins have multiple hydrophilic regions, each of which can be used as an immunogen. In one embodiment, a
contemplated GPCR2a-c epitope is from about amino acids 55 to 65. In another embodiment, a GPCR2a-c epitope is from about amino acids 120 to 130. In additional embodiments, GPCR2a-c epitopes are from about amino acids 165 to 185, from about amino acids 220 to 240 and from about amino acids 285 to 310.
GPCR3
GPCR3 includes two nucleic acids disclosed below. The disclosed nucleic acids encode a GPCR-like protein.
GPCR3a
The disclosed GPCR3a (also referred to herein as 645i8A) is encoded by a nucleic acid, 1011 nucleotides long. An open reading frame was identified beginning with an ATG initiation codon at nucleotides 20-22 and ending with a TAA codon at nucleotides 977-979. Putative untranslated regions upstream from the initiation codon and downstream from the termination codon are underlined in Table 3 A, and the start and stop codons are in bold letters.
Table 3A. GPCR3a Nucleotide Sequence (SEQ ID NO:ll).
AGACTGAGGCACCCATTCCATGGATACCTCCACCAGTGTTACCTATGACTCCAGCCTCCAGATTTCCCAGTTCAT CCTGATGGGATTACCAGGCATTCATGAGTGGCAGCACTGGCTCTCCCTGCCCCTGACTCTGCTCTACCTCTTAGC TCTTGGTGCCAACCTCCTCATCATAATCACCATTCAACATGAGACCGTGCTACATGAACCCATGTACCATTTGCT GGGCATATTAGCAGTGGTGGACATTGGCCTGGCCACCACCATCATGCCCAAGATCCTGGCCATCTTCTGGTTTGA TGCCAAGGCCATTAGCCTCCCCATGTGTTTTGCTCAGATCTATGCCATCCACTGCTTCTTCTGCATAGAGTCAGG CATCTTTCTCTGCATGGCAGTAGACAGATACATAGCCATCTGTCGCCCTCTTCAGTACCCCTCCATAGTCACTAA AGCTTTTGTCTTCAAAGCCACAGGGTTCATCATGCTCAGGAATGGCCTGTTGACCATCCCAGTGCCTATACTGGC TGCCCAGAGACACTACTGTTCCAGGAATGAAATCGAGCACTGCCTCTGCTCTAACTTGGGGGTTATCAGCCTGGC TTGTGATGACATCACTGTGAACAAATTTTACCAACTGATGCTAGCATGGGTCTTGGTTGGGAGTGATATGGCTCT GGTATTTTCTTCCTATGCTGTAATCCTTCACTCTGTGCTGAGGCTGAACTCAGCAGAAGCAATGTCCAAGGCTCT GAGCACTTGTAGCTCCCACCTCATCCTCATCCTCTTCCACACAGGTATCATTGTGCTGTCTGTCACACACCTTGC AGAGAAAAAGATTCCCCTTATTCCTGTGTTCCTTAATGTGCTGCACAATGTCATCCCCCCTGCACTCAACCCCCT GGCCTGTGCACTCAGGATGCACAAACTCAGACTGGGCTTTCAGAGACTGCTTGGACTGGGTCAGGACGTGTCCAA GTAACTCCAGCTTTAGGAATCCAGTAAGATGTTAGG
The GPCR3a polypeptide (SEQ ID NO:12) encoded by SEQ ID NO:l l has 319 amino acid residues, referred to as GPCR3a protein, is presented using the one-letter amino acid code in Table 3B. The Psort profile for GPCR3a predicts that this sequence has a signal peptide and is likely to be localized at the plasma membrane with a certainty of 0.6000. The most likely cleavage site for a GPCR3a peptide is between amino acids 46 and 47, at: ALG-AN., based on the SignalP result. GPCR3a has a molecular weight of 35453.0.
Table 3B. GPCR3a protein sequence (SEQ ID NO:12)
MDTSTSVTYDSSLQISQFILMGLPGIHE QHWLSLPLTLLYLLALGANLLIIITIQHETVLHEPMYHLLGILAWD IGLATTIMPKILAIFWFDAKAISLP CFAQIYAIHCFFCIESGIFLCMAVDRYIAICRPLQYPSIVTKAFVFKATG
FI LRNGLLTIPVPILAAQRHYCSRNEIEHCLCSNLGVISLACDDITVNKFYQLMLA VLVGSDMALVFSSYAVIL HSVLRLNSAEAMSKALSTCSSHLILILFHTGIIVLSVTHLAEKKIPLIPVFLNVLHNVIPPALNPLACALRMHKLR LGFQRLLGLGQDVSK
The amino acid sequence of GPCR3a had high homology to other proteins as shown in Table 3C.
Table 3C. BLASTX results for GPCR3a
Smallest Sum
Reading High Prob
Sequences producing High-scoring Segment Pairs: Frame Score P(N) N patp:AAY92365 G protein-coupled receptor protein 5 - H +2 565 5e-54 patp:AA 01730 Human G-protein receptor HPRAJ70 - Homo +2 515 3e-48 patp:AAW56641 G-protein coupled prostate tissue recept +2 515 3e-48 patp:AAR27872 Odorant receptor clone 17 - Rattus rattu +2 394 5e-36 patp:AAY90873 Human G protein-coupled receptor GTAR14- +2 390 3e-35 patp:AAY90874 Human G protein-coupled receptor GTAR14- +2 387 7e-35 patp:AAR27868 Odorant receptor clone F5 - Rattus rattu +2 375 8e-34 patp:AAY83389 Olfactory receptor protein OLF-4 - Homo +2 374 le-33
GPCR3b
The disclosed GPCR3b (also referred to herein as CG52784-02) is encoded by a nucleic acid, 973 nucleotides long. An open reading frame was identified beginning with an ATG initiation codon at nucleotides 5-7 and ending with a TAA codon at nucleotides 962-964. Putative untranslated regions upstream from the initiation codon and downstream from the termination codon are underlined in Table 3D, and the start and stop codons are in bold letters.
Table 3D. GPCR3b Nucleotide Sequence (SEQ ID NO:13)
TTCCATGGATACCTCCACCAGTGTTACCTATGACTCCAGCCTCCAGATTTCCCAGTTCATCCTGATG GGATTACCAGGCATTCATGAGTGGCAGCACTGGCTCTCCCTGCCCCTGACTCTGCTCTACCTCTTAG CTCTTGGTGCCAACCTCCTCATCATAATCACCATTCAACATGAGACCGTGCTACATGAACCCATGTA CCATTTGCTGGGCATATTAGCAGTGGTGGACATTGGCCTGGCCACCACCATCATGCCCAAGATCCTG GCCATCTTCTGGTTTGATGCCAAGGCCATTAGCCTCCCCATGTGTTTTGCTCAGATCTATGCCATCC ACTGCTTCTTCTGCATAGAGTCAGGCATCTTTCTCTGCATGGCAGTAGACAGATACATAGCCATCTG TCGCCCTCTTCAGTACCCCTCCATAGTCACTAAAGCTTTTGTCTTCAAAGCCACAGGGTTCATCATG CTCAGGAATGGCCTGTTGACCATCCCAGTGCCTATACTGGCTGCCCAGAGACACTACTGTTCCAGGA ATGAAATCGAGCACTGCCTCTGCTCTAACTTGGGGGTTATCAGCCTGGCTTGTGATGACATCACTGT GAACAAATTTTACCAACTGATGCTAGCATGGGTCTTGGTTGGGAGTGATATGGCTCTGGTATTTTCT TCCTATGCTGTAATCCTTCACTCTGTGCTGAGGCTGAACTCAGCAGAAGCAATGTCCAAGGCTCTGA GCACTTGTAGCTCCCACCTCATCCTCATCCTCTTCCACACAGGTATCATTGTGCTGTCTGTCACACA CCTTGCAGAGAAAAAGATTCCCCTTATTCCTGTGTTCCTTAATGTGCTGCACAATGTCATCCCCCCT GCACTCAACCCCCTGGCCTGTGCACTCAGGATGCACAAACTCAGACTGGGCTTTCAGAGACTGCTTG GACTGGGTCAGGACGTGTCCAAGTAACTCCAGCTT
The disclosed nucleic acid sequence has 504 of 808 bases (62%) identical to a Mus musculus odorant receptor S18 mRNA (gb:GENBANK-ID:AF121975|acc: AF121975).
The GPCR3b polypeptide (SEQ ID NO:14) encoded by SEQ ID NO:13 is presented using the one-letter amino acid code in Table 3E. The SignalP, Psort and/or Hydropathy profile for the disclosed GPCR3b GPCR-like protein predicts that this sequence has a signal peptide and is likely to be localized at the plasma membrane with a certainty of 0.6000. The most likely cleavage site for a GPCR3b peptide is between amino acids 46 and 47, at: ALG-AN., based on the SignalP result.
Table 3E. GPCR3b Amino Acid Sequence (SEQ ID NO:14)
MDTSTSVTYDSSLQISQFILMGLPGIHEWQH LSLPLTLLYLLALGANLLIIITIQHETVLHEPMYHLLGILAV VDIGLATTIMPKILAIF FDAKAISLPMCFAQIYAIHCFFCIESGIFLCMAVDRYIAICRPLQYPSIVTKAFVF KATGFIMLRNGLLTIPVPILAAQRHYCSRNEIEHCLCSNLGVISLACDDITVNKFYQLMLAVLVGSDMALVFS SYAVILHSVLRLNSAEAMSKALSTCSSHLILILFHTGIIVLSVTHLAEKKIPLIPVFLNVLHNVIPPALNPLAC ALRMHKLRLGFQRLLGLGQDVS
BLASTP analysis of the best hits for alignments with GPCR3b are listed in Table 3F.
Table 3F. BLASTP results for GPCR3b
Gene Index/ Protein/ Organism Length Identity Positives Expect Identifier (aa) (%) (%)
SPTREMBL- ODORANT RECEPTOR S18 ■ 321 123/298 175/298 6.6e-56 ACC:Q9WU89 Mus musculus (Mouse) (41%) (58%)
GPCR3a also has 184 of 290 residues (63%) identical to and 219 of 290 residues (75%) positive with olfactory receptor S83 from mouse [Mus musculus] (GenBank Ace. No.:AAK00590.1).
Table 3G. BLASTP alignment of GPCR3a against olfactory receptor S83 [Mus musculus] (SEQ ID NO:51)
Score = 337 bits (865), Expect = 2e-91
Identities = 184/290 (63%), Positives = 219/290 (75%), Gaps = 1/290 (0%)
310 320
I.... |....|
GPCR 3A KgRLgFQR ffl [lGØGQDVSK gi 1127045411 NKE EJaKQgLYKvg: DVKEG-
Table 3H lists the domain description from DOMAIN analysis results against GPCR3a. This indicates that the GPCR3a sequence has properties similar to those of other proteins known to contain this domain as well as to the 377 amino acid 7tm domain itself.
70 80 90 100 110 120
GPCR 3A FAQIYAIHCFFCIESG^FLCMAV KAFVFKATGFJMlRNGffllTI
7 transmembrane receptor (rhod KLVGA FVVNGYAΞIL LTAlSl IBSBiJ. JVHl iPRRAKV ILLy V ALmSL
130 140 150 160 170 180 I I I
GPCR 3A raVPI AACjHYCSRNEIEHCLCSra GffilSliACDEHTfflNKFHQljMLAWfflLVGSDMAI.
7 transmembrane receptor (rhod gP LFSj/ΪLgTV EEG 1 TJl c IDFPBEsBKRSgvlkSTLjl GFV PJjLV
190 200 210 220 230 240
GPCR 3A VFSSWAVI
7 transmembrane receptor (rhod tLVCHT iRTLRKRARSQ-
250 260 270 280 290 300 ..| .. ..|..
GPCR 3A
7 transmembrane receptor (rhod
310 320 330 340 350 360
....|....|....|....|....|....|
GPCR 3A SV AEAMSI gLSTCSSHj ILlffl FgTGlIVL|VTHfflAEKKIP IPVFIjNV HN
7 transmembrane receptor (rhod RΞ SSERK? 3κML VVV VFVaC LPY3ι lVl. LD| C aSIff VLPTALLI iW A
370 • I.
GPCR 3A VIPP. jIiAC
7 transmembrane receptor (rhod YVNSCl toiY
The GPCR3b is expressed in at least the following tissues: Apical microvilli of the retinal pigment epithelium, arterial (aortic), basal forebrain, brain, Burkitt lymphoma cell lines, corpus callosum, cardiac (atria and ventricle), caudate nucleus, CNS and peripheral tissue, cerebellum, cerebral cortex, colon, cortical neurogemc cells, endothelial (coronary artery and
umbilical vein) cells, palate epithelia, eye, neonatal eye, frontal cortex, fetal hematopoietic cells, heart, hippocampus, hypothalamus, leukocytes, liver, fetal liver, lung, lung lymphoma cell lines, fetal lymphoid tissue, adult lymphoid tissue, Those that express MHC II and ITI nervous, medulla, subthalamic nucleus, ovary, pancreas, pituitary, placenta, pons, prostate, putamen, serum, skeletal muscle, small intestine, smooth muscle (coronary artery in aortic) spinal cord, spleen, stomach, taste receptor cells of the tongue, testis, thalamus, and thymus tissue. This information was derived by determining the tissue sources of the sequences that were included in the invention including but not limited to SeqCalling sources, Public EST sources, Literature sources, and/or RACE sources. The nucleic acids and proteins of GPCR3a and GPCR3b are useful in potential therapeutic applications implicated in various GPCR-related pathological disorders and/or OR- related pathological disorders, described further below. For example, a cDNA encoding the olfactory receptor-like protein may be useful in gene therapy, and the olfactory receptor-like protein may be useful when administered to a subject in need thereof. The protein similarity information, expression pattern, and map location for the Olfactory Receptor-like protein and nucleic acid disclosed herein suggest that this Olfactory Receptor may have important structural and/or physiological functions characteristic of the Olfactory Receptor family. Therefore, the nucleic acids and proteins of the invention are useful in potential diagnostic and therapeutic applications and as a research tool. These include serving as a specific or selective nucleic acid or protein diagnostic and/or prognostic marker, wherein the presence or amount of the nucleic acid or the protein are to be assessed, as well as potential therapeutic applications such as the following: (i) a protein therapeutic, (ii) a small molecule drug target, (iii) an antibody target (therapeutic, diagnostic, drug targeting/cytotoxic antibody), (iv) a nucleic acid useful in gene therapy (gene delivery/gene ablation), and (v) a composition promoting tissue regeneration in vitro and in vivo (vi) biological defense weapon.
The nucleic acids and proteins of the invention are useful in potential diagnostic and therapeutic applications implicated in various diseases and disorders described below and/or other pathologies. For example, the compositions of the present invention will have efficacy for treatment of patients suffering from: developmental diseases, MHCII and III diseases (immune diseases), taste and scent detectability disorders, Burkitt's lymphoma, corticoneurogenic disease, signal transduction pathway disorders, retinal diseases including those involving photoreception, cell growth rate disorders, cell shape disorders, feeding disorders, potential obesity due to over-eating, potential disorders due to starvation (lack of appetite), noninsulin-dependent diabetes mellitus (NTDDMl), bacterial, fungal, protozoal and
viral infections (particularly infections caused by HTV-l or HTV-2), pain, cancer (including but not limited to neoplasm; adenocarcinoma; lymphoma; prostate cancer; uterus cancer), anorexia, bulimia, asthma, Parkinson's disease, acute heart failure, hypotension, hypertension, urinary retention, osteoporosis, Crohn's disease; multiple sclerosis; and Treatment of Albright Hereditary Ostoeodystrophy, angina pectoris, myocardial infarction, ulcers, allergies, benign prostatic hypertrophy, and psychotic and neurological disorders, including anxiety, schizophrenia, manic depression, delirium, dementia, severe mental retardation, dentatorubro- pallidoluysian atrophy (DRPLA), hypophosphatemic rickets, autosomal dominant (2) acrocallosal syndrome and dyskinesias, such as Huntington's disease or Gilles de la Tourette and/or other pathologies and disorders of the like. The polypeptides can be used as immunogens to produce antibodies specific for the invention, and as vaccines. They can also be used to screen for potential agonist and antagonist compounds. For example, a cDNA encoding the OR -like protein may be useful in gene therapy, and the OR-like protein may be useful when administered to a subject in need thereof. By way of nonlimiting example, the compositions of the present invention will have efficacy for treatment of patients suffering from bacterial, fungal, protozoal and viral infections (particularly infections caused by HIV-1 or HIV-2), pain, cancer (including but not limited to neoplasm; adenocarcinoma; lymphoma; prostate cancer; uterus cancer), anorexia, bulimia, asthma, Parkinson's disease, acute heart failure, hypotension, hypertension, urinary retention, osteoporosis, Crohn's disease; multiple sclerosis; and treatment of Albright Hereditary Ostoeodystrophy, angina pectoris, myocardial infarction, ulcers, allergies, benign prostatic hypertrophy, and psychotic and neurological disorders, including anxiety, schizophrenia, manic depression, delirium, dementia, severe mental retardation and dyskinesias, such as Huntington's disease or Gilles de la Tourette syndrome. Other GPCR-3 diseases and disorders are contemplated. The novel nucleic acid encoding the GPCR-like protein of the invention, or fragments thereof, may further be useful in diagnostic applications, wherein the presence or amount of the nucleic acid or the protein are to be assessed. These materials are further useful in the generation of antibodies that bind immunospecifically to the novel substances of the invention for use in therapeutic or diagnostic methods. These antibodies may be generated according to methods known in the art, using prediction from hydrophobicity charts, as described in the "Anti-GPCRX Antibodies" section below. The disclosed GPCR3a and 3b proteins have multiple hydrophilic regions, each of which can be used as an immunogen. In one embodiment, a contemplated GPCR3a and 3b epitope is from about amino acids 165 to 185. In another
embodiment, a GPCR3a and 3b epitope is from about amino acids 235 to 245. In an additional embodiment, GPCR3a and 3b epitopes are from amino acids 310 to 320.
GPCR4
The disclosed novel GPCR4 nucleic acid of 1103 nucleotides (also referred to as 645i8_B) is shown in Table 4A. An ORF begins with an ATG initiation codon at nucleotides 42-44 and ends with a TGA codon at nucleotides 1014-1016. A putative untranslated region upstream from the initiation codon and downstream from the termination codon is underlined in Table 4A, and the start and stop codons are in bold letters.
Table 4A. GPCR4 Nucleotide Sequence (SEQ ID NO:15)
CTTTTAGTGGAACTTACAACTGGCTCTGCTGACTAACTCTCATGTTCTTTAGTCTTCATAGAGAGACTCCAAAAG AAGGCATGTTTGGTGCTAATCTCACCACCTTCCATCCCACTCTATTCATTCTCCTTGGCATCCCAGGACTGGAGC AATACCACATCTGGCTTTCCATTCCTTTCTACCTTATGTACATCACTGCAGTCTTGGGAAATGGAGCCCTCATCC TAGTTGTCCTCAGTGAACACACCCTCCATGTCTTCCTATCCATGCTGGCTGGCACTGATATCCTGCTATCCACCA CCACTGTGCCTAAGGCCTTGGCGATCTTCTGGGTCCACGCTGGGGAGATAGCCTTTGATGCCTGCATTACTCAGA TGTTTTTCATTCATGTTGCCTTTGTGGCTGAGTCAGGAATCCTGCTGGCCATGGCATTTGACAGTTATGTAGCCA TTTGTACTCCCTTGAGATACACTACCATCTTAACTTCTATGGTAAATGGAAAAATGACCCTGACAATCTGGGGAC AAAGCATTGGGACAATTTTTCCTGTCATATTCCTGCTGAAGAGGCTGCCATACTGTCAGACCAATATCATCCCCC ACTCATACTGTGAGCACATTGGGGTGGCCCAATTGGCCTGTGCTGACATAACTGTCAATATCTGGTATGGCTTTT CAGTGCCAATGGCATCGGTTTTGGTAGATGTTGCATTCATTGGTTTTTCCTACACTTTGATCCTCCAGGCTGTGT TTAGACTTCCTTCCCAGGAGTCCCAGCACAAAGCTCTTAACACCTGTGGTTCATACATTGGAGTTGTTCTCCTCT TCTTCATCCCATCATTTTTTACTTTCCTGACCCACCGCTTTGGCAAGAATATCCCCCATCATGTCCACATACTTC TGGCAAATCTCTACTTGCTTGTTCCCCCCATGCTTAACCCCATTATCTACGGAGAGAAGACCAAGCAAATCAGGG ACAGTATGGCTCATATGTTATCTGTGGTGGGGAAGTCTTGAGACATCATGGTCTCTTCACAGTTTTCTCTTACCA GTAGGAGAAAAGAGAAGTTGCCAATCAAGTTCCCAAGTTTAGGCCCTATAGTG
The GPCR4 protein encoded by SEQ ID NO: 15 has 324 amino acid residues and is presented using the one-letter code in Table 4B. The Psort profile for GPCR4 predicts that this sequence has a signal peptide and is likely to be localized at the plasma membrane with a certainty of 0.6000. GPCR4 has a molecular weight of 36125.5.
Table 4B. Encoded GPCR4 protein sequence (SEQ ID NO:16)
MFFSLHRETPKEGMFGANLTTFHPTLFILLGIPGLEQYHIWLSIPFYLMYITAVLGNGALILWLSEHTLHVF LSMLAGTDILLSTTTVPKALAIF VHAGEIAFDACITQMFFIHVAFVAESGILLAMAFDSYVAICTPLRYTTI LTSMVNGKMTLTIWGQSIGTIFPVIFLLKRLPYCQTNIIPHSYCEHIGVAQLACADITVNIWYGFSVPMASVL VDVAFIGFSYTLILQAVFRLPSQESQHKALNTCGSYIGVVLLFFIPSFFTFLTHRFGKNIPHHVHILLANLYL LVPPMLNPIIYGEKTKQIRDSMAHMLSVVGKS
The amino acid sequence of GPCR4 had high homology to other proteins as shown in Table 4C.
Table 4C. BLASTX results for GPCR4
Smallest
Sum
Reading High Prob
Sequences producing High-scoring Segment Pairs: Frame Score P(N) N patp:AAW01730 Human G-protein receptor HPRAJ70 - Homo ... +3 708 4.5e-69 1 patp:AA 56641 G-protein coupled prostate tissue recept ... +3 708 4.5e-69 1 patp:AAY92365 G protein-coupled receptor protein 5 - H I... +3 702 2.0e-68 1 patp:AAR27875 Odorant receptor clone 114 - Rattus ratt ... +3 447 2.1e-41 1 patp:AAR27876 Odorant receptor clone 115 - Rattus ratt ... +3 414 6.5e-38 1 patp:AAR27874 Odorant receptor clone 19 - Rattus rattui... +3 409 2.2e-37 1 patp:AAR27869 Odorant receptor clone F6 - Rattus rattui... +3 398 3.2e-36 1
GPCR4 also has homology to the proteins shown in the BLASTP data in Table 4D.
This BLASTP data is displayed graphically in the Clustal W in Table 4E.
Table 4E. ClustalW Analysis of GPCR4
1) Novel GPCR4 (SEQ ID NO: 16)
2) gi|99380141ref1NP 064686.1| odorant receptor S18 gene [Mus musculus] (SEQ ID NO:52)
3) gill 190821 llgb|AAG41676.11 (AF137396) HOR 5'Betal4 [Homo sapiens] (SEQ ID NO:53)
4) gi|7305349|ref|NP 038647.1[ olfactory receptor 67 [Mus musculus] (SEQ ID NO:54)
5) gil6532001|gp|AAD27596.2lAF121976 1 (AF121976) odorant receptor S19 [Mus musculus] (SEQ ID NO:55)
6) gi|9935442|ref]NP 064688.11 odorant receptor S46 gene [Mus musculus] (SEQ ID NO:56)
Table 4F lists the domain description from DOMAIN analysis results against GPCR4. This indicates that the GPCR4 sequence has properties similar to those of other proteins known to contain this domain as well as to the 377 amino acid 7tm domain itself.
Table 4F Domain Analysis of GPCR4 gnl I PfamlpfamOOOOl, 7tm_l, 7 transmembrane receptor (rhodopsin family) (SEQ ID NO: 65) Length: 254 Score = 78.2 bits (191), Expect = 6e-16
70 80 90 100 110 120
....| ....| .... | ....|....| ....] ....|....| ....|.... | ....| ....|
GPCR 4 QMFFIH^FVBESGl{ΪLr χ[AF!Ss^OTcτB^TTπLπSMVNGK τ[jTlfflGQΞIGTIFJ3v
7 transmembrane receptor (rhod VGALFVmHGYasiLlfflTlTRT'RBl^VHBMRRn nPRRAKV lil Vl V ALLI.RLlaP
130 140 150 160 170 180
....l....|....l....|....l....|....|....|....|.... I....I....I
GPCR 4 ΪFL KR PYCQTNII HSYCEHI GEAQJJACA ITHN IW0GFSVPtøASVl<VD
7 transmembrane receptor (rhod XiFSWLR TVEEGNT TBC IDF BESSK RS0VLLSTi,VGFVLP
GPCR 4
7 transmembrane receptor (rhod
250 260 270 280 290 300
....|....|....|....|....|....|....|....|....|....|....|....| GPCR 4 |QHHLN
7 transmembrane receptor (rhod RSLKRRSSgERggAK
310 320 330 340 350
....1.... I .... I .... I .... I .... I .... I .... I .... I .... I .. ,.|.... GPCR 4 TCGSYlGt vlfflFFl|SFFTFgTHRFGKNIPH HJlJH ifflLA L LLfflPPMffl^TO
7 transmembrane receptor (rhod ML VV^g vBclTOgYHIVLg DSLC SIW RM PTAliffllTLW AYmscHBaBH
The nucleic acids and proteins of GPCR4 are useful in potential therapeutic applications implicated in various GPCR-related pathological disorders and/or OR-related pathological disorders, described further below. For example, a cDNA encoding the GPCR (or olfactory- receptor) like protein may be useful in gene therapy, and the receptor -like protein may be useful when administered to a subject in need thereof. The nucleic acids and proteins of the invention are also useful in potential therapeutic applications used in the treatment of developmental diseases, MHCπ and III diseases (immune diseases), taste and scent detectability disorders, Burkitt's lymphoma, corticoneurogenic disease, signal transduction pathway disorders, retinal diseases including those involving photoreception, cell growth rate disorders, cell shape disorders, feeding disorders, potential obesity due to over-eating, potential disorders due to starvation (lack of appetite), noninsulin-dependent diabetes mellirus (NiDDMl), bacterial, fungal, protozoal and viral infections (particularly infections caused by HIV-1 or HIV-2), pain, cancer (including but not limited to neoplasm; adenocarcinoma; lymphoma; prostate cancer; uterus cancer), anorexia, bulimia, asthma, allergies, Parkinson's disease, acute heart failure, hypotension, hypertension, urinary retention, osteoporosis, Crohn's disease, multiple sclerosis, Albright hereditary ostoeodystrophy, angina pectoris, myocardial infarction, ulcers, benign prostatic hypertrophy, psychotic and neurological disorders (including anxiety, schizophrenia, manic depression, delirium, dementia, and severe mental retardation), dentatorubro-pallidoluysian atrophy (DRPLA), hypophosphatemic rickets, autosomal dominant (2) acrocallosal syndrome and dyskinesias, such as Huntington's disease or Gilles de la Tourette syndrome and/or other pathologies and disorders. Other GPCR-related diseases and disorders are contemplated. The polypeptides can be used as immunogens to produce antibodies specific for the invention, and as vaccines. They can also be used to screen for potential agonist and antagonist compounds. For example, a cDNA encoding the GPCR-like protein may be useful in gene therapy, and the GPCR-like protein may be useful when administered to a subject in need
thereof. By way of nonlimiting example, the compositions of the present invention will have efficacy for treatment of patients suffering from developmental diseases, MHCII and III diseases (immune diseases), taste and scent detectability disorders, Burkitt's lymphoma, corticoneurogenic disease, signal transduction pathway disorders, retinal diseases including those involving photoreception, cell growth rate disorders, cell shape disorders, feeding disorders, potential obesity due to over-eating, potential disorders due to starvation (lack of appetite), noninsulin-dependent diabetes mellitus (NTDDM1), bacterial, fungal, protozoal and viral infections (particularly infections caused by HIV-1 or HIV-2), pain, cancer (including but not limited to neoplasm; adenocarcinoma; lymphoma; prostate cancer; uterus cancer), anorexia, bulimia, asthma, allergies, Parkinson's disease, acute heart failure, hypotension, hypertension, urinary retention, osteoporosis, Crohn's disease, multiple sclerosis, Albright hereditary ostoeodystrophy, angina pectoris, myocardial infarction, ulcers, benign prostatic hypertrophy, psychotic and neurological disorders (including anxiety, schizophrenia, manic depression, delirium, dementia, and severe mental retardation), dentatorubro-pallidoluysian atrophy (DRPLA), hypophosphatemic rickets, autosomal dominant (2) acrocallosal syndrome and dyskinesias, such as Huntington's disease or Gilles de la Tourette syndrome and/or other pathologies and disorders. The novel nucleic acid encoding GPCR-like protein, and the GPCR- like protein of the invention, or fragments thereof, may further be useful in diagnostic applications, wherein the presence or amount of the nucleic acid or the protein are to be assessed. These materials are further useful in the generation of antibodies that bind immunospecifically to the novel substances of the invention for use in therapeutic or diagnostic methods..
The novel nucleic acid encoding the GPCR-like protein of the invention, or fragments thereof, may further be useful in diagnostic applications, wherein the presence or amount of the nucleic acid or the protein are to be assessed. These materials are further useful in the generation of antibodies that bind immunospecifically to the novel substances of the invention for use in therapeutic or diagnostic methods. These antibodies may be generated according to methods known in the art, using prediction from hydrophobicity charts, as described in the "Anti-GPCRX Antibodies" section below. The disclosed GPCR4 protein has multiple hydrophilic regions, each of which can be used as an immunogen. In one embodiment, a contemplated GPCR4 epitope is from about amino acids 10 to 20. In another embodiment, a
GPCR4 epitope is from about amino acids 175 to 190. In additional embodiments, GPCR4 epitopes are from about amino acids 230 to 255, from about amino acids 270 to 280 and from about amino acids 300 to 320.
GPCR5
GPCR5a
The disclosed novel GPCR5a nucleic acid of 1241 nucleotides (also referred to as 645i8_C) is shown in Table 5A. An open reading frame begins with an ATG initiation codon at nucleotides 129-131 and ends with a TAA codon at nucleotides 1089-1091. A putative untranslated region upstream from the initiation codon and downstream from the termination codon are underlined in Table 5A, and the start and stop codons are in bold letters.
Table 5A. GPCR5a Nucleotide Sequence (SEQ ID NO:17)
CCCTGCACTACTCCTACACGTAT6CCATG6ATGGGTTTGTGCCACTGTGAGACCCCCAa7AATCTCTCCCCTTATC TTCCCTGTCAGGAGTCATGCCCCCATGAGCCCTCAAGTTGTGCCCACCACAGA&TGGCAGAAACTCTACAACTCA ATTCCACCTTCCTACACCCAAACTTCTTCATACTGACTGGCTTTCCAGGGCTAGGAAGTGCCCAGACTTGGCTGA CACTGGTCTTTGGGCCCATTTATCTGCTGGCCCTGCTGGGCAATGGAGCACTGCCGGCAGTGGTGTGGATAGACT CCACACTGCACCAGCCCATGTTTCTACTGTTGGCCATCCTGGCAGCCACAGACCTGGGCTTAGCCACATCTKTAG CCCCAGGGTTGCTGGCTGTGCTGTGGCTTGGGCCCCGATCTGTGCCATATGCTGTGTGCCTGGTCCAGATGTTCT TTGTACATGCACTGACTGCCATGGAATCAGGTGTGCTTTTGGCCATGGCCTGTGATCGTGCTGCGGCAATAGGGC GTCCACTGCACTACCCTGTCCTGGTCACCAAAGCCTGTGTGGGTTATGCAGCCTTGGCCCTGGCACTGAAAGCTG TGGCTATTGTTGTACCTTTCCCACTGCTGGTGGCAAAGTTTGAGCACTTCCAAGCCAAGACCATAGGCCATACCT ATTGTGCACACATGGCAGTGGTAGAACTGGTGGTGGGTAACACACAGGCCACCAACTTATATGGTCTGGCACTTT CACTGGCCATCTCAGGTATGGATATTCTGGGTATCACTGGCTCCTATGGACTCATTGCCCATGCTGTGCTGCAGC TACCTACCCGGGAGGCCCATGCCAAGGCCTTTGGTACATGTAGTTCTCACATCTGTGTCATTCTGGCCTTCTACA TACCTGGTCTCTTCTCCTACCTCACACACCGCTTTGGTCATCACACTGTCCCAAAGCCTGTGCACATCCTTCTCT CCAACATCTACTTGCTGCTGCCACCTGCCCTCAACCCCCTCATCTATGGGGCCCGCACCAAGCAGATCAGAGACC GACTCCTGGAAACCTTCACATTCAGAAAAAGCCCGTTGTAA.TGTCCAGTGGTAACAATGGAGCCTAAGAGTGGAG GTGAGGGGACAATCGGAGGGGAGTCTGGGGGTGTGGATCATGTTATTTTCATCCCACTGCATATGACTGTTATCA TCATTTAACAGGTACTTGCTGTGGAGTCCCTATGTGCAACA
The GPCR5a protein encoded by SEQ ID NO: 17 has 320 amino acid residues, and is presented using the one-letter code in Table 5B (SEQ ID NO: 18). The Psort profile for GPCR5a predicts that this sequence has a signal peptide and is likely to be localized at the plasma membrane with a certainty of 0.6000. The most likely cleavage site for a GPCR5a peptide is between amino acids 51 and 52, at: ALP-AV., based on the SignalP result. GPCR5a has a molecular weight of 34413.5.
Table 5B. Encoded GPCR5a protein sequence (SEQ ID NO:18).
MAETLQLNSTFLHPNFFILTGFPGLGSAQTWLTLVFGPIYLLALLGNGALPAW IDSTLHQPMFLLLAILAATDL GLATSIAPGLLAVL LGPRSVPYAVCLVQ FFVHALTA ESGVLLAMACDRAAAIGRPLHYPVLVTKACVGYAALA LALKAVAIWPFPLLVAKFEHFQAKTIGHTYCAHMAWELWGNTQATNLYGLALSLAISGMDILGITGSYGLIAH AVLQLPTREAHAKAFGTCSSHICVILAFYIPGLFSYLTHRFGHHTVPKPVHILLSNIYLLLPPALNPLIYGARTKQ IRDRLLETFTFRKSPL
The amino acid sequence of GPCR5a had high homology to other proteins as shown in Table 5C.
Table 5C. BLASTX results for GPCR5a
Smallest
Sum
Reading High Prob
Sequences producing High-scoring Segment Pairs : Frame Score P (N) N patp :AAW01730 Human G-protein receptor HPRAJ70 - Homo . . . +3 632 5. 1e-61 1 patp :AAW56641 G-protein coupled prostate tissue recept . +3 632 5. 1e-61 1 patp :AAY92365 G protein-coupled receptor protein 5 - H. . +3 624 3. 6e-60 1 patp :AAR27874 Odorant receptor clone 19 - Rattus rattu. . +3 362 2. 1e-32 1 patp :AAB68889 Human RECAP polypeptide, SEQ ID NO: 19 - . . +3 348 6. 4e-31 1 patp :AAY90873 Human G protein-coupled receptor GTAR14- . . +3 347 8 .2e-31 1 patp :AAY90874 Human G protein-coupled receptor GTAR14- . . +3 343 2 .2e-30 1 patp :AAW75960 Human olfactory 0LRCC15 receptor - Homo . . +3 334 1. 9e-29 1
Possible SNPs found for GPCR5a are listed in Table5D.
GPCR5b
In the present invention, the target sequence identified previously, Accession Number 645i8_C, was subjected to the exon linking process to confirm the sequence. PCR primers were designed by starting at the most upstream sequence available, for the forward primer, and at the most downstream sequence available for the reverse primer. In each case, the sequence was examined, walking inward from the respective termini toward the coding sequence, until a suitable sequence that is either unique or highly selective was encountered, or, in the case of the reverse primer, until the stop codon was reached. Such suitable sequences were then employed as the forward and reverse primers in a PCR amplification based on a wide range of cDNA libraries. The resulting amplicon was gel purified, cloned and sequenced to high redundancy to provide the sequence reported below, winch is designated Accession Number 645i8C_dal . The disclosed novel GPCR5b nucleic acid of 974 nucleotides (also referred to as 645i8C_dal) is shown in Table 5E. An open reading frame begins with an ATG initiation codon at nucleotides 6-8 and ends with a TAA codon at nucleotides 966-968. A putative untranslated region upstream from the initiation codon and downstream from the termination codon are underlined in Table 5E, and the start and stop codons are in bold letters.
Table 5E. GPCR5b Nucleotide Sequence (SEQ ID NO:19)
ACAGAATGGCAGAAACTCTACAACTCAATTCCACCTTCCTACACCCAAACTTCTTCATACTGACTGGCTTTCCAG GACTAGGAAGTGCCCAGACTTGGCTGACACTGGTCTTTGGGCCCATTTATCTGCTGGCCCTGCTGGGCAATGGAG CACTGCCGGCAGTGGTGTGGATAGACTCCACACTGCACCAGCCCATGTTTCTACTGTTGGCCATCCTGGCAGCCA GAGACCTGGGCTTAGCCACATCTATAGCCCCAGGGTTGCTGGCTGTGCTGTGGCTTGGGCCCCGATCTGTGCCAT ATGCTGTGTGCCTGGTCCAGATGTTCTTTGTACATGCACTGACTGCCATGGAATCAGGTGTGCTTTTGGCCATGG CCTGTGATCGTGCTGCGGCAATAGGGCGTCCACTGCACTACCCTGTCCTGGTCACCAAAGCCTGTGTGGGTTATG CAGCCTTGGCCCTGGCACTGAAAGCTGTGGCTATTGTTGTACCTTTCCCACTGCTGGTGGCAAAGTTTGAGCACT TCCAAGCCAAGACCATAGGCCATACCTATTGTGCACACATGGCAGTGGTAGAACTGGTGGTGGGTAACACACAGG CCACCAACTTATATGGTCTGGCACTTTCACTGGCCATCTCAGGTATGGATATTCTGGGTATCACTGGCTCCTATG GACTCATTGCCCATGCTGTGCTGCAGCTACCTACCCGGGAGGCCCATGCCAAGGCCTTTGGTACATGTAGTTCTC ACATCTGTGTCATTCAGGCCTTCTACATACCTGGTCTCTTCTCCTACCTCACACACCGCTTTGGTCATCACACTG TCCCAAAGCCTGTGCACATCCTTCTCTCCAACATCTACTTGCTGCTGCCACCTGCCCTCAACCCCCTCATCTATG GGGCCCGCACCAAGCAGATCAGAGACCGACTCCTGGAAACCTTCACATTCAGAAAAAGCCCGTTGTAATGTCCA
The GPCR5b protein encoded by SEQ ID NO: 19 has 320 amino acid residues, and is presented using the one-letter code in Table 5F (SEQ ID NO:20). The Psort profile for GPCR5b predicts that this sequence has a signal peptide and is likely to be localized at the plasma membrane with a certainty of 0.6000. The most likely cleavage site for a peptide is between amino acids 51 and 52, at: ALP-AV, based on the SignalP result. The molecular weight of GPCR5b is 34483.6 Daltons.
Table 5F. Encoded GPCR5b protein sequence (SEQ ID NO:20).
MAETLQLNSTFLHPNFFILTGFPGLGSAQTWLTLVFGPIYLLALLGNGALPAVV IDSTLHQPMFLLLAILAARDL GLATSIAPGLLAVL LGPRSVPYAVCLVQMFFVHALTAESGVLLAMACDRAAAIGRPLHYPVLVTACVGYAALA LALKAVAIWPFPLLVAKFEHFQAKTIGHTYCAHMAVVELVVGNTQATNLYGLALSLAISGMDILGITGSYGLIAH AVLQLPTREAHAKAFGTCSSHICVIQAFYIPGLFSYLTHRFGHHTVPKPVHILLSNIYLLLPPALNPLIYGARTKQ IRDRLLETFTFRKSPL
BLASTP analysis of the best hits for alignments with GPCR5b are listed in Table 5G.
GPCR5c
In the present invention, the target sequence identified previously, Accession Number 645i8_C, was subjected to the exon linking process to confirm the sequence. PCR primers were designed by starting at the most upstream sequence available, for the forward primer, and at the most downstream sequence available for the reverse primer. In each case, the sequence was examined, walking inward from the respective termini toward the coding sequence, until a
suitable sequence that is either unique or highly selective was encountered, or, in the case of the reverse primer, until the stop codon was reached. Such suitable sequences were then employed as the forward and reverse primers in a PCR amplification based on a wide range of cDNA libraries. The resulting amplicon was gel purified, cloned and sequenced to high redundancy to provide the sequence reported below, which is designated Accession Number 645i8Cl.
The disclosed novel GPCR5c nucleic acid of 1241 nucleotides (also referred to as 645i8Cl) is shown in Table 5H. An open reading frame begins with an ATG initiation codon at nucleotides 129-131 and ends with a TAA codon at nucleotides 1089-1091. Aputative untranslated region upstream from the initiation codon and downstream from the termination codon are underlined in Table 5H, and the start and stop codons are in bold letters.
Table 5H. GPCR5c Nucleotide Sequence (SEQ ID NO:21)
CCCTGCACTACTCCTACACGTATGCCATGGATGGGTTTGTGCCACTGTGAGACCCCCAAAATCTCTCCCCTTATC TTCCCTGTCAGGAGTCATGCCCCCATGAGCCCTCAAGTTGTGCCCACCACAGAATGGCAGAAACTCTACAACTCA ATTCCACCTTCCTACACCCAAACTTCTTCATACTGACTGGCTTTCCAGGACTAGGAAGTGCCCAGACTTGGCTGA CACTGGTCTTTGGGCCCATTTATCTGCTGGCCCTGCTGGGCAATGGAGCACTGCCGGCAGTGGTGTGGATAGACT CCACACTGCACCAGCCCATGTTTCTACTGTTGGCCATCCTGGCAGCCACAGACCTGGGCTTAGCCACATCTATAG CCCCAGGGTTGCTGGCTGTGCTGTGGCTTGGGCCCCGATCTGTGCCATATGCTGTGTGCCTGGTCCAGATGTTCT TTGTACATGCACTGACTGCCATGGAATCAGGTGTGCTTTTGGCCATGGCCTGTGATCGTGCTGCGGCAATAGGGC GTCCACTGCACTACCCTGTCCTGGTCACCAAAGCCTGTGTGGGTTATGCAGCCTTGGCCCTGGCACTGAAAGCTG TGGCTATTGTTGTACCTTTCCCACTGCTGGTGGCAAAGTTTGAGCACTTCCAAGCCAAGACCATAGGCCATACCT ATTGTGCACACATGGCAGTGGTAGAACTGGTGGTGGGTAACACACAGGCCACCAACTTATATGGTCTGGCACTTT CACTGGCCATCTCAGGTATGGATATTCTGGGTATCACTGGCTCCTATGGACTCATTGCCCATGCTGTGCTGCAGC TACCTACCCGGGAGGCCCATGCCAAGGCCTTTGGTACATGTAGTTCTCACATCTGTGTCATTCTGGCCTTCTACA TACCTGGTCTCTTCTCCTACCTCGCACACCGCTTTGGTCATCACACTGTCCCAAAGCCTGTGCACATCCTTCTCT CCAACATCTACTTGCTGCTGCCACCTGCCCTCAACCCCCTCATCTATGGGGCCCGCACCAAGCAGATCAGAGACC GACTCCTGGAAACCTTCACATTCAGAAAAAGCCCGTTGTAA.TGTCCAGTGGTAACAATGGAGCCTAAGAGTGGAG GTGAGGGGACAATCGGAGGGGAGTCTGGGGGTGTGGATCATGTTATTTTCATCCCACTGCATATGACTGTTATCA TCATTTAACAGGTACTTGCTGTGGAGTCCCTATGTGCAACA
The GPCR5c protein encoded by SEQ ID NO:21 has 320 amino acid residues, and is presented using the one-letter code in Table 51 (SEQ ID NO:22). The Psort profile for GPCR5c predicts that this sequence has a signal peptide and is likely to be localized at the plasma membrane with a certainty of 0.6000. The most likely cleavage site for a peptide is between amino acids 51 and 52, at: ALP-AV, based on the SignalP result. The molecular weight of GPCR5c is 34383.5 Daltons.
Table 51. Encoded GPCR5c protein sequence (SEQ ID NO:22).
MAETLQLNSTFLHPNFFILTGFPGLGSAQTWLTLVFGPIYLLALLGNGALPAVVWIDSTLHQP FLLLAILAATDL GLATSIAPGLLAVL LGPRSVPYAVCLVQMFFVHALTAMESGVLLAMACDRAAAIGRPLHYPVLVTKACVGYAALA LALKAVAIWPFPLLVAKFEHFQAKTIGHTYCAHMAVVELWGNTQATNLYGLALSLAISGMDILGITGSYGLIAH VLQLPTREAHAKAFGTCSSHICVILAFYIPGLFSYLAHRFGHHTVPKPVHILLSNIYLLLPPALNPLIYGARTKQ IRDRLLETFTFRKSPL
BLASTP analysis of the best hits for alignments with GPCR5c are listed in Table 5J. BLASTX analysis was also performed to determine which proteins have significant identity with GPCR5c, as shown in Table 5K.
Table 5K. BLASTX results for GPCR5c
Smallest Sum Reading High Prob
Sequences producing High-scoring Segment Pairs: Frame Score P(N) N ptnr:SPTREMBL-ACC:Q9WU90 ODORANT RECEPTOR S19 - Mus mu.+3 731 2 Oe-71 1 ptnr:SPTREMBL-ACC:Q9 U93 ODORANT RECEPTOR S46 - Mus mu.+3 702 2 4e-68 1 ptnr:SPTREMBL-ACC:Q9WU89 ODORANT RECEPTOR S18 - Mus mu.+3 695 1 3e-67 1 ptnr:SPTREMBL-ACC:Q9WVD7 MOR 3ΕETA3 - Mus musculus (M.+3 684 1 9e-66 1 ptnr:SPTREMBL-ACC:Q9UKL2 OLFACTORY RECEPTOR HPFHIOR - +3 683 2 5e-66 1 ptnr:SPTREMBL-ACC:Q9WVD8 MOR 3ΕETA2 - Mus musculus (M.+3 676 1 4e-65 1 ptnr : SPTREMBL-ACC : Q9WVD9 MOR 3'BETAl - Mus musculus (M.+3 668 9 6e-65 1
GPCR5d
In the present invention, the target sequence identified previously, Accession Number 645i8_C, was subjected to the exon linking process to confirm the sequence. PCR primers were designed by starting at the most upstream sequence available, for the forward primer, and at the most downstream sequence available for the reverse primer. In each case, the sequence was examined, walking inward from the respective termini toward the coding sequence, until a suitable sequence that is either unique or highly selective was encountered, or, in the case of the reverse primer, until the stop codon was reached. Such suitable sequences were then employed as the forward and reverse primers in a PCR amplification based on a wide range of cDNA libraries. The resulting amplicon was gel purified, cloned and sequenced to high redundancy to provide the sequence reported below, which is designated Accession Number AL078595_da2.
The disclosed novel GPCR5d nucleic acid of 1240 nucleotides (also referred to as AL078595_da2) is shown in Table 5L. An open reading frame begins with an ATG initiation codon at nucleotides 129-131 and ends with a TAA codon at nucleotides 1089-1091. A putative untranslated region upstream from the initiation codon and downstream from the termination codon are underlined in Table 5L, and the start and stop codons are in bold letters.
Table 5L. GPCR5d Nucleotide Sequence (SEQ ID NO:23)
CCCTGCACTACTCCTACACGTATGCCATGGATGGGTTTGTGCCACTGTGACACCCCCAAAATCTCTCCCCTTATC TTCCCTGTCAGGAGTCATGCCCCCATGAGCCCTCAAGTTGTGCCCACCACAGAATGGCAGAAACTCTACAACTCA ATTCCACCTTCCTACACCCAAACTTCTTCATACTGACTGGCTTTCCAGGACTAGGAAGTGCCCAGACTTGGCTGA CACTGGTCTTTGGGCCCATTTATCTGCTGGCCCTGCTGGGCAATGGAGCACTGCCGGCAGTGGTGTGGATAGACT CCACACTGCACCAGCCCATGTTTCTACTGTTGGCCATCCTGGCAGCCATAGACCTGGGCTTAGCCACATCTATAG CCCCAGGGTTGCTGGCTGTGCTGTGGCTTGGGCCCCGATCTGTGCCATATGCTGTGTGCCTGGTCCAGATGTTCT TTGTACATGCACTGACTGCCATGGAATCAGGTGTGCTTTTGGCCATGGCCTGTGATCGTGCTGCGGCAATAGGGC GTCCACTGCACTACCCTGTCCTGGTCACCAAAGCCTGTGTGGGTTATGCAGCCTTGGCCCTGGCACTGAAAGCTG TGGCTATTGTTGTACCTTTCCCACTGCTGGTGGCAAAGTTTGAGCACTTCCAAGCCAAGACCATAGGCCATACCT ATTGTGCACACATGGCAGTGGTAGAACTGGTGGTGGGTAACACACAGGCCACCAACTTATATGGTCTGGCACTTT CACTGGCCATCTCAGGTATGGATATTCTGGGTATCACTGGCTCCTATGGACTCATTGCCCATGCTGTGCTGCAGC TACCTACCCGGGAGGCCCGTGCCAAGGCCTTTGGTACATGTAGTTCTCACATCTGTGTCATTCTGGCCTTCTACA TACCTGGTCTCTTCTCCTACCTCGCACACCGCTTTGGTCATCACACTGTCCCAAAGCCTGTGCACATCCTTCTCT CCAACATCTACTTGCTGCTGCCACCTGCCCTCAACCCCCTCATCTATGGGGCCCGCACCAAGCAGATCAGAGACC GACTCCTGGAAACCTTCACATTCAGAAAAAGCCCGTTGTAA.TGTCCAGTGGTAACAATGGAGCCTAAGAGTGGAG GTGAGGGGACAATCGGAGGGGAGTCTGGGGGTGTGGATCATGTTATTTTCATCCCACTGCATATGACTGTTATCA TCATTTAACAGGTACTTGCTGTGGAGTCCCTATGTGCAAC
The disclosed nucleic acid sequence for GPCR5d has 582 of 943 bases (61%) identical to a Mus musculus odorant receptor S19 mRNA (gb:GENBANK- ID:AF121976|acc:AF121976.2).
The GPCR5d protein encoded by SEQ ID NO:23 has 320 amino acid residues, and is presented using the one-letter code in Table 5M (SEQ ID NO:24). The Psort and Hydropathy profile for GPCR5d predicts that this sequence has a signal peptide and is likely to be localized at the plasma membrane with a certainty of 0.6000. The most likely cleavage site for a peptide is between amino acids 51 and 52, at: ALP-AV, based on the SignalP result.
Table 5M. Encoded GPCR5d protein sequence (SEQ ID NO:24).
MAETLQLNSTFLHPNFFILTGFPGLGSAQT LTLVFGPIYLLALLGNGALPAW IDSTLHQPMFLLLAILAAIDL GLATSIAPGLLAVLWLGPRSVPYAVCLVQMFFVHALTAMESGVLLAMACDRAAAIGRPLHYPVLVTKACVGYAALA LALKAVAIVVPFPLLVAKFEHFQAKTIGHTYCAHMAVVELVVGNTQATNLYGLALSLAISGMDILGITGSYGLIAH AVLQLPTREARAKAFGTCSSHICVILAFYIPGLFSYLAHRFGHHTVPKPVHILLSNIYLLLPPALNPLIYGARTKQ IRDRLLETFTFRKSPL
BLASTP analysis of the best hits for alignments with GPCR5d are listed in Table 5N.
Possible SNPs found for GPCR5d are listed in Table 50.
GPCR5a also has homology to the proteins shown in the BLASTP data in Table 5P.
The homology data shown above is represented graphically in a Clustal W shown in Table 5Q.
Table 5Q. ClustalW Analysis of GPCR5a
1) Novel GPCR5a (SEQ ID NO: 18)
2) ill 190821 l|gblAAG41676.1l (AF137396) HOR 5'Betal4 [Homo sapiens] (SEQ ID NO:53)
3) gi|6532001jgblAAD27596.2lAF121976 1 (AF121976) odorant receptor S19 [Mus musculus] (SEQ ID NO:55)
4) gi|9935442lreι]NP 064688.1| odorant receptor S46 gene [Mus musculus] (SEQ ID NO:56)
5) gill l991867lgblAAG42368.ll (AF289204) odorant receptor HOR3'beta5 [Homo sapiens] (SEQ ID NO:57)
6) gill l908221 |gb|AAG41685.1l (AF133300) MOR 3'Beta6 [Mus musculus] (SEQ ID NO:58)
Table 5R lists the domain description from DOMAIN analysis results against GPCR5a. This indicates that the GPCR5a sequence has properties similar to those of other proteins known to contain this domain as well as to the 377 amino acid 7tm domain itself.
Table 5R. Domain Analysis of GPCR5a qnl I PfamlpfamOOOOl, 7tm_l, 7 transmembrane receptor (rhodopsin family) (SEQ ID NO: 65) Length: 254 Score = 55.1 bits (131), Expect = 6e-09 10 20 30 40 50 60
....|....| ....| ....| ....|....| ....| ....| ....] ....|.... | ....| GPCR 5A ^GaTιPΑ^WID5THH-O^ L^iroΑτffijl LATSIΑHGLfflAvHwT,S-PRRVPYgvSTι
7 transmembrane receptor (rhod ^Ii VILtSlLR KKgR-TgTNI g NEv Si LF TLPgW gY aVGg-DWVFGDltHK 70 80 90 100 110 120
....|....|....|....|....1....|....|....1....|....|....| ....| GPCR 5A Q F VH T MESGVffl gMACgAft^GRlMHHpVJvnK CVGY Affl ljALKgV IVVra
7 transmembrane receptor (rhod IιVGA FVVNGYASIL " "lSliBγL™VH™RHRRΪRPpRRAKVLl" V VL"lιLιΞϊιl9 130 140 150 160 170 180
....I....I ....|....|....|....|....|....|....|....|....| ....1
GPCR 5A FPffl VAKFEHFQAKTIGH πYHAHK&VVraLgVGNTQATNLHGπALSfflAISGMDlir
7 transmembrane receptor ( hod P JP SW RTVBEGNT @VSLIDFPE@SI3 KRsSvιJ SiaVGFVI,PI,g 190 200 210 220 230 240
....|....|....|....|....|....|....|....|....1....|....|....|
GPCR 5A TGSHGLH H
7 transmembrane receptor (rhod LVCgTRgLRTIiRKRARSQ 250 260 270 280 290 300
....|....|....|....| ....|....| ....|....|....|....|....|....|
GPCR 5A fflQLPTREAHAKJjJFGTCS
7 transmembrane receptor (rhod RSgKRRSSSERKAgKMLLV 310 320 330 340 350
|....|....l.. l....|....|. .N...|....|....l. GPCR 5A 7 transmembrane receptor (rhod _
The GPCR5 family members have strong homology for each other as shown in Table
5S.
Table 5S Clustal W of GPCR5 family
10 20 30 40 50 60 !_. • • • I_ - _- 1 • ■ ■ ■ 1 1.... I ....I.... I .... I ...^
GPCR 5A Protein Sequence (645i lAETLQLNSTFLHPNFFILTGFPGLGSAQTWLTLVFGPIYLLALLGNGALPAVVWIDST] GPCR 5B Protein Sequence (645i ΪAET QLNSTFLHPNFFILTGFPGLGSAQTWLTLVFGPIY LALLGNGALPAVV IDST1 GPCR 5C Protein Sequence (6451 IAETLQNSTFLHPNFFI TGFPG GSAQTWLTLVFGPIY AL GNGALPAVVWIDSTJ GPCR 5D Protein Sequence (AL07 1AETLQLNSTFLHPNFFILTGFPGLGSAQTWLTLVFGPIYLLALLGNGALPAVVWIDST]
70 80 90 100 110 120 I I I I 1 I I I I .... I I I
GPCR 5A Protein Sequence (645i IQPMFLLLAILAAΠDLGLATSIAPGLLAVLWLGPRSVPYAVCLVQMFFVHALTAMESGVL GPCR 5B Protein Sequence (645i ΪQPMFLLLAILAASDLGLATSIAPGLLAVLWLGPRSVPYAVCLVQMFFVHALTAMESGVL GPCR 5C Protein Sequence (6451 HQPMFL LAILAΆBD G ATSIAPGLLAVLW GPRSVPYAVC VQMFFVHALTAMESGVL GPCR 5D Protein Sequence (AL07 HQPMFLLLAILAAΠDLGLATSIAPGLLAVLWLGPRSVPYAVCLVQMFFVHALTAMESGVI
130 140 150 160 170 180
■ ■■•|....|....|....|....|....|....|....|.... I .... I .... I .... I. GPCR 5A Protein Sequence (645i FPLLVA FEHFQAKTIG GPCR 5B Protein Sequence (645i JAMACDRAAAIGRP HYPVLVTKACVGYAALALALKAVAIVVPFPL VAKFEHFQAKTIG GPCR 5C Protein Sequence (6451 .AMACDRAAAIGRPLHYPV VTKACVGYAALALALKAVAIVVPFPLLVAKFEHFQAKTIG GPCR 5D Protein Sequence (AL07 AMACDRAAAIGRPLHYPVLVTKACVGYAALALALKAVAIVVPFPLLVAKFEHFQAKTIG
190 200 210 220 230 240
■ ■■•I....I....I....I. ■■■)....1....I .. ,.| ■■■■I....I ... ,|.... L GPCR 5A Protein Sequence (645i [TYCAHMAVVELVVGNTQATNLYGLALSLAISGMDILGITGSYG IAHAV Q PTREAHJ GPCR 5B Protein Sequence (645i HTYCAHMAVVELVVGNTQATNLYGLALSLAIΞGMDILGITGSYGLIAHAVLQ PTREAHΪ GPCR 5C Protein Sequence (6451 HTYCAHMAVVELVVGNTQATN YG ALSLAISGMDILGITGSYGLIAHAVLQLPTREAHJ GPCR 5D Protein Sequence (A 07 HTYCAHMAVVELVVGNTQATNLYGLALSLAISGMDILGITGSYGLIAHAVLQLPTREAE?
250 260 270 280 290 300
■ •■■I....I....1....I ....I....I- ...|....1....I....1.... I .... I GPCR 5A Protein Sequence (645i GPCR 5B Protein Sequence (645i CAFGTCSSHICVI@AFYIPG FSYLMHRFGHHTVPKPVHILLSNIY LLPPALNPLIYG2 GPCR 5C Protein Sequence (6451 CAFGTCSSHICVILAFYIPGLFSYL0HRFGHHTVPKPVHILLSNIYLLLPPALNPLIYGA GPCR 5D Protein Sequence (A 07 CAFGTCSSHICVILAFYIPGLFSYLSHRFGHHTVPKPVHILLSNIYLLLPPALNPLIYGA
310 320
■•■•I....I....I....I
GPCR 5A Protein Sequence (645i RTKQIRDRLLETFTFRKSPL
GPCR 5B Protein Sequence (645i RTKQIRDRLLETFTFRKSPL
GPCR 5C Protein Sequence (6451 RTKQIRDRLLETFTFRKSPL
GPCR 5D Protein Sequence (A 07 RTKQIRDRLLETFTFRKSPL
GPCR5d is expressed in at least the following tissues: Apical microvilli of the retinal pigment epithelium, arterial (aortic), basal forebrain, brain, Burkitt lymphoma cell lines, corpus callosum, cardiac (atria and ventricle), caudate nucleus, CNS and peripheral tissue, cerebellum, cerebral cortex, colon, cortical neurogenic cells, endothelial (coronary artery and umbilical vein) cells, palate epithelia, eye, neonatal eye, frontal cortex, fetal hematopoietic cells, heart, hippocampus, hypothalamus, leulcocytes, liver, fetal liver, lung, lung lymphoma cell lines, fetal lymphoid tissue, adult lymphoid tissue, Those that express MHC II and III nervous, medulla, subthalamic nucleus, ovary, pancreas, pituitary, placenta, pons, prostate, putamen, serum, skeletal muscle, small intestine, smooth muscle (coronary artery in aortic) spinal cord, spleen, stomach, taste receptor cells of the tongue, testis, thalamus, and thymus tissue. This
information was derived by determining the tissue sources of the sequences that were included in the invention including but not limited to SeqCalling sources, Public EST sources, Literature sources, and/or RACE sources.
The similarity information for the GPCR5 proteins and nucleic acids disclosed herein suggest that GPCR5a-d may have important structural and/or physiological functions characteristic of the Olfactory Receptor family and the GPCR family. Therefore, the nucleic acids and proteins of the invention are useful in potential diagnostic and therapeutic applications and as a research tool. These include serving as a specific or selective nucleic acid or protein diagnostic and/or prognostic marker, wherein the presence or amount of the nucleic acid or the protein are to be assessed, as well as potential therapeutic applications such as the following: (i) a protein therapeutic, (ii) a small molecule drug target, (iii) an antibody target (therapeutic, diagnostic, drug targeting/cytotoxic antibody), (iv) a nucleic acid useful in gene therapy (gene delivery/gene ablation), and (v) a composition promoting tissue regeneration in vitro and in vivo (vi) biological defense weapon. The nucleic acids and proteins of the invention are useful in potential diagnostic and therapeutic applications implicated in various diseases and disorders described below and/or other pathologies. For example, the compositions of the present invention will have efficacy for treatment of patients suffering from: developmental diseases, MHCII and III diseases (immune diseases), Taste and scent detectability Disorders, Burkitt's lymphoma, Corticoneurogenic disease, Signal Transduction pathway disorders, Retinal diseases including those involving photoreception, Cell Growth rate disorders; Cell Shape disorders, Feeding disorders; control of feeding; potential obesity due to over-eating; potential disorders due to starvation (lack of appetite), noninsulin-dependent diabetes mellitus (NTDDMl), bacterial, fungal, protozoal and viral infections (particularly infections caused by HIV-1 or HIV-2), pain, cancer (including but not limited to Neoplasm; adenocarcinoma; lymphoma; prostate cancer; uterus cancer), anorexia, bulimia, asthma, Parkinson's disease, acute heart failure, hypotension, hypertension, urinary retention, osteoporosis, Crohn's disease; multiple sclerosis; and Treatment of Albright Hereditary Ostoeodystrophy, angina pectoris, myocardial infarction, ulcers, asthma, allergies, benign prostatic hypertrophy, and psychotic and neurological disorders, including anxiety, schizophrenia, manic depression, delirium, dementia, severe mental retardation.
Dentatorubro-pallidoluysian atrophy (DRPLA) Hypophosphatemic rickets, autosomal dominant
(2) Acrocallosal syndrome and dyskinesias, such as Huntington's disease or Gilles de la
Tourette syndrome and/or other pathologies and disorders of the like. The polypeptides can be used as immunogens to produce antibodies specific for the invention, and as vaccines. They
can also be used to screen for potential agonist and antagonist compounds. For example, a cDNA encoding the OR -like protein may be useful in gene therapy, and the OR-like protein may be useful when administered to a subject in need thereof. By way of nonlimiting example, the compositions of the present invention will have efficacy for treatment of patients suffering from bacterial, fungal, protozoal and viral infections (particularly infections caused by HIV-l or HIV-2), pain, cancer (including but not limited to Neoplasm; adenocarcinoma; lymphoma; prostate cancer; uterus cancer), anorexia, bulimia, asthma, Parkinson's disease, acute heart failure, hypotension, hypertension, urinary retention, osteoporosis, Crohn's disease; multiple sclerosis; and Treatment of Albright Hereditary Ostoeodystrophy, angina pectoris, myocardial infarction, ulcers, asthma, allergies, benign prostatic hypertrophy, and psychotic and neurological disorders, including anxiety, schizophrenia, manic depression, delirium, dementia, severe mental retardation and dyskinesias, such as Huntington's disease or Gilles de la Tourette syndrome and/or other pathologies and disorders.
The novel nucleic acid encoding OR-like protein, and the OR-like protein of the invention, or fragments thereof, may further be useful in diagnostic applications, wherein the presence or amount of the nucleic acid or the protein are to be assessed. The novel nucleic acid encoding GPCR-like protein, and the GPCR-like protein of the invention, or fragments thereof, may further be useful in diagnostic applications, wherein the presence or amount of the nucleic acid or the protein are to be assessed. These materials are further useful in the generation of antibodies that bind immunospecifically to the novel substances of the invention for use in therapeutic or diagnostic methods. These antibodies may be generated according to methods known in the art, using prediction from hydrophobicity charts, as described in the "Anti- GPCRX Antibodies" section below. For example the disclosed GPCR5a and 5d proteins have multiple hydrophilic regions, each of which can be used as an immunogen. In one embodiment, a contemplated GPCR5a and 5d epitope is from about amino acids 170 to 180. h another embodiment, a GPCR5a and 5d epitope is from about amino acids 230 to 245. In additional embodiments, GPCR5a and 5d epitopes are from about amino acids 260 to 280 and about amino acids 290-320. This novel protein also has value in development of powerful assay system for functional analysis of various human disorders, which will help in understanding of pathology of the disease and development of new drug targets for various disorders.
GPCR6
GPCR6a
A disclosed novel GPCR6a nucleic acid of 1201 nucleotides (also referred to as 645i_8D) is shown in Table 6A. An open reading frame begins with an ATG initiation codon at nucleotides 111-113 and ends with a TGA codon at nucleotides 1104-1106. A putative untranslated region upstream from the initiation codon and downstream from the termination codon are underlined in Table 6 A, and the start and stop codons are in bold letters.
Table 6A. GPCR6a Nucleotide Sequence (SEQ ID NO:25)
6AGAGTCCAGCAATTTAGAAATCCAT6CTGTCTTTTAT6ATATTTATACATATTCGGGAGTGCTTTCTGTTTCTATCA CTTTCAGTAGACTCATCAGCTTCTAAGTCATTATGGAGAGGACCATAAAGTGCATCATGAGTCACACCAATGTTACCA TCTTCCATCCTGCAGTTTTTGTCCTTCCTGGCATCCCTGGGTTGGAGGCTTATCACATTTGGCTGTCAATACCTCTTT GCCTCATTTACATCACTGCAGTCCTGGGAAACAGCATCCTGATAGTGGTTATTGTCATGGAACGTAACCTTCATGTGC CCATGTATTTCTTCCTCTCAATGCTGGCCGTCATGGACATCCTGCTGTCTACCACCACTGTGCCCAAGGCCCTAGCCA TCTTTTGGCTTCAAGCACATAACATTGCTTTTGATGCCTGTGTCACCCAAGGCTTCTTTGTCCATATGATGTTTGTGG GGGAGTCAGCTATCCTGTTAGCCATGGCCTTTGATCGCTTTGTGGCCATTTGTGCCCCACTGAGATATACAACAGTGC TAACATGGCCTGTTGTGGGGAGGATTGCTCTGGCCGTCATCACCCGAAGCTTCTGCATCATCTTCCCAGTCATATTCT TGCTGAAGCGGCTGCCCTTCTGCCTAACCAACATTGTTCCTCACTCCTACTGTGAGCATATTGGAGTGGCTCGTTTAG CCTGTGCTGACATCACTGTTAACATTTGGTATGGCTTCTCAGTGCCCATTGTCATGGTCATCTTGGATGTTATCCTCA TCGCTGTGTCTTACTCACTGATCCTCCGAGCAGTGTTTCGTTTGCCCTCCCAGGATGCTCGGCACAAGGCCCTCAGCA CTTGTGGCTCCCACCTCTGTGTCATCCTTATGTTTTATGTTCCATCCTTCTTTACCTTATTGACCCATCATTTTGGGC GTAATATTCCTCAACATGTCCATATCTTGCTGGCCAATCTTTATGTGGCAGTGCCACCAATGCTGAACCCCATTGTCT ATGGTGTGAAGACTAAGCAGATACGTGAGGGTGTAGCCCACCGGTTCTTTGACATCAAGACTTGGTGCTGTACCTCCC CTCTGGGCTCATGAATCTTCATGTCCAGCTCCTTCAAGGAAGCTAGATGTGAAATACAGAGATTTCCTGGACTGAGAA GGCTTTTCTTCCTCTTCTTTCTATTTCATCT
The GPCR6a protein encoded by SEQ ID NO:25 has 331 amino acid residues, and is presented using the one-letter code in Table 6B (SEQ ID NO:26). The Psort profile for GPCR6a predicts that this sequence has a signal peptide and is likely to be localized at the plasma membrane with a certainty of 0.6400. The most likely cleavage site for a GPCR6a peptide is between amino acids 51 and 52, at: VLG-NS., based on the SignalP result. GPCR6a has a molecular weight of 37159.3.
Table 6B. Encoded GPCR6a protein sequence (SEQ ID NO:26). ERTIKCIMSHTNVTIFHPAVFVLPGIPGLEAYHIWLSIPLCLIYITAVLGNSILIWIVMERNLHVPMYFFLSM LAVMDILLSTTTVPKALAIF LQAHNIAFDACVTQGFFVHMMFVGESAILLAMAFDRFVAICAPLRYTTVLTWPV VGRIALAVITRSFCIIFPVIFLLKRLPFCLTNIVPHSYCEHIGVARLACADITVNIWYGFSVPIVMVILDVILIA SYSLILRAVFRLPSQDARHKALSTCGSHLCVILMFYVPSFFTLLTHHFGRNIPQHVHILLANLYVAVPP LNPI VYGVKTKQIREGVAHRFFDIKTWCCTSPLGS
The amino acid sequence of GPCR6a had high homology to other proteins as shown in Table 6C.
Table 6C. BLASTX results for GPCR6a
Smallest
Sum
Reading High Prob
Sequences producing High-scoring Segment Pairs: Frame Score P(N) N patp:AAY92365 G protein-coupled receptor protein 5 - H... +3 766 3.2e-75 1 patp:AA 01730 Human G-protein receptor HPRAJ70 - Homo . . +3 751 1.3e-73 1 patp:AA 56641 G-protein coupled prostate tissue recept. . +3 751 1.3e-73 1 patp:AAR27876 Odorant receptor clone 115 - Rattus ratt. . +3 485 1.9e-45 1 patp:AAR27875 Odorant receptor clone 114 - Rattus ratt. . +3 477 1.4e-44 1 patp:AAR27874 Odorant receptor clone 19 - Rattus rattu. . +3 465 2.6e-43 1 patp:AAB68885 Human RECAP polypeptide, SEQ ID NO: 15 -. . +3 454 3.7e-42 1
GPCR6b, c and d
In the present invention, the target sequence identified previously, Accession Number 645i8_D, was subjected to the exon linking process to confirm the sequence. PCR primers were designed by starting at the most upstream sequence available, for the forward primer, and at the most downstream sequence available for the reverse primer. In each case, the sequence was examined, walking inward from the respective termini toward the coding sequence, until a suitable sequence that is either unique or highly selective was encountered, or, in the case of the reverse primer, until the stop codon was reached. Such suitable sequences were then employed as the forward and reverse primers in PCR amplifications based on a library containing a wide range of cDNA species. The resulting two amplicons were gel purified, cloned and sequenced to high redundancy to provide the sequences reported below, which are designated GPCR6b (Accession Numbers 6458iD_dal), GPCR6c (CG50231-01) and GPR6d (CG50171-01).
GPCR6b
A disclosed novel GPCR6b nucleic acid of 1004 nucleotides (also referred to as 6458iD_dal) is shown in Table 6D. An open reading frame begins with an ATG initiation codon at nucleotides 5-7 and ends with a TGA codon at nucleotides 998-1000. A putative untranslated region upstream from the initiation codon and downstream from the termination codon are underlined in Table 6D, and the start and stop codons are in bold letters.
Table 6D. GPCR6b Nucleotide Sequence (SEQ ID NO-.27)
CATTATGGAGAGGACCATAAAGTGCATCATGAGTCACACCAATGTTACCATCTTCCATCCTGCAGTTTTTGTCCTTCC TGGCATCCCTGGGGTGGGGGCTTATCACATTTGGCTGTCAATACCTCTTTGCCTCATTTACATCACTGCAGTCCTGGG AAACAGCATCCTGATAGTGGTTATTGTCATGGAACGTAACCTTCATGTGCCCATGTATTTCTTCCTCTCAATGCTGGC CGTCATGGACATCCTGCTGTCTACCACCACTGTGCCCAAGGCCCTAGCCATCTTTTGGCTTCAAGCACATAACATTGC TTTTGATGCCTGTGTCACCCAAGGCTTCTTTGTCCATATGATGTTTGTGGGGGAGTCAGCTATCCTGTTAGCCATGGC CTTTGATCGCTTTGTGGCCTTTTGTGCCCCACTGAGATATACAACAGTGCTAACATGGCCTGTTGTGGGGAGGATTGC TCTGGCCGTCATCACCCGAAGCTTCTGCATCATCTTCCCAGTCATATTCTTGCTGAAGCGGCTGCCCTTCTGCCTAAC CAACATTGTTCCTCACTCCTACTGTGAGCATATTGGAGTGGCTCGTTTAGCCTGTGCTGACATCACTGTTAACATTTG GTATGGCTTCTCAGTGCCCATTGTCATGGTCATCTTGGATGTTATCCTCATCGCTGTGTCTTACTCACTGATCCTCCG AGCAGTGTTTCGTTTGCCCTCCCAGGATGCTCGGCACAAGGCCCTCAGCACTTGTGGCTCCCACCTCTGTGTCATCCT TATGTTTTATGTTCCATCCTTCTTTACCTTATTGACCCATCATTTTGGGCATAATATTCCTCAACATGTCCATATCTT GCTGGCCAATCTTTATGTGGCAGTGCCACCAATGCTGAACCCCATTGTCTATGGTGTGAAGACTAAGCAGATACGTGA GGGTGTAGCCCACCGGTTCTTTGACATCAAGACTTGGTGCTGTACCTCCCCTCTGGGCTCATGAATCT
The GPCR6b protein encoded by SEQ ID NO:27 has 331 amino acid residues, and is presented using the one-letter code in Table 6E (SEQ ID NO:28). The SignalP, Psort and/or Hydropathy profile for GPCR6b predict that GPCRόb has a signal peptide and is likely to be localized at the plasma membrane with a certainty of 0.6400. SignalP results predict that GPCRόb is cleaved between position 51 and 52 of SEQ ID NO:28, i.e., in the amino acid sequence VLG-NS. This is typical of this type of membrane protein. The molecular weight of GPCRόb is 37088.2 Daltons.
Table 6E. Encoded GPCR6b protein sequence (SEQ ID NO:28).
MERTIKCI SHTNVTIFHPAVFVLPGIPGVGAYHIWLSIPLCLIYITAVLGNSILIVVIVMERNLHVP YFFLSM LAV DILLSTTTVPKALAIF LQAHNIAFDACVTQGFFVH MFVGESAILLAMAFDRFVAFCAPLRYTTVLTWPV VGRIALAVITRSFCIIFPVIFLLKRLPFCLTNIVPHSYCEHIGVARLACADITVNIWYGFSVPIVMVILDVILIA VSYSLILRAVFRLPSQDARHKALSTCGSHLCVILMFYVPSFFTLLTHHFGHNIPQHVHILLANLYVAVPP LNPI VYGVKTKQIREGVAHRFFDIKT CCTSPLGS
BLASTP analysis of the best hits for alignments with GPCRόb are listed in Table 6F.
GPCRόc
The disclosed, novel GPCRδc nucleic acid of 1124 nucleotides (also referred to as
CG50231-01) is shown in Table 6G. An open reading frame begins with an ATG initiation
codon at nucleotides 90-92 and ends with a TGA codon at nucleotides 1059-1061. A putative untranslated region upstream from the initiation codon and downstream from the termination codon are underlined in Table 6G, and the start and stop codons are in bold letters.
Table 6G. GPCRόc Nucleotide Sequence (SEQ ID NO:29)
ATACATATTCGGGAGTGCTTTCTGTTTCTATCACTTTCAGTAGACTCATCAGCTTCTAAGTCATTATAGAGAGGACCA TAAAGTGCATCATGAGTCACACCAATGTTACCATCTTCCATCCTGCAGTTTTTGTCCTTCCTGGCATCCCTGGGTTGG AGGCTTATCACATTTGGCTGTCAATACCTCTTTGCCTCATTTACATCACTGCAGTCCTGGGAAACAGCATCCTGATAG TGGTTATTGTCATGGAACGTAACCTTCATGTGCCCATGTATTTCTTCCTCTCAATGCTGGCCGTCATGGACATCCTGC TGTCTACCACCACTGTGCCCAAGGCCCTAGCCATCTTTTGGCTTCAAGCACATAACATTGCTTTTGATGCCTGTGTCA CCCAAGGCTTCTTTGTCCATATGATGTTTGTGGGGGAGTCAGCTATCCTGTTAGCCATGGCCTTTGATCGCTTTGTGG CCTTTTGTGCCCCACTGAGATATACAACAGTGCTAACATGGCCTGTTGTGGGGAGGATTGCTCTGGCCGTCATCACCC GAAGCTTCTGCATCATCTTCCCAGTCATATTCTTGCTGAAGCGGCTGCCCTTCTGCCTAACCAACATTGTTCCTCACT CCTACTGTGAGCATATTGGAGTGGCTCGTTTAGCCTGTGCTGACATCACTGTTAACATTTGGTATGGCTTCTCAGTGC CCATTGTCATGGTCATCTTGGATGTTATCCTCATCGCTGTGTCTTACTCACTGATCCTCCGAGCAGTGTTTCGTTTGC CCTCCCAGGATGCTCGGCACAAGGCCCTCAGCACTTGTGGCTCCCACCTCTGTGTCATCCTTATGTTTTATGTTCCAT CCTTCTTTACCTTATTGACCCATCATTTTGGGCGTAATATTCCTCAACATGTCCATATCTTGCTGGCCAATCTTTATG TGGCAGTGCCACCAATGCTGAACCCCATTGTCTATGGTGTGAAGACTAAGCAGATACGTGAGGGTGTAGCCCACCGGT TCTTTGACATCAAGACTTGGTGCTGTACCTCCCCTCTGGGCTCATGAATCTTCATGTCCAGCTCCTTCAAGGAAGCTA GATGTGAAATACAGAGATTTCCTGGACTGAGA
The disclosed nucleic acid sequence for GPCRόc has 688 of 996 bases (69%) identical to a Mus musculus odorant receptor S18 mRNA (gb:GENBANK- ID:AF121975|acc:AF121975.1).
The GPCRόc protein encoded by SEQ ID NO:29 has 323 amino acid residues, and is presented using the one-letter code in Table 6H (SEQ ID NO:30). The SignalP, Psort and/or Hydropathy profile for GPCRόc predict that GPCRόc has a signal peptide and is likely to be localized at the plasma membrane with a certainty of 0.6400. The SignalP shows a signal sequence is coded for in the first 43 amino acids with a cleavage site between amino acids 43 and 44 of SEQ ID NO:30, i.e., in the amino acid sequence VLG-NS. This is typical of this type of membrane protein.
Table 6H. Encoded GPCR6c protein sequence (SEQ ID NO:30).
MSHTNVTIFHPAVFVLPGIPGLEAYHI LSIPLCLIYITAVLGNSILIWIVMERNLHVPMYFFLSMLAVMDILL STTTVPKALAIF LQAHNIAFDACVTQGFFVHMMFVGESAILLAMAFDRFVAFCAPLRYTTVLT PWGRIALAV ITRSFCIIFPVIFLLKRLPFCLTNIVPHSYCEHIGVARLACADITVNIWYGFSVPIVMVILDVILIAVSYSLILR AVFRLPSQDARHKALSTCGSHLCVILMFYVPSFFTLLTHHFGRNIPQHVHILLANLYVAVPPMLNPIVYGVKTKQ IREGVAHRFFDIKTWCCTSPLGS
The full amino acid sequence of the protein of the invention was found to have 220 of 315 amino acid residues (69%) identical to, and 254 of 315 amino acid residues (80%) similar to, the 321 amino acid residue ODORANT RECEPTOR SI 8 from Mus musculus (SPTREMBL-ACC:Q9 U89)
BLASTP results include those listed in Table 61.
Possible SNPs found for GPCRόc are listed in Table 6J.
GPCRόd
The disclosed novel GPCRόd nucleic acid of 1006 nucleotides (also referred to as CG50171-01) is shown in Table 6K. An open reading frame begins with an ATG initiation codon at nucleotides 6-8 and ends with a TGA codon at nucleotides 999-1001. A putative untranslated region upstream from the initiation codon and downstream from the termination codon are underlined in Table 6K, and the start and stop codons are in bold letters.
Table 6K. GPCRόd Nucleotide Sequence (SEQ ID NO:31)
TCATTATGGAGAGGACCATAAAGTGCATCATGAGTCACACCAATGTTACCATCTTCCATCCTGCAGTTTTTGTCCTTC CTGGCATCCCTGGGTTGGAGGCTTATCACATTTGGCTGTCAATACCTCTTTGCCTCATTTACATCACTGCAGTCCTGG GAAACAGCATCCTGATAGTGGTTATTGTCATGGAACGTAACCTTCATGTGCCCATGTATTTCTTCCTCTCAATGCTGG CCGTCATGGACATCCTGCTGTCTACCACCACTGTGCCCAAGGCCCTAGCCATCTTTTGGCTTCAAGCACATAACATTG CTTTTGATGCCTGTGTCACCCAAGGCTTCTTTGTCCATATGATGTTTGTGGGGGAGTCAGCTATCCTGTTAGCCATGG CCTTTGATCGCTTTGTGGCCTTTTGTGCCCCACTGAGATATACAACAGTGCTAACATGGCCTGTTGTGGGGAGGATTG CTCTGGCCGTCATCACCCGAAGCTTCTGCATCATCTTCCCAGTCATATTCTTGCTGAAGCGGCTGCCCTTCTGCCTAA CCAACATTGTTCCTCACTCCTACTGTGAGCATATTGGAGTGGCTCGTTTAGCCTGTGCTGACATCACTGTTAACATTT GGTATGGCTTCTCAGTGCCCATTGTCATGGTCATCTTGGATGTTATCCTCATCGCTGTGTCTTACTCACTGATCCTCC GAGCAGTGTTTCGTTTGCCCTCCCAGGATGCTCGGCACAAGGCCCTCAGCACTTGTGGCTCCCACCTCTGTGTCATCC
TTATGTTTTATGTTCCATCCTTCTTTACCTTATTGACCCATCATTTTGGGCATAATATTCCTCAACATGTCCATATCT TGCTGGCCAATCTTTATGTGGCAGTGCCACCAATGCTGAACCCCATTGTCTATGGTGTGAAGACTAAGCAGATACGTG AGGGTGTAGCCCACCGGTTCTTTGACATCAAGACTTGGTGCTGTACCTCCCCTCTGGGCTCATGAATCTA
The disclosed nucleic acid sequence for GPCRόd has 680 of 984 bases (69%) identical to a Mus musculus odorant receptor S18 mRNA (gb:GENBANK- ID:AF121975|acc:AF121975.1).
The GPCRόd protein encoded by SEQ ID NO:31 has 331 amino acid residues, and is presented using the one-letter code in Table 6L (SEQ ID NO:32). The SignalP, Psort and/or Hydropathy profile for GPCRόd predict that GPCRόd has a signal peptide and is likely to be localized at the plasma membrane with a certainty of 0.6400. The most likely cleavage site for a peptide is between amino acids 51 and 52, at: VLG-NS, based on the SignalP result. This is typical of this type of membrane protein.
Table 6L. Encoded GPCRόd protein sequence (SEQ ID NO:32).
MERTIKCIMSHTNVTIFHPAVFVLPGIPGLEAYHI LSIPLCLIYITAVLGNSILIWIVMERNLHVPMYFFLSM LAVMDILLSTTTVPKALAIF LQAHNIAFDACVTQGFFVHMMFVGESAILLAMAFDRFVAFCAPLRYTTVLT PV VGRIALAVITRSFCIIFPVIFLLKRLPFCLTNIVPHSYCEHIGVARLACADITVNI YGFSVPIVMVILDVILIA VSYSLILRAVFRLPSQDARHKALSTCGSHLCVILMFYVPSFFTLLTHHFGHNIPQHVHILLANLYVAVPPMLNPI VYGVKTKQIREGVAHRFFDIKTWCCTSPLGS
The full amino acid sequence of the protein of the invention was found to have 221 of 318 amino acid residues (69%>) identical to, and 254 of 318 amino acid residues (79%) similar to, the 321 amino acid residue ODORANT RECEPTOR S 18 from Mus musculus (SPTREMBL-ACC:Q9WU89)
BLASTP results include those listed in Table 6M.
Possible SNPs found for GPCRόd are listed in Table 6N.
GPCRόa also has homology to the proteins shown in the BLASTP results in Table 6O.
The homology data shown above is represented graphically in a Clustal W shown in Table 6P.
Table 6P. ClustalW Analysis of GPCRόa
1) Novel GPCRόa (SEQ ID NO:26)
2) gi|9938014|reflNP_064686.1| odorant receptor S18 gene [Mus musculus] (SEQ ID NO:52)
3) gil7305349|reflNP_038647.1| olfactory receptor 61 [Mus musculus] (SEQ ID NO:54)
4) gi|l 190821 l|gb|AAG41676.1| (AF137396) HOR 5'Betal4 [Homo sapiens] (SEQ ID NO:53)
5) gi|6532001|gb|AAD27596.2|AF121976_l (AF121976) odorant receptor S19 [Mus musculus] (SEQ ID NO:55)
6) gi|9935442|reflNP )64688.1| odorant receptor S46 gene [Mus musculus] (SEQ ID NO:56)
Table 6Q lists the domain description from DOMAIN analysis results against GPCRόa. Tins indicates that the GPCRόa sequence has properties similar to those of other proteins known to contain this domain as well as to the 377 amino acid 7tm domain itself.
Table 6Q. Domain Analysis of GPCRόa crnllPfamlpfamOOOOl, 7tm_l, 7 transmembrane receptor (rhodopsin family) (SEQ ID NO: 65) Length: 254 Score = 114 bits (284), Expect = le-26
10 20 30 40 50 60
....|....)....|....|....|....|....)....|....|....|....|....| GPCR 6A raSlL^OTV ER fflHV|M Fffl|SMI^Λl| lffl S πτvra ^AIF LQAH I FD fflvTQ
7 transmembrane receptor (rhod IMτ, H fflLR Kκ'Rτ"τ T™τ, l^ABL" T "τ, ^ιw™YYLV6 D VF DAL"κLV
70 80 90 100 110 120
....|....|....|....|....|....1....|....|....|....|....|....| GPCR 6A BFFVHMMFVGESAlBLΘlXlAFraFVSHCΑj^^TTVLSjWPVVGRΪAfflδ lTRSFClIFraVΪ
7 transmembrane receptor (rhod ^L VWGYASI lBTgISlExi.EvH,^SRRIR|3 RRAKVlιlB] VLA LLS gPI(
130 140 150 160 170 180
....|....|....|....|....|....|....|....|....1....|....|....|
GPCR 6A F KRLPFCLTNIVPHSYCEHI GJBAKEIACADITHN — IWHGFSVPIB VIBDVIIIH
7 transmembrane receptor (rhod LFS R TVEEGHT TgCLIDFPEESgK — RSgVL STIiHGF gPIiL g
GPCR 6A
7 transmembrane receptor (rhod
250 260 270 280 290 300
....|....|....|....|....|....|....|....|....1....|....|....|
GPCR 6A D Rlβ STCGS
7 transmembrane receptor (rhod RSLKRRΞs F.RlSWAKMT .v
GPCR 6A
7 transmembrane receptor (rhod
Homologies between the GPCRό variants is shown in a Clustal W in Table 6R.
Table 6R. Clustal W of GPCRό family
10 20 30 40 50 60 • I.. • I..
GPCR 6A Protein Sequence (645i ΪERTIKCIMSHTNVTIFHPAVFVLPGIPG EAYHIW SIPLC IYITAVLGNSI IVVIV GPCR 6B Protein Sequence (6458 IERTIKCIMSHTNVTIFHPAVFVLPGIPGS5@AYHIWLSIP CLIYITAVLGNSILIVVIV GPCR 6C Protein Sequence (CG50 4SHTNVTIFHPAVFVLPGIPG EAYHIWLSIP CLIYITAV GNSI IVVI1 GPCR 6D Protein Sequence (CG50 IERTIKCIMSHTNVTIFHPAVFV PGIPG EAYHIWLSIP CLIYITAVLGNSILIVVI\I
70 80 90 100 110 120 .1.. .. I .. .. I ..
GPCR 6A Protein Sequence (645i 1ERNLHVPMYFFLSMLAVMDILLSTTTVPKALAIFWLQAHNIAFDACVTQGFFVHMMFV
GPCR 6B Protein Sequence (6458 lERNLHVPMYFFLSMLAVMDILLSTTTVPKALAIFWLQAHNIAFDACVTQGFFVHMMFVl
GPCR 6C Protein Sequence (CG50 iffiRN HVPMYFFLSMLAVMDI LSTTTVP A AIFWLQAHNIAFDACVTQGFFVHMMFVC
GPCR 6D Protein Sequence (CG50 4ERNLHVPMYFFLSMLAVMDILLSTTTVPKALAIFWLQAHNIAFDACVTQGFFVHMMFVC
130 140 150 160 170 180 ..|.. ..|.. ..|.. ..I
GPCR 6A Protein Sequence (645i JSAIL AMAFDRFVftJJCAP RXTTV T PVVGRIALAVITRSFCIIFPVIFLLKR PFC
GPCR 6B Protein Sequence (6458 ΪSAI LAMAFDRFVAFCAPLRYTTV TWPVVGRIALAVITRSFCIIFPVIFLLKR PFCL
GPCR 6C Protein Sequence (CG50 5SAILLAMAFDRFVAFCAPLRYTTVLTWPVVGRIALAVITRSFCIIFPVIF LKRLPFCL
GPCR 6D Protein Sequence (CG50 ΪSAI LAMAFDRFVAFCAPLRYTTVLTWPVVGRIALAVITRSFCIIFPVIFLLKRLPFCL
190 200 210 220 230 240
1 ..|.. ..1.. ..I .. ..I .. ..I
GPCR 6A Protein Sequence (645i TNIVPHSYCEHIGVARACADITVNIWYGFSVPIVMVI DVILIAVSYS ILRAVFRLPΞ
GPCR 6B Protein Sequence (6458 TNIVPHSYCEHIGVARLACADITVNIWYGFSVPIVMVILDVILIAVSYSLILRAVFRLPΞ
GPCR 6C Protein Sequence (CG50 TNIVPHSYCEHIGVARACADITVNIWYGFSVPIVMVILDVILIAVSYS ILRAVFR PS
GPCR 6D Protein Sequence (CG50 ΓNIVPHSYCEHIGVARLACADITVNIWYGFSVPIVMVILDVILIAVSYSLI RAVFR PS
250 260 270 280 290 300 ..|.. ..| .. ..| ..
GPCR 6A Protein Sequence (645i QDARH ALSTCGSHLCVILMFYVPSFFTL THHFG3NIPQHVHILI.ANLYVAVPPMLNPI
GPCR 6B Protein Sequence (6458 QDARHKALSTCGSH CVILMFYVPSFFT LTHHFGGNIPQHVHI AN YVAVPPM NPI
GPCR 6C Protein Sequence (CG50 QDARHKALSTCGSHLCVILMFYVPSFFTL THHFG|NIPQHVHILLANLYVAVPPMLNPI
GPCR 6D Protein Sequence (CG50 QDARHKALSTCGSHLCVI MFYVPSFFTLLTHHFGSNIPQHVHILLΆNLYVAVPPMLNPI
310 320 330 ..|.. ..I.
GPCR 6A Protein Sequence (645i fYGVKTKQIREGVAHRFFDIKTWCCTSPLGS
GPCR 6B Protein Sequence (6458 nTGVKTKQIREGVAHRFFDIKTWCCTSPLGS
GPCR 6C Protein Sequence (CG50 7YGVKTKQIREGVAHRFFDIKTWCCTSPLGS
GPCR 6D Protein Sequence (CG50 7YGVKTKQIREGVAHRFFDIKT CCTSPLGS
Because of the close homology among the members of the GPCRό family, proteins that are homologous to any one member of the family are also largely homologous to the other members except where the sequences are different as shown above in Table 6R.
GPCRό is expressed in at least the following tissues: Apical microvilli of the retinal pigment epithelium, arterial (aortic), basal forebrain, brain, Burkitt lymphoma cell lines, corpus callosum, cardiac (atria and ventricle), caudate nucleus, CNS and peripheral tissue, cerebellum, cerebral cortex, colon, cortical neurogenic cells, endothelial (coronary artery and umbilical vein) cells, palate epithelia, eye, neonatal eye, frontal cortex, fetal hematopoietic cells, heart, hippocampus, hypothalamus, leukocytes, liver, fetal liver, lung, lung lymphoma cell lines, fetal lymphoid tissue, adult lymphoid tissue, Those that express MHC II and III nervous, medulla, subthalamic nucleus, ovary, pancreas, pituitary, placenta, pons, prostate, putamen, serum, skeletal muscle, small intestine, smooth muscle (coronary artery in aortic) spinal cord, spleen, stomach, taste receptor cells of the tongue, testis, thalamus, and thymus tissue. This information was derived by determining the tissue sources of the sequences that were included in the invention including but not limited to SeqCalling sources, Public EST sources, Literature sources, and/or RACE sources.
The similarity information for the GPCRό proteins and nucleic acids disclosed herein suggest that GPCR6a-d may have important structural and/or physiological functions characteristic of the Olfactory Receptor family and the GPCR family. Therefore, the nucleic acids and proteins of the invention are useful in potential diagnostic and therapeutic applications and as a research tool. These include serving as a specific or selective nucleic acid or protein diagnostic and/or prognostic marker, wherein the presence or amount of the nucleic acid or the protein are to be assessed, as well as potential therapeutic applications such as the following: (i) a protein therapeutic, (ii) a small molecule drug target, (iii) an antibody target (therapeutic, diagnostic, drug targeting/cytotoxic antibody), (iv) a nucleic acid useful in gene therapy (gene delivery/gene ablation), and (v) a composition promoting tissue regeneration in vitro and in vivo (vi) biological defense weapon.
The nucleic acids and proteins of the invention are useful in potential diagnostic and therapeutic applications implicated in various diseases and disorders described below and/or other pathologies. For example, the compositions of the present invention will have efficacy for treatment of patients suffering from: developmental diseases, MHCII and III diseases
(immune diseases), Taste and scent detectability Disorders, Burkitt's lymphoma, Corticoneurogenic disease, Signal Transduction pathway disorders, Retinal diseases including those involving photoreception, Cell Growth rate disorders; Cell Shape disorders, Feeding disorders; control of feeding; potential obesity due to over-eating; potential disorders due to starvation (lack of appetite), noninsulin-dependent diabetes mellitus (NIDDMl), bacterial, fungal, protozoal and viral infections (particularly infections caused by HIV-l or HIV-2), pain, cancer (including but not limited to Neoplasm; adenocarcinoma; lymphoma; prostate cancer; uterus cancer), anorexia, bulimia, asthma, Parkinson's disease, acute heart failure, hypotension, hypertension, urinary retention, osteoporosis, Crohn's disease; multiple sclerosis; and Treatment of Albright Hereditary Ostoeodystrophy, angina pectoris, myocardial infarction, ulcers, asthma, allergies, benign prostatic hypertrophy, and psychotic and neurological disorders, including anxiety, schizophrenia, manic depression, delirium, dementia, severe mental retardation. Dentatorubro-pallidoluysian atrophy (DRPLA) Hypophosphatemic rickets, autosomal dominant (2) Acrocallosal syndrome and dyskinesias, such as Huntington's disease or Gilles de la Tourette syndrome and/or other pathologies and disorders of the like. The polypeptides can be used as immunogens to produce antibodies specific for the invention, and as vaccines. They can also be used to screen for potential agonist and antagonist compounds. For example, a cDNA encoding the OR -like protein may be useful in gene therapy, and the OR-like protein may be useful when administered to a subject in need thereof. By way of nonlimiting example, the compositions of the present invention will have efficacy for treatment of patients suffering from bacterial, fungal, protozoal and viral infections (particularly infections caused by HIV-l or HIV-2), pain, cancer (including but not limited to Neoplasm; adenocarcinoma; lymphoma; prostate cancer; uterus cancer), anorexia, bulimia, asthma, Parkinson's disease, acute heart failure, hypotension, hypertension, urinary retention, osteoporosis, Crohn's disease; multiple sclerosis; and Treatment of Albright Hereditary Ostoeodystrophy, angina pectoris, myocardial infarction, ulcers, asthma, allergies, benign prostatic hypertrophy, and psychotic and neurological disorders, including anxiety, schizophrenia, manic depression, delirium, dementia, severe mental retardation and dyskinesias, such as Huntington's disease or Gilles de la Tourette syndrome and/or other pathologies and disorders. The novel nucleic acid encoding GPCR-like protein, and the GPCR-like protein of the invention, or fragments thereof, may further be useful in diagnostic applications, wherein the presence or amount of the nucleic acid or the protein are to be assessed. These materials are further useful in the generation of antibodies that bind immunospecifically to the novel substances of the invention for use in therapeutic or diagnostic methods. These antibodies may
be generated according to methods known in the art, using prediction from hydrophobicity charts, as described in the "Anti-GPCRX Antibodies" section below. For example the disclosed GPCR6a-d proteins have multiple hydrophilic regions, each of which can be used as an immunogen. h one embodiment, contemplated GPCR6a-d epitopes are from about amino acids 225 to 245. In another embodiment, GPCR6a-d epitopes are from about amino acids 260 to 270. In additional embodiments, GPCR6a-d epitopes are from amino acids 275 to 280 and 290 to 320. These novel proteins also have value in the development of powerful assay system for functional analysis of various human disorders, which will help in understanding of pathology of the disease and development of new drug targets for various disorders.
GPCR7
A disclosed novel GPCR7 nucleic acid of 1102 nucleotides (also referred to as 645i8_E) is shown in Table 7 A. An open reading frame begins with an ATG initiation codon at nucleotides 69-71 and ends with a TAG codon at nucleotides 954-956. A putative untranslated region upstream from the initiation codon and downstream from the termination codon are underlined in Table 7A, and the start and stop codons are in bold letters.
Table 7A. GPCR7 Nucleotide Sequence (SEQ ID NO:33)
CTAACCACAGTCTTATTCT6TTTCCACTTAATCCACAGGCA6ATGCTGA6AACAAAACATCTCAT6ATATGGATAT TGGCCTGAGTATAGCCAATAGCTCAGGGTTTCAACTGTCTGAGTTCATTCTGATAGGGTTCCCAGGCATTCATGAG TGGCAGCACTGGCTCTCCCTGCCCTTAGCTCTTGGTGCCAATCTCCTCATCATAATCACCATTCAACATGAGACCA TGCTACATGAACCCATGTACCATTTGCTGGGCATATTAGCAGTGGTGGACATTGGCCTGGCCACCACCATCATGCC CAAGATCCTGGCCATCTTCTGGTTTGATGCCAAGGCCATCAGCCTCCCTGAGTGTTTTGCTCAGATCTATGCCATC CACTCTTTCATGTGCATGGAGTCAGGCATCTTCCTCTGCATGGCAGTGGATAGATATATGGCCATTTGTTATCCCC TTCAGTACACTTCCATAGTTACTGAAGCTTTTGTCATCAAAGCCACACTGTCAGTAGTGCTCAGGAATGGCCTGTT GACCATCCCAGTGCCAGTATTGGCTGCCCAGCGACACTACTGCTCCAGGAATGAGATTGATCAGTGCCTCTGCTCT AACTTGGGGGTCACAAGTCTGGCCTGTGATGACACCACTATTAACAGGTTTTACCAGCTGGCCTTGGTCTGGGTTG TGGTTGGGAGTGACATGGGTCTGGTCTTTGCTTCCTATTCTTTGATTATTCACTCAGTGCTGAAGCTGAACTCTGC TAAAGCAACATCTAAGGCCCTGAATACCTGCAGCTCTCACCTTATCCTCATTCTCTTTTTCTACACAGCCTATTAT TGTAGTATCTGTCACCACCTGGCAGGAAGAAGGGCTCCCCGCATCCCTGTTCTCCTCAATGTGCTGCATATTGTCA TCCCCTCAGCCCTTAACCCCATAGTATATGCCCTTAGGACCTAGGAGCTGAGAGCGGGCTTCCAGAAGCTGCTTGG TTTGGGCGAGTATGTGTCCAGGAAGTGAGCTAACTTTAAATAGCAGAATATTATATGCAATGCTGAAGAAAGAGCA CAGTCTTTGGAGTCCAACACACCTTGGTTAAAATTCCA
The GPCR7 protein encoded by SEQ ID NO:33 has 295 amino acid residues, and is presented using the one-letter code in Table 7B (SEQ ID NO:34). The Psort profile for GPCR7 predicts that this sequence has a signal peptide and is likely to be localized at the plasma membrane with a certainty of 0.6000. GPCR7 has a molecular weight of 32704.3.
Table 7B. Encoded GPCR7 protein sequence (SEQ ID NO:34).
MDIGLSIANSSGFQLSEFILIGFPGIHE QHWLSLPLALGANLLIIITIQHETM HEPMYHLLGILAWDIG ATT
I PKILAIFWFDAKAISLPECFAQIYAIHSFMCMESGIFLCMAVDRYMAICYPLQYTSIVTEAFVIKATLSVVLRN GLLTIPVPVLAAQRHYCSRNEIDQC CSNLGVTSLACDDTTINRFYQLALV VWGSDMGLVFASYSLIIHSVLK NSAKATSKALNTCSSHLILILFFYTAYYCSICHHLAGRRAPRIPVLLNVLHIVIPSALNPIVYALRT
The amino acid sequence of GPCR7 had high homology to other proteins as shown in Table 7C.
Table 7C. BLASTX results for GPCR7
Smallest
Sum
Reading High Prob
Sequences producing High-scoring Segment Pairs: Frame Score P(N) N patp:AAY92365 G protein-coupled receptor protein 5 - H +3 568 le-54 patp:AAW01730 Human G-protein receptor HPRAJ70 - Homo +3 520 8e-49 patp:AAW56641 G-protein coupled prostate tissue recept +3 520 8e-49 patp:AAR27868 Odorant receptor clone F5 - Rattus rattu +3 375 8e-34 patp:AAY90874 Human G protein-coupled receptor GTAR14- +3 373 4e-33 patp:AAR27872 Odorant receptor clone 17 - Rattus rattu +3 369 8e-33 patp:AAY90873 Human G protein-coupled receptor GTAR14- +3 368 9e-33
GPCR7 also has strong homology to the following proteins shown in the BLASTP data in Table 7D.
The homology is displayed graphically in the Clustal W shown in Table 7E.
Table 7E. ClustalW Analysis of GPCR7
1) Novel GPCR7 (SEQ ID NO:34)
2) gill2704541|gblAAK00590.11 (AY026034) olfactory receptor S83 [Mus musculus] (SEQ ID NO:51)
3). gil73053491reflNP_038647.ll olfactory receptor 67 [Mus musculus] (SEQ ID NO:54) 4) gi|99354421reflNP_064688.11 odorant receptor S46 gene [Mus musculus] (SEQ ID NO:56)
5) gi|9938014|reflNP 064686.11 odorant receptor Si 8 gene [Mus musculus] (SEQ ID NO:52)
6) gi|3420759]gbl AAD 12761.11 (AF079864) putative G-protein coupled receptor RAlc [Rattus norvegicus] (SEQ ID NO:59)
Table 7F lists the domain description from DOMAIN analysis results against GPCR7.
This indicates that the GPCR7 sequence has properties similar to those of other proteins known to contain this domain as well as to the 377 amino acid 7tm domain itself.
Table 7F. Domain Analysis of GPCR7 gnl I PfamlpfamOOOOl, 7tm_l, 7 transmembrane receptor (rhodopsin family) (SEQ ID NO: 65) Length: 254 Score = 67.4 bits (163), Expect = le-12
10 20 30 40 50 60
....|....|....|....|....|....|....|....|....|....|....|....| GPCR 7 ABBIJJITHGHETMHH — EBMYHLfflGlH^BvffllGLATHIMHκifflΑIFWFDAKAIS -PEffl
7 transmembrane receptor (rhod GIMVBIΛ/HT.RTKKHR — τla lBfl MlB6 9lι Ll tιpB flBγγ VGGD V GD-ALB
70 80 90 100 110 120
....|....|....|....|....|....|....|....|....|....|....|....| GPCR 7 FAQIYAIHSFMCMESGlFLCMAraMigC glQHTSnvnE FVIKATBsfflviiRNG[gTI
7 transmembrane receptor (rhod K VG LFVVNGY SILiLT ISlHLιSVHS^R^^PRI^κvtIBLiawVLALEsi<
130 140 150 160 170 180
....|....|....|....|....|....|....1....|....|....|....|....|
GPCR 7 [vPVLAAQgHYCSRlSEI-DQCLJ8sN GVTSLACDDTTΪNRF YQlTflfflvWVVfflGSD Gffl
7 transmembrane receptor (rhod gPLlJFSWLgTVEEGl TT HLIDFPEESVKRSYVliLSTLVGFVap|lι I BcYTRIH
190 200 210 220 230 240
....|....|....1....|....|....|....|....|....|....|....|....|
GPCR 7 VFASYSLIIH
7 transmembrane receptor (rhod RTLRKRARSQ
310 320 330 340 350 360
....|....1....|....|....|....|....1....| ....|....|....|....|
GPCR 7 ATSKgLNTCSSHIiILlfflFFY TAYYC|ϊfflHHffl&GRgAPRIPVι lιNV' HIVIP&
7 transmembrane receptor (rhod ERJC lKM LVVVV^FVHC^ PYHIVL LDgLS-iall Sv PT lBl W YVNgCji
GPCR 7
7 transmembrane receptor (rhod jjl£
The similarity information for the GPCR7 protein and nucleic acid disclosed herein suggest that GPCR7 may have important structural and/or physiological functions characteristic of the Olfactory Receptor family and the GPCR family. Therefore, the nucleic acids and proteins of the invention are useful in potential diagnostic and therapeutic applications and as a research tool. These include serving as a specific or selective nucleic acid or protein diagnostic and/or prognostic marker, wherein the presence or amount of the nucleic acid or the protein are to be assessed, as well as potential therapeutic applications such as the following: (i) a protein therapeutic, (ii) a small molecule drug target, (iii) an antibody target (therapeutic, diagnostic, drug targeting/cytotoxic antibody), (iv) a nucleic acid useful in gene therapy (gene delivery/gene ablation), and (v) a composition promoting tissue regeneration in vitro and in vivo (vi) biological defense weapon. The novel nucleic acid encoding GPCR7, and the GPCR7 protein of the invention, or fragments thereof, may further be useful in diagnostic applications, wherein the presence or amount of the nucleic acid or the protein are to be assessed.
The disclosed GPCR7 nucleic acids and proteins of the invention are useful in potential therapeutic applications implicated in bacterial, fungal, protozoal and viral infections
(particularly infections caused by HTV-l or HIV-2), pain, cancer (including but not limited to
Neoplasm; adenocarcinoma; lymphoma; prostate cancer; uterus cancer), anorexia, bulimia, asthma, Parkinson's disease, acute heart failure, hypotension, hypertension, urinary retention,
osteoporosis, Crohn's disease; multiple sclerosis; and treatment of Albright Hereditary Ostoeodystrophy, angina pectoris, myocardial infarction, ulcers, asthma, allergies, benign prostatic hypertrophy, and psychotic and neurological disorders, including anxiety, schizophrenia, manic depression, delirium, dementia, severe mental retardation and dyskinesias, such as Huntington's disease or Gilles de la Tourette syndrome and/or other pathologies and disorders. For example, a cDNA encoding the GPCR-like protein may be useful in gene therapy, and the GPCR-like protein may be useful when administered to a subject in need thereof. By way of nonlimiting example, the compositions of the present invention will have efficacy for treatment of patients suffering from bacterial, fungal, protozoal and viral infections (particularly infections caused by HIV-l or HTV-2), pain, cancer (including but not limited to Neoplasm; adenocarcinoma; lymphoma; prostate cancer; uterus cancer), anorexia, bulimia, asthma, Parkinson's disease, acute heart failure, hypotension, hypertension, urinary retention, osteoporosis, Crohn's disease; multiple sclerosis; and Treatment of Albright Hereditary Ostoeodystrophy, angina pectoris, myocardial infarction, ulcers, asthma, allergies, benign prostatic hypertrophy, and psychotic and neurological disorders, including anxiety, schizophrenia, manic depression, delirium, dementia, severe mental retardation and dyskinesias, such as Huntington's disease or Gilles de la Tourette syndrome. The novel nucleic acid encoding GPCR-like protein, and the GPCR-like protein of the invention, or fragments thereof, may further be useful in diagnostic applications, wherein the presence or amount of the nucleic acid or the protein are to be assessed. These materials are further useful in the generation of antibodies that bind immunospecifically to the novel substances of the invention for use in therapeutic or diagnostic methods.
The novel nucleic acid encoding the GPCR-like protein of the invention, or fragments thereof, may further be useful in diagnostic applications, wherein the presence or amount of the nucleic acid or the protein are to be assessed. These materials are further useful in the generation of antibodies that bind immunospecifically to the novel substances of the invention for use in therapeutic or diagnostic methods. These antibodies may be generated according to methods known in the art, using prediction from hydrophobicity charts, as described in the "Anti-GPCRX Antibodies" section below. The disclosed GPCR7 protein has multiple hydrophilic regions, each of which can be used as an immunogen. In one embodiment, a contemplated GPCR7 epitope is from about amino acids 160 to 180. In another embodiment, a GPCR7 epitope is from about amino acids 185 to 195. hi additional embodiments, GPCR7 epitopes are from about amino acids 230 to 240 and from about amino acids 260 to 270.
GPCR8
GPCR8a
A disclosed novel GPCR8a nucleic acid of 1201 nucleotides (also referred to as 17e20) is shown in Table 8A. An open reading frame begins with an ATG initiation codon at nucleotides 105-107 and ends with a TGA codon at nucleotides 1053-1055. Aputative untranslated region upstream from the initiation codon and downstream from the termination codon are underlined in Table 8A, and the start and stop codons are in bold letters.
Table 8A. GPCR8a Nucleotide Sequence (SEQ ID NO:35)
AGTATTTTCCTAACTAAATGAATATCACACTTTATTCATTTCTTTATGTCCTCAGATTAAATGAAGGGAAT AGTACTGGAAGAAATATAGATGAAAGAAAGAAAATGCAAGGAGAAAACTTCACCATTTGGAGCATTTTTTT CTTGGAGGGATTTTCCCAGTACCCAGGGTTAGAAGTGGTTCTCTTCGTCTTCAGCCTTGTAATGTATCTGA CAACGCTCTTGGGCAACAGCACTCTTATTTTGATCACTATCCTAGATTCACGCCTTAAAACCCCCATGTAC TTATTCCTTGGAAATCTCTCTTTCATGGATATTTGTTACACATCTGCCTCTGTTCCTACTTTGCTGGTGAA CTTGCTGTCATCCCAGAAAACCATTATCTTTTCTGGGTGTGCTGTACAGATGTATCTGTCCCTTGCCATGG GCTCCACAGAGTGTGTGCTCCTGGCCGTGATGGCATATGACCGTTATGTGGCCATTTGTAACCCGCTGAGA TACTCCATCATCATGAACAGGTGCGTCTGTGCACGGATGGCCACGGTCTCCTGGGTGACGGGTTGCCTGAC CGCTCTGCTGGAAACCAGTTTTGCCCTGCAGATACCCCTCTGTGGGAATCTCATCGATCACTTCACGTGTG AAATTCTGGCGGTGCTAAAGTTAGCTTGCACAAGTTCACTGCTCATGAACACCATCATGCTGGTGGTCAGC ATTCTCCTCTTGCCAATTCCAATGCTCTTAGTTTGCATCTCTTACATCTTCATCCTTTCCACTATTCTGAG AATCACCTCAGCAGAGGGAAGAAACAAGGCTTTTTCTACCTGTGGTGCCCATTTGACTGTGGTGATTTTGT ATTATGGGGCTGCCCTCTCTATGTACCTAAAGCCTTCTTCATCAAATGCACAAAAAATAGACAAAATCATC TCGTTGCTTTACGGAGTGCTTACCCCTATGTTGAACCCCATAATTTACAGTTTAAGAAACAAGGAAGTCAA AGATGCTATGAAGAAATTGCTGGGCAAAATAACATTGCATCAAACACACGAACATCTCTGATTGGGTCCCT ATGGTTTTACCAGAGATGTGCCCCTGGCAGAGCTCATCAGAGAAATTCGAGACAACATACAACCTCTTAGA ACTCTGATCGGATCTTATCTCTATATAATTTCACAGTTATGAGCTGCATACACAGAGTGATTGCC
The GPCR8a protein encoded by SEQ ID NO:35 has 345 amino acid residues, and is presented using the one-letter code in Table 8B (SEQ ID NO:36). The Psort profile for GPCR8a predicts that this sequence has a signal peptide and is likely to be localized at the plasma membrane with a certainty of 0.8000. The most likely cleavage site for a GPCR8a peptide is between amino acids 21 and 22, at: STG-RN., based on the SignalP result. GPCRδa has a molecular weight of 38472.5.
Table 8B. Encoded GPCR8a protein sequence (SEQ ID NO:36)
MNITLYSFLYVLRLNEGNSTGRNIDERKKMQGENFTIWSIFFLEGFSQYPGLEWLFVFSLV YLTTLLGN STLILITILDSRLKTPMYLFLGNLSFMDICYTSASVPTLLV LLSSQKTIIFSGCAVQMYLSLAMGSTECV LLAVMAYDRYVAICNPLRYSIIMNRCVCARMATVSWVTGCLTALLETSFALQIPLCGNLIDHFTCEILAVL KLACTSSLLMNTI LWSILLLPIPMLLVCISYIFILSTILRITSAEGRNKAFSTCGAHLTVVILYYGAAL SMYLKPSSSNAQKIDKIISLLYGVLTPMLNPIIYSLRNKEVKDAMKKLLGKITLHQTHEHL
The amino acid sequence of GPCR8a had high homology to other proteins as shown in Table 8C.
Table 8C. BLASTX results for GPCR8a
Smallest
Sum
Reading High Prob
Sequences producing High-scoring Segment Pairs: Frame Score P(N) N patp:AAB43266 Human ORFX ORF3030 polypeptide sequence . . +3 787 1.9e-77 1 patp:AAB68885 Human RECAP polypeptide, SEQ ID NO: 15 -. . +3 702 2.0e-68 1 patp:AAY83390 Olfactory receptor protein 0LF-5 - Homo . . +3 683 2.0e-66 1 patp:AA 21665 Rat spermatid chemoreceptor D-9 - Rattus. . +3 680 4.2e-66 1 patp:AAY83389 Olfactory receptor protein OLF-4 - Homo . . +3 680 4.2e-66 1 patp:AAB30873 Amino acid sequence of D class sperm rec. . +3 680 4.2e-66 1 patp:AAW21662 Rat spermatid chemoreceptor D-2 - Rattus . . +3 679 5.4e-66 1 patp:AAB30870 Amino acid sequence of D class sperm rec. . +3 679 5.4e-66 1
GPCR8b
In the present invention, the target sequence identified previously, Accession Number 17e20, was subjected to the exon linking process to confirm the sequence. PCR primers were designed by starting at the most upstream sequence available, for the forward primer, and at the most downstream sequence available for the reverse primer. In each case, the sequence was examined, walking inward from the respective termini toward the coding sequence, until a suitable sequence that is either unique or highly selective was encountered, or, in the case of the reverse primer, until the stop codon was reached. Such primers were designed based on in silico predictions for the full length cDNA, part (one or more exons) of the DNA or protein sequence of the target sequence, or by translated homology of the predicted exons to closely related human sequences sequences from other species. These primers were then employed in PCR amplification based on the following pool of human cDNAs: adrenal gland, bone marrow, brain - amygdala, brain - cerebellum, brain - hippocampus, brain - substantia nigra, brain - thalamus, brain -whole, fetal brain, fetal kidney, fetal liver, fetal lung, heart, kidney, lymphoma - Raji, mammary gland, pancreas, pituitary gland, placenta, prostate, salivary gland, skeletal muscle, small intestine, spinal cord, spleen, stomach, testis, thyroid, trachea, uterus. Usually the resulting amplicons were gel purified, cloned and sequenced to high redundancy. The resulting sequences from all clones were assembled with themselves, with other fragments in CuraGen Corporation's database and with public ESTs. Fragments and ESTs were included as components for an assembly when the extent of their identity with another component of the assembly was at least 95% over 50 bp. In addition, sequence traces were evaluated manually and edited for corrections if appropriate. These procedures provide the sequence reported below, which is designated Accession Number 17e20_A_dal. The disclosed novel GPCR8b nucleic acid of 961 nucleotides (also referred to as
17e20_A_dal) is shown in Table 8D. An open reading frame begins with an ATG initiation
codon at nucleotides 5-7 and ends with a TGA codon at nucleotides 953-955. A putative untranslated region upstream from the initiation codon and downstream from the termination codon are underlined in Table 8D, and the start and stop codons are in bold letters.
Table 8D. GPCR8b Nucleotide Sequence (SEQ ID NO:37)
GAAAATGCAAGGAGAAAACTTCACCATTTGGAGCATTTTTTTCTTGGAGGGATTTTCCCAGTACCCAGGTT TAGAAGTGGTTCTCTTCGTCTTCAGCCTTGTAATGTATCTGACAACGCTCTTGGGCAACAGCACTCTTATT TTGATCACTATCCTAGATTCACGCCTTAAAACCCCCATGTACTTATTCCTTGGAAATCTCTCTTTCATGGA TATTTGTTACACATCTGCCTCTGTTCCTACTTTGCTGGTGAACTTGCTGTCATCCCAGAAAACCATTATCT TTTCTGGGTGTGCTGTACAGATGTATCTGTCCCTTGCCATGGGCTCCACAGAGTGTGTGCTCCTGGCCGTG ATGGCATATGACCGTTATGTGGCCATTTGTAACCCGCTGAGATACTCCATCATCATGAACAGGTGCGTCTG TGCACGGATGGCTACGGTCTCCTGGGTGACGGGTTGCCTGACCGCTCTGCTGGAAACCAGTTTTGCCCTGC AGATACCCCTCTGTGGGAATCTCATCGATCACTTCACGTGTGAAATTCTGGCGGTGCTAAAGTTAGCTTGC ACAAGTTCACTGCTCATGAACACCATCATGCTGGTGGTCAGCATTCTCCTCTTGCCAATTCCAATGCTCTT AGTTTGCATCTCTTACATCTTCATCCTTTCCACTATTCTGAGAATCACCTCAGCAGAGGGAAGAAACAAGG CTTTTTCTACCTGTGGTGCCCATTTGACTGTGGTGATTTTGTATTATGGGGCTGCCCTCTCTATGTACCCA AAGCCTTCTTCATCAAATGCACAAAAAATAGACAAAATCATCTCGTTGCTTTACGGAGTGCTTACCCCTAT GTTGAACCCCATAATTTACAGTTTAAGAAACAAGGAAGTCAAAGATGCTATGAAGAAATTGCTGGGCAAAA TAACATTGCATCAAACACACGAACATCTCTGATTGGGT
The GPCR8b protein encoded by SEQ ID NO:37 has 316 amino acid residues, and is presented using the one-letter code in Table 8E (SEQ ID NO:38). The SignalP, Psort and/or Hydropathy profile for GPCR8b predict that GPCR8b has a signal peptide and is likely to be localized at the plasma membrane with a certainty of 0.6000. The SignalP predicts a cleavage site at the sequence LLG-NS between amino acids 41 and 42. The Molecular weight of GPCR8b is 35028.5 Daltons.
Table 8E. Encoded GPCR8b protein sequence (SEQ ID NO:38)
MQGENFTIWSIFFLEGFSQYPGLEVVLFVFSLVMYLTTLLGNSTLILITILDSRLKTPMYLFLGNLSFMDI CYTSASVPTLLVNLLSSQKTIIFSGCAVQMYLSLAMGSTECVLLAVMAYDRYVAICNPLRYSII NRCVCA R ATVSWVTGCLTALLETSFALQIPLCGNLIDHFTCEILAVLKLACTSSLL NTIMLVVSILLLPIPMLLV CISYIFILSTILRITSAEGRNKAFSTCGAHLTWILYYGAALSMYPKPSSSNAQKIDKIISLLYGVLTPML NPIIYSLRNKEVKDA KKLLGKITLHQTHEHL
BLASTP analysis of the best hits for alignments with GPCRδb are listed in Table 8F.
PAGE INTENTIONALLY LEFT BLANK
Possible SNPs found for GPCR8b are listed in Table 8G.
GPCR8c In the present invention, the target sequence identified previously, Accession Number
17e20, was subjected to the exon linking process to confirm the sequence. PCR primers were designed by starting at the most upstream sequence available, for the forward primer, and at the most downstream sequence available for the reverse primer. In each case, the sequence was examined, walking inward from the respective termini toward the coding sequence, until a suitable sequence that is either unique or highly selective was encountered, or, in the case of the reverse primer, until the stop codon was reached. Such primers were designed based on in silico predictions for the full length cDNA, part (one or more exons) of the DNA or protein sequence of the target sequence, or by translated homology of the predicted exons to closely related human sequences sequences from other species. These primers were then employed in PCR amplification based on the following pool of human cDNAs: adrenal gland, bone marrow, brain - amygdala, brain - cerebellum, brain - hippocampus, brain - substantia nigra, brain - thalamus, brain -whole, fetal brain, fetal kidney, fetal liver, fetal lung, heart, kidney, lymphoma - Raji, mammary gland, pancreas, pituitary gland, placenta, prostate, salivary gland, skeletal muscle, small intestine, spinal cord, spleen, stomach, testis, thyroid, trachea, uterus. Usually the resulting amplicons were gel purified, cloned and sequenced to high redundancy. The resulting sequences from all clones were assembled with themselves, with other fragments in CuraGen Corporation's database and with public ESTs. Fragments and ESTs were included as components for an assembly when the extent of their identity with another component of the assembly was at least 95% over 50 bp. hi addition, sequence traces were evaluated manually and edited for corrections if appropriate. These procedures provide the sequence reported below, which is designated Accession Number ba386d8_dal .
The disclosed novel GPCR8c nucleic acid of 963 nucleotides (also referred to as ba386d8_dal) is shown in Table 8H. An open reading frame begins with an ATG initiation codon at nucleotides 6-8 and ends with a TGA codon at nucleotides 954-956. A putative untranslated region upstream from the initiation codon and downstream from the termination codon are underlined in Table 8H, and the start and stop codons are in bold letters.
Table 8H. GPCR8c Nucleotide Sequence (SEQ ID NO:39)
TGAA ATGCAAGGAGAAAACTTCACCATTTGGAGCATTTTTTTCTTGGAGGGATTTTCCCAGTACCCAGGT TTAGAAGTGGTTCTCTTCGTCTTCAGCCTTGTAATGTATCTGACAACGCTCTTGGGCAACAGCACTCTTAT TTTGATCACTATCCTAGATTCACGCCTTAAAACCCCCATGTACTTATTCCTTGGAAATCTCTCTTTCATGG ATATTTGTTACACATCTGCCTCTGTTCCTACTTTGCTGGTGAACTTGCTGTCATCCCAGAAAACCATTATC TTTTCTGGGTGTGCTGTACAGATGTATCTGTCCCTTGCCATGGGCTCCACAGAGTGTGTGCTCCTGGCCGT GATGGCATATGACCGTTATGTGGCCATTTGTAACCCGCTGAGATACTCCATCATCATGAACAGGTGCGTCT GTGCACGGATGGCCACGGTCTCCTGGGTGACGGGTTGCCTGACCGCTCTGCTGGAAACCAGTTTTGCCCTG CAGATACCCCTCTGTGGGAATCTCATCGATCACTTCACGTGTGAAATTCTGGCGGTGCTAAAGTTAGCTTG CACAAGTTCACTGCTCATGAACACCATCATGCTGGTGGTCAGCATTCTCCTCTTGCCAATTCCAATGCTCT TAGTTTGCATCTCTTACATCTTCATCCTTTCCACTATTCTGAGAATCACCTCAGCAGAGGGAAGAAACAAG GCTTTTTCTACCTGTGGTGCCCATTTGACTGTGGTGATTTTGTATTATGGGGCTGCCCTCTCTATGTACCT AAAGCCTTCTTCATCAAATGCACAAAAAATAGACAAAATCATCTCGTTGCTTTACGGAGTGCTTACCCCTA TGTTGAACCCCATAATTTACAGTTTAAGAAACAAGGAAGTCAAAGATGCTATGAAGAAATTGCTGGGCAAA ATAACATTGCATCAAACACACGAACATCTCTGATTGGGTA
The GPCR8c protein encoded by SEQ ID NO:39 has 316 amino acid residues, and is presented using the one-letter code in Table 81 (SEQ ID NO:40). The SignalP, Psort and/or Hydropathy profile for GPCR8c predict that GPCR8c has a signal peptide and is likely to be localized at the plasma membrane with a certainty of 0.6000. The SignalP predicts a cleavage site at the sequence LLG-NS between amino acids 41 and 42. The Molecular weight of GPCR8c is 35044.6 Daltons.
Table 81. Encoded GPCR8c protein sequence (SEQ ID NO:40) QGENFTIWSIFFLEGFSQYPGLEVVLFVFSLVMYLTTLLGNSTLILITILDSRLKTPMYLFLGNLSFMDI CYTSASVPTLLVNLLSSQKTIIFSGCAVQMYLSLAMGSTECVLLAVMAYDRYVAICNPLRYSIIMNRCVCA RMATVS VTGCLTALLETSFALQIPLCGNLIDHFTCEILAVLKLACTSSLLMNTIMLWSILLLPIPMLLV CISYIFILSTILRITSAEGRNKAFSTCGAHLTVVILYYGAALSMYLKPSSSNAQKIDKIISLLYGVLTP L NPIIYSLRNKEVKDAMKKLLGKITLHQTHEHL
BLASTP analysis ofthe best hits for alignments with GPCR8c are listed in Table 8J.
Possible SNPs found for GPCR8c are listed in Table 8K.
GPCR8d
In the present invention, the target sequence identified previously, Accession Number 17e20, was subjected to the exon linking process to confirm the sequence. PCR primers were designed by starting at the most upstream sequence available, for the forward primer, and at the most downstream sequence available for the reverse primer. In each case, the sequence was examined, walking inward from the respective termini toward the coding sequence, until a suitable sequence that is either unique or highly selective was encountered, or, in the case of the reverse primer, until the stop codon was reached. Such primers were designed based on in silico predictions for the full length cDNA, part (one or more exons) of the DNA or protein sequence of the target sequence, or by translated homology of the predicted exons to closely related human sequences sequences from other species. These primers were then employed in PCR amplification based on the following pool of human cDNAs: adrenal gland, bone marrow, brain - amygdala, brain - cerebellum, brain - hippocampus, brain - substantia nigra, brain - thalamus, brain -whole, fetal brain, fetal kidney, fetal liver, fetal lung, heart, kidney, lymphoma - Raji, mammary gland, pancreas, pituitary gland, placenta, prostate, salivary gland, skeletal muscle, small intestine, spinal cord, spleen, stomach, testis, thyroid, trachea, uterus. Usually the resulting amplicons were gel purified, cloned and sequenced to high redundancy. The resulting sequences from all clones were assembled with themselves, with other fragments in CuraGen Corporation's database and with public ESTs. Fragments and ESTs were included as components for an assembly when the extent of their identity with another component of the assembly was at least 95% over 50 bp. In addition, sequence traces were evaluated manually
and edited for corrections if appropriate. These procedures provide the sequence reported below, which is designated Accession Number CG50343-01.
The disclosed novel GPCR8d nucleic acid of 963 nucleotides (also referred to as CG50343-01) is shown in Table 8L. An open reading frame begins with an ATG initiation codon at nucleotides 6-8 and ends with a TGA codon at nucleotides 954-956. A putative untranslated region upstream from the initiation codon and downstream from the termination codon are underlined in Table 8L, and the start and stop codons are in bold letters.
Table 8L. GPCRSd Nucleotide Sequence (SEQ ID NO:41)
TGAΆAATGCAAGGAGAAAACTTCACCATTTGGAGCATTTTTTTCTTGGAGGGATTTTCCCAGTACCCAGGG TTAGAAGTGGTTCTCTTCGTCTTCAGCCTTGTAATGTATCTGACAΆCGCTCTTGGGCAACAGCACTCTTAT TTTGATCACTATCCTAGATTCACGCCTTAAAACCCCCATGTACTTATTCCTTGGAAATCTCTCTTTCATGG ATATTTGTTACACATCTGCCTCTGTTCCTACTTTGCTGGTGAACTTGCTGTCATCCCAGAAAACCATTATC TTTTCTGGGTGTGCTGTACAGATGTATCTGTCCCTTGCCATGGGCTCCACAGAGTGTGTGCTCCTGGCCGT GATGGCATATGACCGTTATGTGGCCATTTGTAACCCGCTGAGATACTCCATCATCATGAACAGGTGCGTCT GTGCACGGATGGCCACGGTCTCCTGGGTGACGGGTTGCCTGACCGCTCTGCTGGAAACCAGTTTTGCCCTG CAGATACCCCTCTGTGGGAATCTCATCGATCACTTCACGTGTGAAATTCTGGCGGTGCTAAAGTTAGCTTG CACAAGTTCACTGCTCATGAACACCATCATGCTGGTGGTCAGCATTCTCCTCTTGCCAATTCCAATGCTCT TAGTTTGCATCTCTTACATCTTCATCCTTTCCACTATTCTGAGAATCACCTCAGCAGAGGGAAGAAACAAG GCTTTTTCTACCTGTGGTGCCCATTTGACTGTGGTGATTTTGTATTATGGGGCTGCCCTCTCTATGTACCT AAAGCCTTCTTCATCAAATGCACAAAAAATAGACAAAATCATCTCGTTGCTTTACGGAGTGCTTACCCCTA TGTTGAACCCCATAATTTACAGTTTAAGAAACAAGGAAGTCAAAGATGCTATGAAGAAATTGCTGGGCAAA ATAACATTGCATCAAACACACGAACATCTCTGATTGGGTA
The disclosed nucleic acid sequence for GPCR8d has 544 of 784 bases (69%) identical to a Mus musculus or37a gene mRNA (gb:GENBANK-ID:MMU133424|acc:AJ133424.1).
The GPCR8d protein encoded by SEQ ID NO:41 has 316 amino acid residues, and is presented using the one-letter code in Table 8M (SEQ ID NO:42). The SignalP, Psort and/or Hydropathy profile for GPCR8d predict that GPCR8d has a signal peptide and is likely to be localized at the plasma membrane with a certainty of 0.6000. The SignalP predicts a cleavage site at the sequence LLG-NS between amino acids 41 and 42.
Table 8M. Encoded GPCR8d protein sequence (SEQ ID NO:42)
MQGENFTIWSIFFLEGFSQYPGLEVVLFVFSLVMYLTTLLGNSTLILITILDSRLKTPMYLFLGNLSFMDI CYTSASVPTLLVNLLSSQKTIIFSGCAVQMYLSLAMGSTECVLLAVMΆYDRYVAICNPLRYSIIMNRCVCA RMATVSWVTGCLTALLETSFALQIPLCGNLIDHFTCEILAVLKLACTSSLLMNTIMLWSILLLPIPMLLV CISYIFILSTILRITSAEGRNKAFSTCGAHLTWILYYGAALSMYLKPSSSNAQKIDKIISLLYGVLTPML NPIIYSLRNKEVKDAMKKLLGKITLHQTHEHL
BLASTP analysis of the best hits for alignments with GPCR8d are listed in Table 8N.
Possible SNPs found for GPCRδd are listed in Table 80.
GPCR8e hi the present invention, the target sequence identified previously, Accession Number
17e20, was subjected to the exon linking process to confirm the sequence. PCR primers were designed by starting at the most upstream sequence available, for the forward primer, and at the most downstream sequence available for the reverse primer. In each case, the sequence was examined, walking inward from the respective termini toward the coding sequence, until a suitable sequence that is either unique or highly selective was encountered, or, in the case of the reverse primer, until the stop codon was reached. Such primers were designed based on in silico predictions for the full length cDNA, part (one or more exons) of the DNA or protein sequence of the target sequence, or by translated homology of the predicted exons to closely related human sequences sequences from other species. These primers were then employed in PCR amplification based on the following pool of human cDNAs: adrenal gland, bone marrow, brain - amygdala, brain - cerebellum, brain - hippocampus, brain - substantia nigra, brain - thalamus, brain -whole, fetal brain, fetal kidney, fetal liver, fetal lung, heart, kidney, lymphoma - Raji, mammary gland, pancreas, pituitary gland, placenta, prostate, salivary gland, skeletal muscle, small intestine, spinal cord, spleen, stomach, testis, thyroid, trachea, uterus. Usually the resulting amplicons were gel purified, cloned and sequenced to high redundancy. The resulting sequences from all clones were assembled with themselves, with other fragments in CuraGen Corporation's database and with public ESTs. Fragments and ESTs were included as components for an assembly when the extent of their identity with another component of the assembly was at least 95% over 50 bp. In addition, sequence traces were evaluated manually and edited for corrections if appropriate. These procedures provide the sequence reported below, which is designated Accession Number 17e20_dal.
The disclosed novel GPCR8e nucleic acid of 1115 nucleotides (also referred to as 17e20_dal) is shown in Table 8P. An open reading frame begins with an ATG initiation codon
at nucleotides 50-52 and ends with a TGA codon at nucleotides 998-1000. A putative untranslated region upstream from the initiation codon and downstream from the termination codon are underlined in Table 8P, and the start and stop codons are in bold letters.
Table 8P. GPCR8e Nucleotide Sequence (SEQ ID NO:43)
ATTAAATGAAGGGAATAGTACTGGAAGAAATATAGATGAAAGAAAGAΆAATGCAAGGAGAAAACTTCACCA TTTGGAGCATTTTTTTCTTGGAGGGATTTTCCCAGTACCCAGGGTTAGAAGTGGTTCTCTTCGTCTTCAGC CTTGTAATGTATCTGACAACGCTCTTGGGCAACAGCACTCTTATTTTGATCACTATCCTAGATTCACGCCT TAAΆACCCCCATGTACTTATTCCTTGGAAATCTCTCTTTCATGGATATTTGTTACACATCTGCCTCTGTTC CTACTTTGCTGGTGAACTTGCTGTCATCCCAGAAAACCATTATCTTTTCTGGGTGTGCTGTACAGATGTAT CTGTCCCTTGCCATGGGCTCCACAGAGTGTGTGCTCCTGGCCGTGATGGCATATGACCGTTATGTGGCCAT TTGTAACCCGCTGAGATACTCCATCATCATGAACAGGTGCGTCTGTGCACGGATGGCCACGGTCTCCTGGG TGACGGGTTGCCTGACCGCTCTGCTGGAAACCAGTTTTGCCCTGCAGATACCCCTCTGTGGGAATCTCATC GATCACTTCACGTGTGAAATTCTGGCGGTGCTAAAGTTAGCTTGCACAAGTTCACTGCTCATGAACACCAT CATGCTGGTGGTCAGCATTCTCCTCTTGCCAATTCCAATGCTCTTAGTTTGCATCTCTTACATCTTCATCC TTTCCACTATTCTGAGAATCACCTCAGCAGAGGGAAGAAACAAGGCTTTTTCTACCTGTGGTGCCCATTTG ACTGTGGTGATTTTGTATTATGGGGCTGCCCTCTCTATGTACCTAAAGCCTTCTTCATCAAATGCACAAAA ATAGACAAAATCATCTCGTTGCTTTACGGAGTGCTTACCCCTATGTTGAACCCCATAATTTACAGTTTAA GAAACAAGGAAGTCAAAGATGCTATGAAGAAATTGCTGGGCAAAATAACATTGCATCAAACACACGAACAT CTCTGATTGGGTCCCTATGGTTTTACCAGAGATGTGCCCCTGGCAGAGCTCATCAGAGAAATTCGAGACAA CATACAACCTCTTAGAACTCTGATCGGATCTTATCTCTATATAATTTCAC
The disclosed nucleic acid sequence for GPCR8e has 551 of 796 bases (69%) identical to a Mus niusculus or37a gene mRNA (gb:GENBANK-ID:MMU133424|acc:AJ133424.1). The GPCR8e gene maps to chromosome 9.
The GPCRδe protein encoded by SEQ ID NO:43 has 316 amino acid residues, and is presented using the one-letter code in Table 8Q (SEQ ID NO:44). The SignalP, Psort and/or Hydropathy profile for GPCR8e predict that GPCR8e has a signal peptide and is likely to be localized at the plasma membrane with a certainty of 0.6000. The SignalP predicts a cleavage site at the sequence LLG-NS between amino acids 41 and 42.
Table 8Q. Encoded GPCR8e protein sequence (SEQ ID NO:44)
MQGENFTIWSIFFLEGFSQYPGLEVVLFVFSLVMYLTTLLGNSTLILITILDSRLKTPMYLFLGNLSFMDI CYTSASVPTLLVNLLSSQKTIIFSGCAVQMYLSLAMGSTECVLLAVMAYDRYVAICNPLRYSIIMNRCVCA MATVS VTGCLTALLETSFALQIPLCGNLIDHFTCEILAVLKLACTSSLLMNTIMLWSILLLPIPMLLV CISYIFILSTILRITSAEGRNKAFSTCGAHLTVVILYYGAALSMYLKPSSSNAQKIDKIISLLYGVLTPML NPIIYSLRNKEVKDAMKKLLGKITLHQTHEHL
BLASTP analysis ofthe best hits for alignments with GPCR8e are listed in Table 8R.
Table 8R. BLASTP results for GPCR8e
Gene Index/ Protein/ Organism Length Identity Positives Expect Identifier (aa) (%) (%)
Possible SNPs found for GPCR8e are listed in Table 8S.
GPCR8a also had high homology to the proteins described in the BLASTP data in
Table 8T.
This BLASTP data is displayed graphically in the Clustal W displayed in Table 8U.
Table 8U ClustalW Analysis of GPCR8a
1) Novel GPC 8a (SEQ ID NO:36)
2) gi|l 1276077|reflNP_062347.1| olfactory receptor 37b [Mus musculus] (SEQ ID NO:60) 3). gijl 146498 ljref|NP_062349.1] olfactory receptor 37e [Mus musculus] (SEQ ID NO:61)
4) gi|l 1276075|re±lNP_062346.1| olfactory receptor 37a [Mus musculus] (SEQ ID NO:62)
5) gi|l 1276079|reflNP_062348.1| olfactory receptor 37c [Mus musculus] (SEQ ID NO:63)
6) gi|10092669|reflNP_063950.1| olfactory receptor, family 2, subfamily S, member 2 [Homo sapiens] (SEQ ID NO:64)
Homology between GPCR8a, b, c, d, and e is shown in a Clustal W presented in Table 8V.
Table 8V. Clustal W of GPCR8 variants
10 20 30 40 50 60 ....|....|....|....|....|....|....|....|....|....|....|....|
GPCR 8A Coding Sequence { 17e20 AGTATTTTCCTAACTAAATGAATATCACACTTTATTCATTTCTTTATGTCCTCAGATTAA GPCR 8B Coding Sequence (17e20 GPCR 8C Coding Sequence (ba386 GPCR 8D Coding Sequence (CG503 GPCR 8E Coding Sequence (17e20 ATTAA
70 80 90 100 110 120 ....1.... I .... I .... I .... I .... ] .... I .... I ..^^| .... I ....1....I
GPCR 8A Coding Sequence (I7e20 ATGAAGGGAATAGTACTGGAAGAAATATAGATGAAAGAAAI GAAAATGCAAGGAGAAAACT,
GPCR 8B Coding Sequence (17e20 GAAAATGCAAGGAGAAAACT
GPCR 8C Coding Sequence (ba386 TgGAAAATGCAAGGAGAAAACT
GPCR 8D Coding Sequence (CG503 T£GAAAATGCAAGGAGAAAACT
GPCR 8E Coding Sequence (17e2o ATGAAGGGAATAGTACTGGAAGAAATATAGATGAAAGA GAAAATGCAAGGAGAAAACT
130 140 150 160 170 180
....I....I....I.... l .... ...j_..■■).... ι....| ....ι.... ι_....ι
GPCR 8A Coding Sequence (17e20 llUft UftlrTlΛiAfciUήT11 riTT TTbbAUbUA.1.TXTCUCAi rAUUUAb ϋTTAbAAbTUb
GPCR 8B Coding Sequence (17e20 TCACCATTTGGAGCATTTTTTTCTTGGAGGGATTTTCCCAGTACCCAGGBTTAGAAGTGG
GPCR 8C Coding Sequence (ba386 TCACCATTTGGAGCATTTTTTTCTTGGAGGGATTTTCCCAGTACCCAGGGTTAGAAGTGG
GPCR 8D Coding Sequence (CG503 CACCATTTGGAGCATTTTTTTCTTGGAGGGATTTTCCCAGTACCCAGGGTTAGAAGTGG
GPCR 8E Coding Sequence (17e20 TCACCATTTGGAGCATTTTTTTCTTGGAGGGATTTTCCCAGTACCCAGGGTTAGAAGTGG
190 200 210 220 230 240 ..I.. ..I .. ..I .. I ..I
GPCR 8A Coding Sequence (17e20 TTCTCTTCGTCTTCAGCCTTGTAATGTATCTGACAACGCTCTTGGGCAACAGCACTCTTA
GPCR 8B Coding Sequence (17e20 TTCTCTTCGTCTTCAGCCTTGTAATGTATCTGACAACGCTCTTGGGCAACAGCACTCTTA
GPCR 8C Coding Sequence (ba386 TTCTCTTCGTCTTCAGCCTTGTAATGTATCTGACAACGCTCTTGGGCAACAGCACTCTTA
GPCR 8D Coding Sequence (CG503 TTCTCTTCGTCTTCAGCCTTGTAATGTATCTGACAACGCTCTTGGGCAACAGCACTCTTA
GPCR 8E Coding Sequence (17e20 TTCTCTTCGTCTTCAGCCTTGTAATGTATCTGACAACGCTCTTGGGCAACAGCACTCTTA
250 260 270 280 290 300
I ..|.. ..I.. I ..| ..
GPCR 8A Coding Sequence (17e20 TTTTGATCACTATCCTAGATTCACGCCTTAAAACCCCCATGTACTTATTCCTTGGAAATC
GPCR 8B Coding Sequence (17e20 TTTTGATCACTATCCTAGATTCACGCCTTAAAACCCCCATGTACTTATTCCTTGGAAATC
GPCR 8C Coding Sequence (ba386 TTTTGATCACTATCCTAGATTCACGCCTTAAAACCCCCATGTACTTATTCCTTGGAAATC
GPCR 8D Coding Sequence (CG503 TTTTGATCACTATCCTAGATTCACGCCTTAAAACCCCCATGTACTTATTCCTTGGAAATC
GPCR 8E Coding Sequence (17e20 TTTTGATCACTATCCTAGATTCACGCCTTAAAACCCCCATGTACTTATTCCTTGGAAATC
310 320 330 340 350 360 ..I.. I
GPCR 8A Coding Sequence (17e20 TCTCTTTCATGGATATTTGTTACACATCTGCCTCTGTTCCTACTTTGCTGGTGAACTTGC
GPCR 8B Coding Sequence (17e20 TCTCTTTCATGGATATTTGTTACACATCTGCCTCTGTTCCTACTTTGCTGGTGAACTTGC
GPCR 8C Coding Sequence (ba386 TCTCTTTCATGGATATTTGTTACACATCTGCCTCTGTTCCTACTTTGCTGGTGAACTTGC
GPCR 8D Coding Sequence (CG503 TCTCTTTCATGGATATTTGTTACACATCTGCCTCTGTTCCTACTTTGCTGGTGAACTTGC
GPCR 8E Coding Sequence (17e20 TCTCTTTCATGGATATTTGTTACACATCTGCCTCTGTTCCTACTTTGCTGGTGAACTTGC
370 380 390 400 410 420
I ..I .. I I ..I .. ..I
GPCR 8A Coding Sequence (17e20 TGTCATCCCAGAAAACCATTATCTTTTCTGGGTGTGCTGTACAGATGTATCTGTCCCTTG
GPCR 8B Coding Sequence (17e20 TGTCATCCCAGAAAACCATTATCTTTTCTGGGTGTGCTGTACAGATGTATCTGTCCCTTG
GPCR 8C Coding Sequence (ba386 TGTCATCCCAGAAAACCATTATCTTTTCTGGGTGTGCTGTACAGATGTATCTGTCCCTTG
GPCR 8D Coding Sequence (CG503 TGTCATCCCAGAAAACCATTATCTTTTCTGGGTGTGCTGTACAGATGTATCTGTCCCTTG
GPCR 8E Coding Sequence (17e20 TGTCATCCCAGAAAACCATTATCTTTTCTGGGTGTGCTGTACAGATGTATCTGTCCCTTG
430 440 450 460 470 480
I ..|.. I ..| .. I ..|.. I ..I
GPCR 8A Coding Sequence (17e20 CCATGGGCTCCACAGAGTGTGTGCTCCTGGCCGTGATGGCATATGACCGTTATGTGGCCA
GPCR 8B Coding Sequence (17e20 CCATGGGCTCCACAGAGTGTGTGCTCCTGGCCGTGATGGCATATGACCGTTATGTGGCCA
GPCR 8C Coding Sequence (ba386 CCATGGGCTCCACAGAGTGTGTGCTCCTGGCCGTGATGGCATATGACCGTTATGTGGCCA
GPCR 8D Coding Sequence (CG503 CCATGGGCTCCACAGAGTGTGTGCTCCTGGCCGTGATGGCATATGACCGTTATGTGGCCA
GPCR 8E Coding Sequence (17e20 CCATGGGCTCCACAGAGTGTGTGCTCCTGGCCGTGATGGCATATGACCGTTATGTGGCCA
490 500 510 520 530 540
I ..|.. I .. I .. I I I ..|.. I ..I
GPCR 8A Coding Sequence (17e20 TTTGTAACCCGCTGAGATACTCCATCATCATGAACAGGTGCGTCTGTGCACGGATGGCCA
GPCR 8B Coding Sequence (17e20 TTTGTAACCCGCTGAGATACTCCATCATCATGAACAGGTGCGTCTGTGCACGGATGGCJ3A
GPCR 8C Coding Sequence (ba386 TTTGTAACCCGCTGAGATACTCCATCATCATGAACAGGTGCGTCTGTGCACGGATGGCCA
GPCR 8D Coding Sequence (CG503 TTTGTAACCCGCTGAGATACTCCATCATCATGAACAGGTGCGTCTGTGCACGGATGGCCA
GPCR 8E Coding Sequence (17e20 TTTGTAACCCGCTGAGATACTCCATCATCATGAACAGGTGCGTCTGTGCACGGATGGCCA
550 560 570 580 590 600
I I ..| .. ..I .. I ..|.. I
GPCR 8A Coding Sequence (17e20 CGGTCTCCTGGGTGACGGGTTGCCTGACCGCTCTGCTGGAAACCAGTTTTGCCCTGCAG?
GPCR 8B Coding Sequence (17e20 CGGTCTCCTGGGTGACGGGTTGCCTGACCGCTCTGCTGGAAACCAGTTTTGCCCTGCAGA
GPCR 8C Coding Sequence (ba386 CGGTCTCCTGGGTGACGGGTTGCCTGACCGCTCTGCTGGAAACCAGTTTTGCCCTGCAGA
GPCR 8D Coding Sequence (CG503 CGGTCTCCTGGGTGACGGGTTGCCTGACCGCTCTGCTGGAAACCAGTTTTGCCCTGCAGA
GPCR 8E Coding Sequence (17e20 CGGTCTCCTGGGTGACGGGTTGCCTGACCGCTCTGCTGGAAACCAGTTTTGCCCTGCAGA
610 620 630 640 650 660
I .. I .. I .. I .. I .. I .. I ..I
GPCR 8A Coding Sequence (17e20 TACCCCTCTGTGGGAATCTCATCGATCACTTCACGTGTGAAAT1] 'CTGGCGGTGCTAAAGT
GPCR 8B Coding Sequence (17e20 TACCCCTCTGTGGGAATCTCATCGATCACTTCACGTGTGAAAT']'CTGGCGGTGCTAAAGT
GPCR 8C Coding Sequence (ba386 TACCCCTCTGTGGGAATCTCATCGATCACTTCACGTGTGAAAT1'CTGGCGGTGCTAAAGT
GPCR 8D Coding Sequence (CG503 TACCCCTCTGTGGGAATCTCATCGATCACTTCACGTGTGAAAm'CTGGCGGTGCTAAAGT
GPCR 8E Coding Sequence (17e20 TACCCCTCTGTGGGAATCTCATCGATCACTTCACGTGTGAAATI'CTGGCGGTGCTAAAGT
670 680 690 700 710 720 .. I .. I ..| .. I .. I .. I I
GPCR 8A Coding Sequence (17e20 TAGCTTGCACAAGTTCACTGCTCATGAACACCATCATGCTGGTGGTCAGCATTCTCCTCT
GPCR 8B Coding Sequence (17e20 ΓAGCTTGCACAAGTTCACTGCTCATGAACACCATCATGCTGGTGGTCAGCATTCTCCTCT
GPCR 8C Coding Sequence (ba386 ΓAGCTTGCACAAGTTCACTGCTCATGAACACCATCATGCTGGTGGTCAGCATTCTCCTCT
GPCR 8D Coding Sequence (CG503 ΓAGCTTGCACAAGTTCACTGCTCATGAACACCATCATGCTGGTGGTCAGCATTCTCCTCT
GPCR 8E Coding Sequence (17e20 ΓAGCTTGCACAAGTTCACTGCTCATGAACACCATCATGCTGGTGGTCAGCATTCTCCTCT
730 740 750 760 770 780 ..|.. I ..|.. I I
GPCR 8A Coding Sequence (17e20 TGCCAATTCCAATGCTCTTAGTTTGCATCTCTTACATCTTCATCCTTTCCACTATTCTGi
GPCR 8B Coding Sequence (17e20 TGCCAATTCCAATGCTCTTAGTTTGCATCTCTTACATCTTCATCCTTTCCACTATTCTGA
GPCR 8C Coding Sequence (ba386 TGCCAATTCCAATGCTCTTAGTTTGCATCTCTTACATCTTCATCCTTTCCACTATTCTGA
GPCR 8D Coding Sequence (CG503 TGCCAATTCCAATGCTCTTAGTTTGCATCTCTTACATCTTCATCCTTTCCACTATTCTGA
GPCR 8E Coding Sequence (17e20 TGCCAATTCCAATGCTCTTAGTTTGCATCTCTTACATCTTCATCCTTTCCACTATTCTGA
790 300 810 320 830 840
I I I I I I ..|.. I
GPCR 8A Coding Sequence (17e20 5AATCACCTCAGCAGAGGGAAGAAACAAGGCTTTTTCTACCTGTGGTGCCCATTTGACTG
GPCR 8B Coding Sequence (17e20 3AATCACCTCAGCAGAGGGAAGAAACAAGGCTTTTTCTACCTGTGGTGCCCATTTGACTG
GPCR 8C Coding Sequence (ba386 GAATCACCTCAGCAGAGGGAAGAAACAAGGCTTTTTCTACCTGTGGTGCCCATTTGACTG
GPCR 8D Coding Sequence (CG503 GAATCACCTCAGCAGAGGGAAGAAACAAGGCTTTTTCTACCTGTGGTGCCCATTTGACTG
GPCR 8E Coding Sequence (17e20 GAATCACCTCAGCAGAGGGAAGAAACAAGGCTTTTTCTACCTGTGGTGCCCATTTGACTG
850 860 870 380 890 900
I I I I I I
GPCR 8A Coding Sequence (17e20 TGGTGATTTTGTATTATGGGGCTGCCCTCTCTATGTACCTAAAGCCTTCTTCATCAAATG
GPCR 8B Coding Sequence (17e20 TGGTGATTTTGTATTATGGGGCTGCCCTCTCTATGTACCgAAAGCCTTCTTCATCAAATG
GPCR 8C Coding Sequence (ba386 TGGTGATTTTGTATTATGGGGCTGCCCTCTCTATGTACCTAAAGCCTTCTTCATCAAATG
GPCR 8D Coding Sequence (CG503 TGGTGATTTTGTATTATGGGGCTGCCCTCTCTATGTACCTAAAGCCTTCTTCATCAAATG
GPCR 8E Coding Sequence (17e20 TGGTGATTTTGTATTATGGGGCTGCCCTCTCTATGTACCTAAAGCCTTCTTCATCAAATG
910 920 930 940 950 960 ..I.. I ..I .. I ..I
GPCR 8A Coding Sequence (17e20 CACAAAAAATAGACAAAATCATCTCGTTGCTTTACGGAGTGCTTACCCCTATGTTGAACC
GPCR 8B Coding Sequence (17e20 CACAAAAAATAGACAAAATCATCTCGTTGCTTTACGGAGTGCTTACCCCTATGTTGAACC
GPCR 8C Coding Sequence (ba386 CACAAAAAATAGACAAAATCATCTCGTTGCTTTACGGAGTGCTTACCCCTATGTTGAACC
GPCR 8D Coding Sequence (CG503 CACAAAAAATAGACAAAATCATCTCGTTGCTTTACGGAGTGCTTACCCCTATGTTGAACC
GPCR 8E Coding Sequence (17e20 CACAAAAAATAGACAAAATCATCTCGTTGCTTTACGGAGTGCTTACCCCTATGTTGAACC
970 980 990 1000 1010 1020 ..|.. I ..| .. I ..| ... I I
GPCR 8A Coding Sequence (17e20 CCATAATTTACAGTTTAAGAAACAAGGAAGTCAAAGATGCTATGAAGAAATTGCTGGGCA
GPCR 8B Coding Sequence (17e20 CCATAATTTACAGTTTAAGAAACAAGGAAGTCAAAGATGCTATGAAGAAATTGCTGGGCA
GPCR 8C Coding Sequence (ba386 CCATAATTTACAGTTTAAGAAACAAGGAAGTCAAAGATGCTATGAAGAAATTGCTGGGCA
GPCR 8D Coding Sequence (CG503 CCATAATTTACAGTTTAAGAAACAAGGAAGTCAAAGATGCTATGAAGAAATTGCTGGGCA
GPCR 8E Coding Sequence (17e20 CCATAATTTACAGTTTAAGAAACAAGGAAGTCAAAGATGCTATGAAGAAATTGCTGGGCA
1030 1040 1050 1060 1070 1080 . . I . . . . I . . . . 1 - . . . | . . . . I . . . . 1 . . . . I . . . . 1 . . . . 1 . . . . | . . . . |
GPCR 8A Coding Sequence ( 17e20 k A^AMATJAΛAiWCAMTTiGtiCΛAMTCMAAMACύAΛCύAMCGύAMACΛAATiCΛTMCTMGAMTTiG^GGATic TRTGGΥTΥTΑC ΑCiAG
GPCR 8B Coding Sequence (17e20 AAATAACATTGCATCAAACACACGAACATCTCTGATTGGGT
GPCR 8C Coding Sequence (ba386 AAATAACATTGCATCAAACACACGAACATCTCTGATTGGGT
GPCR 8D Coding Sequence (CG503 AAATAACATTGCATCAAACACACGAACATCTCTGATTGGGT
GPCR 8E Coding Sequence ( 17e20 feAWAA'WTAAAWCAΛT*TrfGeWCAΛTιhChAWAA-TCtMAC»MAC^GAACATCTCTGATTGGGT
1090 1100 1110 1120 1130 1140 ....|....|....|....|....I....]....|....|....|....|....|....|
GPCR 8A Coding Sequence ( 17e20 ATGTGCCCCTGGCAGAGCTCATCAGAGAAATTCGAGACAACATACAACCTCTTAGAACTC GPCR 8B Coding Sequence (17e20 GPCR 8C Coding Sequence (ba386 GPCR 8D Coding Sequence (CG503 GPCR 8E Coding Sequence ( 17e20 ATGTGCCCCTGGCAGAGCTCATCAGAGAAATTCGAGACAACATACAACCTCTTAGAACTC
1150 1160 1170 1180 1190 1200 ....|....1....|....|....|....|....|....|....|....|....|....| GPCR 8A Coding Sequence (17e20 TGATCGGATCTTATCTCTATATAATTTCACAGTTATGAGCTGCATACACAGAGTGATTGC
GPCR 8B Coding Sequence (17e20
GPCR 8C Coding Sequence (ba386
GPCR 8D Coding Sequence (CG503
GPCR 8E Coding Sequence (17e20 TGATCGGATCTTATCTCTATATAATTTCAC
GPCR 8A Coding Sequence (17e20 C
GPCR 8B Coding Sequence (17e20 -
GPCR 8C Coding Sequence (ba386 -
GPCR 8D Coding Sequence (CG503 -
GPCR 8E Coding Sequence (17e20 -
As shown above, these proteins share close homology, and so would also have close homology with the sequences displayed in Tables 8T and U.
Table 8W lists the domain description from DOMAIN analysis results against GPCR8a. This indicates that the GPCR8a sequence has properties similar to those of other proteins known to contain this domain as well as to the 377 amino acid 7tm domain itself.
Table 8W. Domain Analysis of GPCR8a gnl I Pfam] pfaitiOOOOl, 7tm_l, 7 transmembrane receptor (rhodopsin family) (SEQ ID NO: 65) Length: 254 Score = 109 bits (273), Expect = 2e-25
GPCR 8A
7 transmembrane receptor (rhod
130 140 150 160 170 180 I I I I I
GPCR 8A TS5!A QIPI.CGNLID--HFTfflEnLAVLKLAC--T|SiI,fflMN!!ll( V|SILLLPl|Mffll,VCl
7 transmembrane receptor (rhod L@S LRTVEEG— H— T VSLØDFPEESVK— RgYJ?3l,Si3L GFa L§LflVI 'Vi
190 200 210 220 230 240
GPCR 8A
7 transmembrane receptor (rhod CH RERH KRARSQ-
310 320 330 340 350 360
....|....|....|....|....|....|....|....|....|....|....|....|
GPCR 8A RlT| EGraNKgFSTCG HIiTVJjrLYYG AAfflSMYLKPHsΞ-NAQKIDKIΪJsfflLYGVJj
7 transmembrane receptor (rhod KRR|SΞE3I KA KMLLVVVVVF3IICMLPYHIV L3DSLCLL@I -RVLPTALIST3WLAYV
GPCR 8A TP]
7 transmembrane receptor (rhod NSC
The similarity information for the GPCR8 proteins and nucleic acids disclosed herein suggest that GPCR8a-e may have important structural and/or physiological functions characteristic of the Olfactory Receptor family and the GPCR family. Therefore, the nucleic acids and proteins of the invention are useful in potential diagnostic and therapeutic applications and as a research tool. These include serving as a specific or selective nucleic acid or protein diagnostic and/or prognostic marker, wherein the presence or amount of the nucleic acid or the protein are to be assessed, as well as potential therapeutic applications such as the following: (i) a protein therapeutic, (ii) a small molecule drug target, (iii) an antibody target (therapeutic, diagnostic, drug targeting/cytotoxic antibody), (iv) a nucleic acid useful in gene therapy (gene delivery/gene ablation), and (v) a composition promoting tissue regeneration in vitro and in vivo (vi) biological defense weapon. The novel nucleic acid encoding GPCR8a-e, and the GPCR8a-e protein of the invention, or fragments thereof, may further be useful in diagnostic applications, wherein the presence or amount of the nucleic acid or the protein are to be assessed. The nucleic acids and proteins of the invention are useful in potential diagnostic and therapeutic applications implicated in various diseases and disorders described below and/or other pathologies. For example, the compositions of the present invention will have efficacy for treatment of patients suffering from: developmental diseases, MHCII and III diseases (immune diseases), Taste and scent detectability Disorders, Burkitt's lymphoma, Corticoneurogenic disease, Signal Transduction pathway disorders, Retinal diseases including those involving photoreception, Cell Growth rate disorders; Cell Shape disorders, Feeding disorders; control of feeding; potential obesity due to over-eating; potential disorders due to starvation (lack of appetite), noninsulin-dependent diabetes mellitus (NTDDMl), bacterial, fungal, protozoal and viral infections (particularly infections caused by HIV-l or HIV-2), pain, cancer (including but not limited to Neoplasm; adenocarcinoma; lymphoma; prostate cancer; uterus cancer), anorexia, bulimia, asthma, Parkinson's disease, acute heart failure, hypotension, hypertension, urinary retention, osteoporosis, Crohn's disease; multiple sclerosis; and Treatment of Albright Hereditary Ostoeodystrophy, angina pectoris, myocardial infarction, ulcers, asthma, allergies, benign prostatic hypertrophy, and psychotic and neurological disorders, including anxiety, schizophrenia, manic depression, delirium, dementia, severe mental retardation.
Dentatorubro-pallidoluysian atrophy (DRPLA) Hypophosphatemic rickets, autosomal dominant
(2) Acrocallosal syndrome and dyskinesias, such as Huntington's disease or Gilles de la
Tourette syndrome and/or other pathologies and disorders of the like. The polypeptides can be used as immunogens to produce antibodies specific for the invention, and as vaccines. They
can also be used to screen for potential agonist and antagonist compounds. For example, a cDNA encoding the OR -like protein may be useful in gene therapy, and the OR-like protein may be useful when administered to a subject in need thereof. By way of nonlimiting example, the compositions of the present invention will have efficacy for treatment of patients suffering from bacterial, fungal, protozoal and viral infections (particularly infections caused by HIV-l or HIV-2), pain, cancer (including but not limited to Neoplasm; adenocarcinoma; lymphoma; prostate cancer; uterus cancer), anorexia, bulimia, asthma, Parkinson's disease, acute heart failure, hypotension, hypertension, urinary retention, osteoporosis, Crohn's disease; multiple sclerosis; and Treatment of Albright Hereditary Ostoeodystrophy, angina pectoris, myocardial infarction, ulcers, asthma, allergies, benign prostatic hypertrophy, and psychotic and neurological disorders, including anxiety, schizophrenia, manic depression, delirium, dementia, severe mental retardation and dyskinesias, such as Huntington's disease or Gilles de la Tourette syndrome and/or other pathologies and disorders.
The novel nucleic acid encoding GPCR-like protein, and the GPCR-like protein of the invention, or fragments thereof, may further be useful in diagnostic applications, wherein the presence or amount of the nucleic acid or the protein are to be assessed. These materials are further useful in the generation of antibodies that bind immunospecifically to the novel substances of the invention for use in therapeutic or diagnostic methods. These antibodies may be generated according to methods known in the art, using prediction from hydrophobicity charts, as described in the "Anti-GPCRX Antibodies" section below. For example the disclosed GPCRδa, d and GPCR8e proteins have multiple hydrophilic regions, each of which can be used as an immunogen. In one embodiment, GPCR8a epitopes are from about amino acids 10 to 40. hi another embodiment, contemplated GPCR8d and GPCR8e epitopes are from about amino acids 125 to 130. In another embodiment, GPCR8a epitopes are from about amino acids 250 to 260. In another embodiment, GPCR8d and GPCR8e epitopes are from about amino acids 225 to 240. In additional embodiments, GPCR8a epitopes are from amino acids 275 to 290 and 320 to 340. hi additional embodiments, GPCR8d and GPCR8e epitopes are from amino acids 255 to 270 and 285 to 310. These novel proteins also have value in the development of powerful assay system for functional analysis of various human disorders, which will help in understanding of pathology of the disease and development of new drug targets for various disorders.
A summary of the GPCRX nucleic acids and proteins of the invention is provided in
Table 9.
TABLE 9: Summary Of Nucleic Acids And Proteins Of The Invention
GPCRX Nucleic Acids and Polypeptides
One aspect of the invention pertains to isolated nucleic acid molecules that encode GPCRX polypeptides or biologically active portions thereof. Also included in the invention are nucleic acid fragments sufficient for use as hybridization probes to identify GPCRX-encoding nucleic acids (e.g., GPCRX mRNAs) and fragments for use as PCR primers for the amplification and/or mutation of GPCRX nucleic acid molecules. As used herein, the term "nucleic acid molecule" is intended to include DNA molecules (e.g., cDNA or genomic DNA), RNA molecules (e.g., mRNA), analogs of the DNA or RNA generated using nucleotide analogs, and derivatives, fragments and homologs thereof. The nucleic acid molecule may be single-stranded or double-stranded, but preferably is comprised double-stranded DNA.
An GPCRX nucleic acid can encode a mature GPCRX polypeptide. As used herein, a "mature" form of a polypeptide or protein disclosed in the present invention is the product of a naturally occurring polypeptide or precursor form or proprotein. The naturally occurring polypeptide, precursor or proprotein includes, by way of nonlimiting example, the full-length gene product, encoded by the corresponding gene. Alternatively, it may be defined as the polypeptide, precursor or proprotein encoded by an ORF described herein. The product "mature" form arises, again by way of nonlimiting example, as a result of one or more naturally occurring processing steps as they may take place within the cell, or host cell, in which the gene product arises. Examples of such processing steps leading to a "mature" form of a polypeptide or protein include the cleavage of the N-terminal methionine residue encoded by the initiation codon of an ORF, or the proteolytic cleavage of a signal peptide or leader sequence. Thus a mature form arising from a precursor polypeptide or protein that has residues 1 to N, where residue 1 is the N-terminal methionine, would have residues 2 through N remaining after removal of the N-terminal methionine. Alternatively, a mature form arising from a precursor polypeptide or protein having residues 1 to N, in which an N-terminal signal sequence from residue 1 to residue M is cleaved, would have the residues from residue M+l to residue N remaining. Further as used herein, a "mature" form of a polypeptide or protein may arise from a step of post-translational modification other than a proteolytic cleavage event. Such additional processes include, byway of non-limiting example, glycosylation, myristoylation or phosphorylation. hi general, a mature polypeptide or protein may result from the operation of only one of these processes, or a combination of any of them.
The term "probes", as utilized herein, refers to nucleic acid sequences of variable length, preferably between at least about 10 nucleotides (nt), 100 nt, or as many as approximately, e.g., 6,000 nt, depending upon the specific use. Probes are used in the detection of identical, similar, or complementary nucleic acid sequences. Longer length probes are generally obtained from a natural or recombinant source, are highly specific, and much slower to hybridize than shorter- length oligomer probes. Probes may be single- or double-stranded and designed to have specificity in PCR, membrane-based hybridization technologies, or ELISA-like technologies. The term "isolated" nucleic acid molecule, as utilized herein, is one, which is separated from other nucleic acid molecules which are present in the natural source of the nucleic acid.
Preferably, an "isolated" nucleic acid is free of sequences which naturally flank the nucleic acid
(i.e., sequences located at the 5'- and 3 '-termini of the nucleic acid) in the genomic DNA of the organism from which the nucleic acid is derived. For example, in various embodiments, the isolated GPCRX nucleic acid molecules can contain less than about 5 kb, 4 kb, 3 kb, 2 kb, 1 kb,
0.5 kb or 0.1 kb of nucleotide sequences which naturally flank the nucleic acid molecule in genomic DNA of the cell/tissue from which the nucleic acid is derived (e.g., brain, heart, liver, spleen, etc.). Moreover, an "isolated" nucleic acid molecule, such as a cDNA molecule, can be substantially free of other cellular material or culture medium when produced by recombinant techniques, or of chemical precursors or other chemicals when chemically synthesized. A nucleic acid molecule of the invention, e.g., a nucleic acid molecule having the nucleotide sequence of SEQ ID NOS: 1, 3, 5, 7, 9, 11, 13, 15, 17, 19, 21, 23, 25, 27, 29, 31, 33, 35, 37, 39, 41 and 43, or a complement of this aforementioned nucleotide sequence, can be isolated using standard molecular biology techniques and the sequence information provided herein. Using all or a portion of the nucleic acid sequence of SEQ ID NOS: 1, 3, 5, 7, 9, 11, 13, 15, 17, 19, 21, 23, 25, 27, 29, 31, 33, 35, 37, 39, 41 and 43 as a hybridization probe, GPCRX molecules can be isolated using standard hybridization and cloning techniques (e.g., as described in Sambrook, et αl, (eds.), MOLECULAR CLONING: A LABORATORY MANUAL 2nd Ed., Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, 1989; and Ausubel, et αl., (eds.), CURRENT PROTOCOLS IN MOLECULAR BIOLOGY, John Wiley & Sons, New York, NY, 1993.)
A nucleic acid of the invention can be amplified using cDNA, mRNA or alternatively, genomic DNA, as a template and appropriate oligonucleotide primers according to standard PCR amplification techniques. The nucleic acid so amplified can be cloned into an appropriate vector and characterized by DNA sequence analysis. Furthermore, oligonucleotides corresponding to GPCRX nucleotide sequences can be prepared by standard synthetic techniques, e.g., using an automated DNA synthesizer.
As used herein, the term "oligonucleotide" refers to a series of linked nucleotide residues, which oligonucleotide has a sufficient number of nucleotide bases to be used in a PCR reaction. A short oligonucleotide sequence may be based on, or designed from, a genomic or cDNA sequence and is used to amplify, confirm, or reveal the presence of an identical, similar or complementary DNA or RNA in a particular cell or tissue. Oligonucleotides comprise portions of a nucleic acid sequence having about 10 nt, 50 nt, or 100 nt in length, preferably about 15 nt to 30 nt in length. In one embodiment of the invention, an oligonucleotide comprising a nucleic acid molecule less than 100 nt in length would further comprise at least 6 contiguous nucleotides of SEQ ID NOS: 1, 3, 5, 7, 9, 11, 13, 15, 17, 19, 21, 23, 25, 27, 29, 31, 33, 35, 37, 39, 41 and 43, or a complement thereof. Oligonucleotides maybe chemically synthesized and may also be used as probes.
hi another embodiment, an isolated nucleic acid molecule of the invention comprises a nucleic acid molecule that is a complement of the nucleotide sequence shown in SEQ ID NOS: 1, 3, 5, 7, 9, 11, 13, 15, 17, 19, 21, 23, 25, 27, 29, 31, 33, 35, 37, 39, 41 and 43, or aportio of this nucleotide sequence (e.g., a fragment that can be used as a probe or primer or a fragment encoding a biologically-active portion of an GPCRX polypeptide). A nucleic acid molecule that is complementary to the nucleotide sequence shown in SEQ ID NOS: 1, 3, 5, 7, 9, 11, 13, 15, 17, 19, 21, 23, 25, 27, 29, 31, 33, 35, 37, 39, 41 and 43 is one that is sufficiently complementary to the nucleotide sequence shown in SEQ ID NOS: 1, 3, 5, 7, 9, 11, 13, 15, 17, 19, 21, 23, 25, 27, 29, 31, 33, 35, 37, 39, 41 and 43 that it can hydrogen bond with little or no mismatches to the nucleotide sequence shown SEQ ID NOS: 1, 3, 5, 7, 9, 11, 13, 15, 17, 19, 21, 23, 25, 27, 29, 31, 33, 35, 37, 39, 41 and 43, thereby forming a stable duplex.
As used herein, the term "complementary" refers to Watson-Crick or Hoogsteen base pairing between nucleotides units of a nucleic acid molecule, and the term "binding" means the physical or chemical interaction between two polypeptides or compounds or associated polypeptides or compounds or combinations thereof. Binding includes ionic, non-ionic, van der Waals, hydrophobic interactions, and the like. A physical interaction can be either direct or indirect. Indirect interactions may be through or due to the effects of another polypeptide or compound. Direct binding refers to interactions that do not take place through, or due to, the effect of another polypeptide or compound, but instead are without other substantial chemical intermediates.
Fragments provided herein are defined as sequences of at least 6 (contiguous) nucleic acids or at least 4 (contiguous) amino acids, a length sufficient to allow for specific hybridization in the case of nucleic acids or for specific recognition of an epitope in the case of amino acids, respectively, and are at most some portion less than a full length sequence. Fragments may be derived from any contiguous portion of a nucleic acid or amino acid sequence of choice. Derivatives are nucleic acid sequences or amino acid sequences formed from the native compounds either directly or by modification or partial substitution. Analogs are nucleic acid sequences or amino acid sequences that have a structure similar to, but not identical to, the native compound but differs from it in respect to certain components or side chains. Analogs may be synthetic or from a different evolutionary origin and may have a similar or opposite metabolic activity compared to wild type. Homologs are nucleic acid sequences or amino acid sequences of a particular gene that are derived from different species.
Derivatives and analogs may be full length or other than full length, if the derivative or analog contains a modified nucleic acid or amino acid, as described below. Derivatives or
analogs of the nucleic acids or proteins of the invention include, but are not limited to, molecules comprising regions that are substantially homologous to the nucleic acids or proteins of the invention, in various embodiments, by at least about 70%, 80%, or 95% identity (with a preferred identity of 80-95%) over a nucleic acid or amino acid sequence of identical size or when compared to an aligned sequence in which the alignment is done by a computer homology program known in the art, or whose encoding nucleic acid is capable of hybridizing to the complement of a sequence encoding the aforementioned proteins under stringent, moderately stringent, or low stringent conditions. See e.g. Ausubel, et al., CURRENT PROTOCOLS IN MOLECULAR BIOLOGY, John Wiley & Sons, New York, NY, 1993, and below. A "homologous nucleic acid sequence" or "homologous amino acid sequence," or variations thereof, refer to sequences characterized by a homology at the nucleotide level or amino acid level as discussed above. Homologous nucleotide sequences encode those sequences coding for isoforms of GPCRX polypeptides. Isoforms can be expressed in different tissues of the same organism as a result of, for example, alternative splicing of RNA. Alternatively, isoforms can be encoded by different genes. In the invention, homologous nucleotide sequences include nucleotide sequences encoding for an GPCRX polypeptide of species other than humans, including, but not limited to: vertebrates, and thus can include, e.g., frog, mouse, rat, rabbit, dog, cat cow, horse, and other organisms. Homologous nucleotide sequences also include, but are not limited to, naturally occurring allelic variations and mutations of the nucleotide sequences set forth herein. A homologous nucleotide sequence does not, however, include the exact nucleotide sequence encoding human GPCRX protein. Homologous nucleic acid sequences include those nucleic acid sequences that encode conservative amino acid substitutions (see below) in SEQ ID NOS: 1, 3, 5, 7, 9, 11, 13, 15, 17, 19, 21, 23, 25, 27, 29, 31, 33, 35, 37, 39, 41 and 43, as well as a polypeptide possessing GPCRX biological activity. Various biological activities of the GPCRX proteins are described below.
An GPCRX polypeptide is encoded by the open reading frame ("ORF") of an GPCRX nucleic acid. An ORF corresponds to a nucleotide sequence that could potentially be translated into a polypeptide. A stretch of nucleic acids comprising an ORF is uninterrupted by a stop codon. An ORF that represents the coding sequence for a full protein begins with an ATG
"start" codon and terminates with one of the three "stop" codons, namely, TAA, TAG, or TGA. For the purposes of this invention, an ORF may be any part of a coding sequence, with or without a start codon, a stop codon, or both. For an ORF to be considered as a good candidate
δ7
for coding for a bonafide cellular protein, a minimum size requirement is often set, e.g., a stretch of DNA that would encode a protein of 50 amino acids or more.
The nucleotide sequences determined from the cloning of the human GPCRX genes allows for the generation of probes and primers designed for use in identifying and/or cloning GPCRX homologues in other cell types, e.g. from other tissues, as well as GPCRX homologues from other vertebrates. The probe/primer typically comprises substantially purified oligonucleotide. The oligonucleotide typically comprises a region of nucleotide sequence that hybridizes under stringent conditions to at least about 12, 25, 50, 100, 150, 200, 250, 300, 350 or 400 consecutive sense strand nucleotide sequence of SEQ ID NOS: 1, 3, 5, 7, 9, 11, 13, 15, 17, 19, 21, 23, 25, 27, 29, 31, 33, 35, 37, 39, 41 and 43; or an anti-sense strand nucleotide sequence of SEQ ID NOS: 1, 3, 5, 7, 9, 11, 13, 15, 17, 19, 21, 23, 25, 27, 29, 31, 33, 35, 37, 39, 41 and 43; or of a naturally occurring mutant of SEQ ID NOS: 1, 3, 5, 7, 9, 11, 13, 15, 17, 19, 21, 23, 25, 27, 29, 31, 33, 35, 37, 39, 41 and 43.
Probes based on the human GPCRX nucleotide sequences can be used to detect transcripts or genomic sequences encoding the same or homologous proteins. In various embodiments, the probe further comprises a label group attached thereto, e.g. the label group can be a radioisotope, a fluorescent compound, an enzyme, or an enzyme co-factor. Such probes can be used as a part of a diagnostic test kit for identifying cells or tissues which mis- express an GPCRX protein, such as by measuring a level of an GPCRX-encoding nucleic acid in a sample of cells from a subject e.g., detecting GPCRX mRNA levels or determining whether a genomic GPCRX gene has been mutated or deleted.
"A polypeptide having a biologically-active portion of an GPCRX polypeptide" refers to polypeptides exhibiting activity similar, but not necessarily identical to, an activity of a polypeptide of the invention, including mature forms, as measured in a particular biological assay, with or without dose dependency. A nucleic acid fragment encoding a "biologically- active portion of GPCRX" can be prepared by isolating a portion SEQ JD NOS: 1, 3, 5, 7, 9, 11, 13, 15, 17, 19, 21, 23, 25, 27, 29, 31, 33, 35, 37, 39, 41 and 43 that encodes a polypeptide having an GPCRX biological activity (the biological activities of the GPCRX proteins are described below), expressing the encoded portion of GPCRX protein (e.g., by recombinant expression in vitro) and assessing the activity of the encoded portion of GPCRX.
GPCRX Nucleic Acid and Polypeptide Variants
The invention further encompasses nucleic acid molecules that differ from the nucleotide sequences shown SEQ ID NOS: 1, 3, 5, 7, 9, 11, 13, 15, 17, 19, 21, 23, 25, 27, 29, δ8
31, 33, 35, 37, 39, 41 and 43due to degeneracy of the genetic code and thus encode the same GPCRX proteins as that encoded by the nucleotide sequences shown in SEQ ID NOS: 1, 3, 5, 7, 9, 11, 13, 15, 17, 19, 21, 23, 25, 27, 29, 31, 33, 35, 37, 39, 41 and 43. In another embodiment, an isolated nucleic acid molecule of the invention has a nucleotide sequence encoding a protein having an amino acid sequence shown in SEQ ID NOS: 2, 4, 6, 8, 10, 12, 14, 16, 18, 20, 22, 24, 26, 28, 30, 32, 34, 36, 38, 40, 42 and 44. h addition to the human GPCRX nucleotide sequences shown in SEQ ID NOS: 1, 3, 5, 7, 9, 11, 13, 15, 17, 19, 21, 23, 25, 27, 29, 31, 33, 35, 37, 39, 41 and 43 it will be appreciated by those skilled in the art that DNA sequence polymorphisms that lead to changes in the amino acid sequences of the GPCRX polypeptides may exist within a population (e.g., the human population). Such genetic polymorphism in the GPCRX genes may exist among individuals within a population due to natural allelic variation. As used herein, the terms "gene" and "recombinant gene" refer to nucleic acid molecules comprising an open reading frame (ORF) encoding an GPCRX protein, preferably a vertebrate GPCRX protein. Such natural allelic variations can typically result in 1-5% variance in the nucleotide sequence of the GPCRX genes. Any and all such nucleotide variations and resulting amino acid polymoφhisms in the GPCRX polypeptides, which are the result of natural allelic variation and that do not alter the functional activity of the GPCRX polypeptides, are intended to be within the scope of the invention. Moreover, nucleic acid molecules encoding GPCRX proteins from other species, and thus that have a nucleotide sequence that differs from the human sequence SEQ ID NOS: 1, 3, 5, 7, 9, 11, 13, 15, 17, 19, 21, 23, 25, 27, 29, 31, 33, 35, 37, 39, 41 and 43 are intended to be within the scope of the invention. Nucleic acid molecules corresponding to natural allelic variants and homologues of the GPCRX cDNAs of the invention can be isolated based on their homology to the human GPCRX nucleic acids disclosed herein using the human cDNAs, or a portion thereof, as a hybridization probe according to standard hybridization techniques under stringent hybridization conditions.
Accordingly, in another embodiment, an isolated nucleic acid molecule of the invention is at least 6 nucleotides in length and hybridizes under stringent conditions to the nucleic acid molecule comprising the nucleotide sequence of SEQ ID NOS: 1, 3, 5, 7, 9, 11, 13, 15, 17, 19,
21, 23, 25, 27, 29, 31, 33, 35, 37, 39, 41 and 43. In another embodiment, the nucleic acid is at least 10, 25, 50, 100, 250, 500, 750, 1000, 1500, or 2000 or more nucleotides in length. In yet another embodiment, an isolated nucleic acid molecule of the invention hybridizes to the coding region. As used herein, the term "hybridizes under stringent conditions" is intended to describe
conditions for hybridization and washing under which nucleotide sequences at least 60% homologous to each other typically remain hybridized to each other.
Homologs (i.e., nucleic acids encoding GPCRX proteins derived from species other than human) or other related sequences (e.g., paralogs) can be obtained by low, moderate or high stringency hybridization with all or a portion of the particular human sequence as a probe using methods well known in the art for nucleic acid hybridization and cloning.
As used herein, the phrase "stringent hybridization conditions" refers to conditions under which a probe, primer or oligonucleotide will hybridize to its target sequence, but to no other sequences. Stringent conditions are sequence-dependent and will be different in different circumstances. Longer sequences hybridize specifically at higher temperatures than shorter sequences. Generally, stringent conditions are selected to be about 5 °C lower than the thermal melting point (Tm) for the specific sequence at a defined ionic strength and pH. The Tm is the temperature (under defined ionic strength, pH and nucleic acid concentration) at which 50% of the probes complementary to the target sequence hybridize to the target sequence at equilibrium. Since the target sequences are generally present at excess, at Tm, 50% of the probes are occupied at equilibrium. Typically, stringent conditions will be those in which the salt concentration is less than about 1.0 M sodium ion, typically about 0.01 to 1.0 M sodium ion (or other salts) at pH 7.0 to 8.3 and the temperature is at least about 30°C for short probes, primers or oligonucleotides (e.g., 10 nt to 50 nt) and at least about 60°C for longer probes, primers and oligonucleotides. Stringent conditions may also be achieved with the addition of destabilizing agents, such as formamide.
Stringent conditions are known to those skilled in the art and can be found in Ausubel, et al., (eds.), CURRENT PROTOCOLS IN MOLECULAR BIOLOGY, John Wiley & Sons, NY. (1989), 6.3.1-6.3.6. Preferably, the conditions are such that sequences at least about 65%, 70%, 75%, 85%, 90%, 95%, 98%, or 99% homologous to each other typically remain hybridized to each other. A non-limiting example of stringent hybridization conditions are hybridization in a high salt buffer comprising 6X SSC, 50 mM Tris-HCl (pH 7.5), 1 mM EDTA, 0.02% PVP, 0.02% Ficoll, 0.02% BSA, and 500 mg/ml denatured salmon sperm DNA at 65°C, followed by one or more washes in 0.2X SSC, 0.01% BSA at 50°C. An isolated nucleic acid molecule of the invention that hybridizes under stringent conditions to the sequences of SEQ ID NOS: 1, 3, 5, 7, 9, 11, 13, 15, 17, 19, 21, 23, 25, 27, 29, 31, 33, 35, 37, 39, 41 and 43 corresponds to a naturally-occurring nucleic acid molecule. As used herein, a "naturally-occurring" nucleic acid
molecule refers to an RNA or DNA molecule having a nucleotide sequence that occurs in nature (e.g., encodes a natural protein).
In a second embodiment, a nucleic acid sequence that is hybridizable to the nucleic acid molecule comprising the nucleotide sequence of SEQ ID NOS: 1, 3, 5, 7, 9, 11, 13, 15, 17, 19, 21, 23, 25, 27, 29, 31, 33, 35, 37, 39, 41 and 43 or fragments, analogs or derivatives thereof, under conditions of moderate stringency is provided. A non-limiting example of moderate stringency hybridization conditions are hybridization in 6X SSC, 5X Denhardt's solution, 0.5% SDS and 100 mg/ml denatured salmon sperm DNA at 55°C, followed by one or more washes in IX SSC, 0.1% SDS at 37°C. Other conditions of moderate stringency that may be used are well-known within the art. See, e.g., Ausubel, et al. (eds.), 1993, CURRENT PROTOCOLS IN MOLECULAR BIOLOGY, John Wiley & Sons, NY, and Kriegler, 1990; GENE TRANSFER AND EXPRESSION, A LABORATORY MANUAL, Stockton Press, NY.
In a third embodiment, a nucleic acid that is hybridizable to the nucleic acid molecule comprising the nucleotide sequences of SEQ ID NOS: 1, 3, 5, 7, 9, 11, 13, 15, 17, 19, 21, 23, 25, 27, 29, 31, 33, 35, 37, 39, 41 and 43 or fragments, analogs or derivatives thereof, under conditions of low stringency, is provided. A non-limiting example of low stringency hybridization conditions are hybridization in 35% formamide, 5X SSC, 50 mM Tris-HCl (pH 7.5), 5 mM EDTA, 0.02% PVP, 0.02% Ficoll, 0.2% BSA, 100 mg/ml denatured salmon sperm DNA, 10% (wt/vol) dextran sulfate at 40°C, followed by one or more washes in 2X SSC, 25 mM Tris-HCl (pH 7.4), 5 mM EDTA, and 0.1 % SDS at 50°C. Other conditions of low stringency that may be used are well known in the art (e.g., as employed for cross-species hybridizations). See, e.g., Ausubel, et al. (eds.), 1993, CURRENT PROTOCOLS IN MOLECULAR BIOLOGY, John Wiley & Sons, NY, and Kriegler, 1990, GENE TRANSFER AND EXPRESSION, A LABORATORY MANUAL, Stockton Press, NY; Shilo and Weinberg, 1981. Proc Natl Acad Sci USA 78: 6789-6792.
Conservative Mutations
In addition to naturally-occurring allelic variants of GPCRX sequences that may exist in the population, the skilled artisan will further appreciate that changes can be introduced by mutation into the nucleotide sequences of SEQ ID NOS: 1, 3, 5, 7, 9, 11, 13, 15, 17, 19, 21, 23,
25, 27, 29, 31, 33, 35, 37, 39, 41 and 43 thereby leading to changes in the amino acid sequences of the encoded GPCRX proteins, without altering the functional ability of said GPCRX proteins. For example, nucleotide substitutions leading to amino acid substitutions at
"non-essential" amino acid residues can be made in the sequence of SEQ ED NOS: 2, 4, 6, 8, 10,
12, 14, 16, 18, 20, 22, 24, 26, 28, 30, 32, 34, 36, 38, 40, 42 and 44. A "non-essential" amino acid residue is a residue that can be altered from the wild-type sequences of the GPCRX proteins without altering their biological activity, whereas an "essential" amino acid residue is required for such biological activity. For example, amino acid residues that are conserved among the GPCRX proteins of the invention are predicted to be particularly non-amenable to alteration. Amino acids for which conservative substitutions can be made are well-known within the art.
Another aspect of the invention pertains to nucleic acid molecules encoding GPCRX proteins that contain changes in amino acid residues that are not essential for activity. Such GPCRX proteins differ in amino acid sequence from SEQ ID NOS:2, 4, 6, 8, 10, 12, 14, 16, 18, 20, 22, 24, 26, 28, 30, 32, 34, 36, 38, 40, 42 and 44 yet retain biological activity. In one embodiment, the isolated nucleic acid molecule comprises a nucleotide sequence encoding a protein, wherein the protein comprises an amino acid sequence at least about 45% homologous to the amino acid sequences of SEQ ID NOS:2, 4, 6, 8, 10, 12, 14, 16, 18, 20, 22, 24, 26, 28, 30, 32, 34, 36, 38, 40, 42 and 44. Preferably, the protein encoded by the nucleic acid molecule is at least about 60% homologous to SEQ ID NOS:2, 4, 6, 8, 10, 12, 14, 16, 18, 20, 22, 24, 26, 28, 30, 32, 34, 36, 38, 40, 42 and 44; more preferably at least about 70% homologous to SEQ ID NOS:2, 4, 6, 8, 10, 12, 14, 16, 18, 20, 22, 24, 26, 28, 30, 32, 34, 36, 38, 40, 42 and 44; still more preferably at least about 80% homologous to SEQ ID NOS:2, 4, 6, 8, 10, 12, 14, 16, 18, 20, 22, 24, 26, 28, 30, 32, 34, 36, 38, 40, 42 and 44; even more preferably at least about 90% homologous to SEQ ID NOS:2, 4, 6, 8, 10, 12, 14, 16, 18, 20, 22, 24, 26, 28, 30, 32, 34, 36, 38, 40, 42 and 44; and most preferably at least about 95% homologous to SEQ ID NOS:2, 4, 6, 8, 10, 12, 14, 16, 18, 20, 22, 24, 26, 28, 30, 32, 34, 36, 38, 40, 42 and 44.
An isolated nucleic acid molecule encoding an GPCRX protein homologous to the protein of SEQ ID NOS:2, 4, 6, 8, 10, 12, 14, 16, 18, 20, 22, 24, 26, 28, 30, 32, 34, 36, 38, 40, 42 and 44 can be created by introducing one or more nucleotide substitutions, additions or deletions into the nucleotide sequence of SEQ ID NOS: 1, 3, 5, 7, 9, 11, 13, 15, 17, 19, 21, 23, 25, 27, 29, 31, 33, 35, 37,' 39, 41 and 43 such that one or more amino acid substitutions, additions or deletions are introduced into the encoded protein. Mutations can be introduced into SEQ ID NOS:2, 4, 6, 8, 10, 12, 14, 16, 18, 20, 22, 24,
26, 28, 30, 32, 34, 36, 38, 40, 42 and 44 by standard techniques, such as site-directed mutagenesis and PCR-mediated mutagenesis. Preferably, conservative amino acid substitutions are made at one or more predicted, non-essential amino acid residues. A "conservative amino acid substitution" is one in which the amino acid residue is replaced with an amino acid residue
having a similar side chain. Families of amino acid residues having similar side chains have been defined within the art. These families include amino acids with basic side chains (e.g., lysine, arginine, histidine), acidic side chains (e.g., aspartic acid, glutamic acid), uncharged polar side chains (e.g., glycine, asparagine, glutamine, serine, threonine, tyrosine, cysteine), nonpolar side chains (e.g., alanine, valine, leucine, isoleucine, proline, phenylalanine, methionine, tryptophan), beta-branched side chains (e.g., threonine, valine, isoleucine) and aromatic side chains (e.g., tyrosine, phenylalanine, tryptophan, histidine). Thus, a predicted non-essential amino acid residue in the GPCRX protein is replaced with another amino acid residue from the same side chain family. Alternatively, in another embodiment, mutations can be introduced randomly along all or part of an GPCRX coding sequence, such as by saturation mutagenesis, and the resultant mutants can be screened for GPCRX biological activity to identify mutants that retain activity. Following mutagenesis of SEQ ID NOS: 1, 3, 5, 7, 9, 11, 13, 15, 17, 19, 21, 23, 25, 27, 29, 31, 33, 35, 37, 39, 41 and 43, the encoded protein can be expressed by any recombinant technology known in the art and the activity of the protein can be determined.
The relatedness of amino acid families may also be determined based on side chain interactions. Substituted amino acids may be fully conserved "strong" residues or fully conserved "weak" residues. The "strong" group of conserved amino acid residues may be any one of the following groups: STA, NEQK, NHQK, NDEQ, QHRK, MILV, MILF, HY, FYW, wherein the single letter amino acid codes are grouped by those amino acids that may be substituted for each other. Likewise, the "weak" group of conserved residues may be any one of the following: CSA, ATV, SAG, STNK, STPA, SGND, SNDEQK, NDEQHK, NEQHRK, VLIM, HFY, wherein the letters within each group represent the single letter amino acid code, hi one embodiment, a mutant GPCRX protein can be assayed for (i) the ability to form protei protein interactions with other GPCRX proteins, other cell-surface proteins, or biologically-active portions thereof, (ii) complex formation between a mutant GPCRX protein and an GPCRX ligand; or (Hi) the ability of a mutant GPCRX protein to bind to an intracellular target protein or biologically-active portion thereof; (e.g. avidin proteins).
In yet another embodiment, a mutant GPCRX protein can be assayed for the ability to regulate a specific biological function (e.g., regulation of insulin release).
Antisense Nucleic Acids
Another aspect of the invention pertains to isolated antisense nucleic acid molecules that are hybridizable to or complementary to the nucleic acid molecule comprising the nucleotide
sequence of SEQ ID NOS: 1, 3, 5, 7, 9, 11, 13, 15, 17, 19, 21, 23, 25, 27, 29, 31, 33, 35, 37, 39, 41 and 43, or fragments, analogs or derivatives thereof. An "antisense" nucleic acid comprises a nucleotide sequence that is complementary to a "sense" nucleic acid encoding a protein (e.g., complementary to the coding strand of a double-stranded cDNA molecule or complementary to an RNA sequence). In specific aspects, antisense nucleic acid molecules are provided that comprise a sequence complementary to at least about 10, 25, 50, 100, 250 or 500 nucleotides or an entire GPCRX coding strand, or to only a portion thereof. Nucleic acid molecules encoding fragments, homologs, derivatives and analogs of an GPCRX protein of SEQ ID NOS:2, 4, 6, 8, 10, 12, 14, 16, 18, 20, 22, 24, 26, 28, 30, 32, 34, 36, 38, 40, 42 and 44, or antisense nucleic acids complementary to an GPCRX nucleic acid sequence of SEQ ID NOS: 1, 3, 5, 7, 9, 11, 13, 15, 17, 19, 21, 23, 25, 27, 29, 31, 33, 35, 37, 39, 41 and 43, are additionally provided.
In one embodiment, an antisense nucleic acid molecule is antisense to a "coding region" of the coding strand of a nucleotide sequence encoding an GPCRX protein. The term "coding region" refers to the region of the nucleotide sequence comprising codons which are translated into amino acid residues. In another embodiment, the antisense nucleic acid molecule is antisense to a "noncoding region" of the coding strand of a nucleotide sequence encoding the GPCRX protein. The term "noncoding region" refers to 5' and 3' sequences which flank the coding region that are not translated into amino acids (i.e., also referred to as 5' and 3' untranslated regions). Given the coding strand sequences encoding the GPCRX protein disclosed herein, antisense nucleic acids of the invention can be designed according to the rules of Watson and Crick or Hoogsteen base pairing. The antisense nucleic acid molecule can be complementary to the entire coding region of GPCRX mRNA, but more preferably is an oligonucleotide that is antisense to only a portion of the coding or noncoding region of GPCRX mRNA. For example, the antisense oligonucleotide can be complementary to the region surrounding the translation start site of GPCRX mRNA. An antisense oligonucleotide can be, for example, about 5, 10, 15, 20, 25, 30, 35, 40, 45 or 50 nucleotides in length. An antisense nucleic acid of the invention can be constructed using chemical synthesis or enzymatic ligation reactions using procedures known in the art. For example, an antisense nucleic acid (e.g., an antisense oligonucleotide) can be chemically synthesized using naturally-occurring nucleotides or variously modified nucleotides designed to increase the biological stability of the molecules or to increase the physical stability of the duplex formed between the antisense and sense nucleic acids (e.g., phosphorothioate derivatives and acridine substituted nucleotides can be used).
Examples of modified nucleotides that can be used to generate the antisense nucleic acid include: 5-fluorouracil, 5-bromouracil, 5-chlorouracil, 5-iodouracil, hypoxanthine, xanthine, 4-acetylcytosine, 5-(carboxyhydroxyhnethyl) uracil, 5-carboxymethylaminomethyl- 2-thiouridine, 5-carboxymethylaminomethyluracil, dihydrouracil, beta-D-galactosylqueosine, inosine, N6-isopentenyladenine, 1-methylguanine, 1-methylinosine, 2,2-dimethylguanine, 2-methyladenine, 2-methylguanine, 3-methylcytosine, 5-methylcytosine, N6-adenine, 7-methylguanine, 5-methylaminomethyluracil, 5-methoxyaminomethyl-2-thiouracil, beta-D-mannosylqueosine, 5'-methoxycarboxymethyluracil, 5-methoxyuracil, 2-methylthio-N6-isopentenyladenine, uracil-5-oxyacetic acid (v), wybutoxosine, pseudouracil, queosine, 2-thiocytosine, 5-methyl-2-tlιiouracil, 2-thiouracil, 4-thiouracil, 5-methyluracil, uracil-5-oxyacetic acid methylester, uracil-5-oxyacetic acid (v), 5-methyl-2-thiouracil, 3-(3-amino-3-N-2-carboxypropyl) uracil, (acp3)w, and 2,6-diaminopurine. Alternatively, the antisense nucleic acid can be produced biologically using an expression vector into which a nucleic acid has been subcloned in an antisense orientation (i.e., RNA transcribed from the inserted nucleic acid will be of an antisense orientation to a target nucleic acid of interest, described further in the following subsection).
The antisense nucleic acid molecules of the invention are typically administered to a subject or generated in situ such that they hybridize with or bind to cellular mRNA and/or genomic DNA encoding an GPCRX protein to thereby inhibit expression of the protein (e.g., by inhibiting transcription and/or translation). The hybridization can be by conventional nucleotide complementarity to form a stable duplex, or, for example, in the case of an antisense nucleic acid molecule that binds to DNA duplexes, through specific interactions in the major groove of the double helix. An example of a route of administration of antisense nucleic acid molecules of the invention includes direct injection at a tissue site. Alternatively, antisense nucleic acid molecules can be modified to target selected cells and then administered systemically. For example, for systemic administration, antisense molecules can be modified such that they specifically bind to receptors or antigens expressed on a selected cell surface (e.g., by linking the antisense nucleic acid molecules to peptides or antibodies that bind to cell surface receptors or antigens). The antisense nucleic acid molecules can also be delivered to cells using the vectors described herein. To achieve sufficient nucleic acid molecules, vector constructs in which the antisense nucleic acid molecule is placed under the control of a strong pol IT or pol III promoter are preferred.
In yet another embodiment, the antisense nucleic acid molecule of the invention is an -anomeric nucleic acid molecule. An α-anomeric nucleic acid molecule forms specific
double-stranded hybrids with complementary RNA in which, contrary to the usual β-units, the strands run parallel to each other. See, e.g., Gaultier, et al., 1987. Nucl. Acids Res. 15: 6625-6641. The antisense nucleic acid molecule can also comprise a 2'-o-methylribonucleotide (see, e.g., frioue, et al. 1987. Nucl. Acids Res. 15: 6131-6148) or a chimeric RNA-DNA analogue (see, e.g., frioue, et al, 1987. FEES Lett. 215: 327-330.
Ribozymes and PNA Moieties
Nucleic acid modifications include, by way of non-limiting example, modified bases, and nucleic acids whose sugar phosphate backbones are modified or derivatized. These modifications are carried out at least in part to enhance the chemical stability of the modified nucleic acid, such that they may be used, for example, as antisense binding nucleic acids in therapeutic applications in a subject. hi one embodiment, an antisense nucleic acid of the invention is a ribozyme. Ribozymes are catalytic RNA molecules with ribonuclease activity that are capable of cleaving a single-stranded nucleic acid, such as an mRNA, to which they have a complementary region. Thus, ribozymes (e.g., hammerhead ribozymes as described in Haselhoff and Gerlach 1988. Nature 334: 585-591) can be used to catalytically cleave GPCRX mRNA transcripts to thereby inhibit translation of GPCRX mRNA. A ribozyme having specificity for an GPCRX-encoding nucleic acid can be designed based upon the nucleotide sequence of an GPCRX cDNA disclosed herein (i.e., SEQ TD NOS: 1, 3, 5, 7, 9, 11, 13, 15, 17, 19, 21, 23, 25, 27, 29, 31, 33, 35, 37, 39, 41 and 43). For example, a derivative of a Tetrahymena L-19 IVS RNA can be constructed in which the nucleotide sequence of the active site is complementary to the nucleotide sequence to be cleaved in an GPCRX-encoding mRNA. See, e.g., U.S. Patent 4,987,071 to Cech, et al. and U.S. Patent 5,116,742 to Cech, et al. GPCRX mRNA can also be used to select a catalytic RNA having a specific ribonuclease activity from a pool of RNA molecules. See, e.g., Bartel et al, (1993) Science 261:1411-1418.
Alternatively, GPCRX gene expression can be inhibited by targeting nucleotide sequences complementary to the regulatory region of the GPCRX nucleic acid (e.g., the GPCRX promoter and/or enhancers) to form triple helical structures that prevent transcription of the GPCRX gene in target cells. See, e.g., Helene, 1991. Anticancer Drug Des. 6: 569-84;
Helene, et al. 1992. Ann. NY. Acad. Sci. 660: 27-36; Maher, 1992. Bioassays 14: 807-15.
In various embodiments, the GPCRX nucleic acids can be modified at the base moiety, sugar moiety or phosphate backbone to improve, e.g., the stability, hybridization, or solubility of the molecule. For example, the deoxyribose phosphate backbone of the nucleic acids can be
modified to generate peptide nucleic acids. See, e.g., Hyrup, et al, 1996. BioorgMed Chem 4: 5-23. As used herein, the terms "peptide nucleic acids" or "PNAs" refer to nucleic acid mimics (e.g., DNA mimics) in which the deoxyribose phosphate backbone is replaced by a pseudopeptide backbone and only the four natural nucleobases are retained. The neutral backbone of PNAs has been shown to allow for specific hybridization to DNA and RNA under conditions of low ionic strength. The synthesis of PNA oligomers can be performed using standard solid phase peptide synthesis protocols as described in Hyrup, et al, 1996. supra; Perry-O'Keefe, et al, 1996. Proc. Natl. Acad. Sci. USA 93: 14670-14675.
PNAs of GPCRX can be used in therapeutic and diagnostic applications. For example, PNAs can be used as antisense or antigene agents for sequence-specific modulation of gene expression by, e.g., inducing transcription or translation arrest or inhibiting replication. PNAs of GPCRX can also be used, for example, in the analysis of single base pair mutations in a gene (e.g., PNA directed PCR clamping; as artificial restriction enzymes when used in combination with other enzymes, e.g., S] nucleases (see, Hyrup, et al, 1996. supra); or as probes or primers for DNA sequence and hybridization (see, Hyrup, et al, 1996, supra; Perry-O'Keefe, et al, 1996. supra).
In another embodiment, PNAs of GPCRX can be modified, e.g., to enhance their stability or cellular uptake, by attaching lipophilic or other helper groups to PNA, by the formation of PNA-DNA chimeras, or by the use of liposomes or other techniques of drug delivery known in the art. For example, PNA-DNA chimeras of GPCRX can be generated that may combine the advantageous properties of PNA and DNA. Such chimeras allow DNA recognition enzymes (e.g., RNase H and DNA polymerases) to interact with the DNA portion while the PNA portion would provide high binding affinity and specificity. PNA-DNA chimeras can be linked using linkers of appropriate lengths selected in terms of base stacking, number of bonds between the nucleobases, and orientation (see, Hyrup, et al., 1996. supra). The synthesis of PNA-DNA chimeras can be performed as described in Hyrup, et al, 1996. supra and Finn, et al, 1996. Nucl Acids Res 24: 3357-3363. For example, a DNA chain can be synthesized on a solid support using standard phosphoramidite coupling chemistry, and modified nucleoside analogs, e.g., 5'-(4-methoxytrityl)amino-5'-deoxy-thymidine phosphoramidite, can be used between the PNA and the 5' end of DNA. See, e.g., Mag, et al, 1989. Nucl Acid Res 17: 5973-5988. PNA monomers are then coupled in a stepwise manner to produce a chimeric molecule with a 5' PNA segment and a 3' DNA segment. See, e.g., Finn, et al, 1996. supra. Alternatively, chimeric molecules can be synthesized with a 5' DNA segment
and a 3' PNA segment. See, e.g., Petersen, et al, 1975. Bioorg. Med. Chem. Lett. 5: 1119-11124.
In other embodiments, the oligonucleotide may include other appended groups such as peptides (e.g., for targeting host cell receptors in vivo), or agents facilitating transport across the cell membrane (see, e.g., Letsinger, et al, 1989. Proc. Natl. Acad. Sci. U.S.A. 86: 6553-6556; Lemaitre, et al, 1987. Proc. Natl. Acad. Sci. 84: 648-652; PCT Publication No. WO88/09810) or the blood-brain barrier (see, e.g., PCT Publication No. WO 89/10134). In addition, oligonucleotides can be modified with hybridization triggered cleavage agents (see, e.g., Krol, et al, 1988. BioTechniques 6:958-976) or intercalating agents (see, e.g., Zon, 1988. Pharm. Res. 5: 539-549). To this end, the oligonucleotide may be conjugated to another molecule, e.g., a peptide, a hybridization triggered cross-linking agent, a transport agent, a hybridization-triggered cleavage agent, and the like.
GPCRX Polypeptides
A polypeptide according to the invention includes a polypeptide including the amino acid sequence of GPCRX polypeptides whose sequences are provided in SEQ ID NOS:2, 4, 6, 8, 10, 12, 14, 16, 18, 20, 22, 24, 26, 28, 30, 32, 34, 36, 38, 40, 42 and 44. The invention also includes a mutant or variant protein any of whose residues may be changed from the corresponding residues shown in SEQ ID NOS:2, 4, 6, 8, 10, 12, 14, 16, 18, 20, 22, 24, 26, 28, 30, 32, 34, 36, 38, 40, 42 and 44 while still encoding a protein that maintains its GPCRX activities and physiological functions, or a functional fragment thereof.
In general, an GPCRX variant that preserves GPCRX-like function includes any variant in which residues at a particular position in the sequence have been substituted by other amino acids, and further include the possibility of inserting an additional residue or residues between two residues of the parent protein as well as the possibility of deleting one or more residues from the parent sequence. Any amino acid substitution, insertion, or deletion is encompassed by the invention, hi favorable circumstances, the substitution is a conservative substitution as defined above.
One aspect of the invention pertains to isolated GPCRX proteins, and biologically- active portions thereof, or derivatives, fragments, analogs or homologs thereof. Also provided are polypeptide fragments suitable for use as immunogens to raise anti-GPCRX antibodies. In one embodiment, native GPCRX proteins can be isolated from cells or tissue sources by an appropriate purification scheme using standard protein purification techniques. In another embodiment, GPCRX proteins are produced by recombinant DNA techniques. Alternative to
recombinant expression, an GPCRX protein or polypeptide can be synthesized chemically using standard peptide synthesis techniques.
An "isolated" or "purified" polypeptide or protein or biologically-active portion thereof is substantially free of cellular material or other contaminating proteins from the cell or tissue source from which the GPCRX protein is derived, or substantially free from chemical precursors or other chemicals when chemically synthesized. The language "substantially free of cellular material" includes preparations of GPCRX proteins in which the protein is separated from cellular components of the cells from which it is isolated or recombinantly-produced. i one embodiment, the language "substantially free of cellular material" includes preparations of GPCRX proteins having less than about 30% (by dry weight) of non-GPCRX proteins (also referred to herein as a "contaminating protein"), more preferably less than about 20% of non-GPCRX proteins, still more preferably less than about 10% of non-GPCRX proteins, and most preferably less than about 5% of non-GPCRX proteins. When the GPCRX protein or biologically-active portion thereof is recombinantly-produced, it is also preferably substantially free of culture medium, i.e., culture medium represents less than about 20%, more preferably less than about 10%, and most preferably less than about 5% of the volume of the GPCRX protein preparation.
The language "substantially free of chemical precursors or other chemicals" includes preparations of GPCRX proteins in which the protein is separated from chemical precursors or other chemicals that are involved in the synthesis of the protein, h one embodiment, the language "substantially free of chemical precursors or other chemicals" includes preparations of GPCRX proteins having less than about 30% (by dry weight) of chemical precursors or non-GPCRX chemicals, more preferably less than about 20% chemical precursors or non-GPCRX chemicals, still more preferably less than about 10% chemical precursors or non-GPCRX chemicals, and most preferably less than about 5% chemical precursors or non-GPCRX chemicals.
Biologically-active portions of GPCRX proteins include peptides comprising amino acid sequences sufficiently homologous to or derived from the amino acid sequences of the GPCRX proteins (e.g., the amino acid sequence shown in SEQ ID NOS:2, 4, 6, 8, 10, 12, 14, 16, 18, 20, 22, 24, 26, 28, 30, 32, 34, 36, 38, 40, 42 and 44) that include fewer amino acids than the full- length GPCRX proteins, and exhibit at least one activity of an GPCRX protein. Typically, biologically-active portions comprise a domain or motif with at least one activity of the GPCRX protein. A biologically-active portion of an GPCRX protein can be a polypeptide which is, for example, 10, 25, 50, 100 or more amino acid residues in length.
Moreover, other biologically-active portions, in which other regions of the protein are deleted, can be prepared by recombinant techniques and evaluated for one or more of the functional activities of a native GPCRX protein.
In an embodiment, the GPCRX protein has an amino acid sequence shown in SEQ ID NOS:2, 4, 6, 8, 10, 12, 14, 16, 18, 20, 22, 24, 26, 28, 30, 32, 34, 36, 38, 40, 42 and 44. In other embodiments, the GPCRX protein is substantially homologous to SEQ ID NOS:2, 4, 6, 8, 10, 12, 14, 16, 18, 20, 22, 24, 26, 28, 30, 32, 34, 36, 38, 40, 42 and 44, and retains the functional activity of the protein of SEQ ID NOS:2, 4, 6, 8, 10, 12, 14, 16, 18, 20, 22, 24, 26, 28, 30, 32, 34, 36, 38, 40, 42 and 44, yet differs in amino acid sequence due to natural allelic variation or mutagenesis, as described in detail, below. Accordingly, in another embodiment, the GPCRX protein is a protein that comprises an amino acid sequence at least about 45% homologous to the amino acid sequence SEQ ID NOS:2, 4, 6, 8, 10, 12, 14, 16, 18, 20, 22, 24, 26, 28, 30, 32, 34, 36, 38, 40, 42 and 44, and retains the functional activity of the GPCRX proteins of SEQ ID NOS:2, 4, 6, 8, 10, 12, 14, 16, 18, 20, 22, 24, 26, 28, 30, 32, 34, 36, 38, 40, 42 and 44.
Determining Homology Between Two or More Sequences
To determine the percent homology of two amino acid sequences or of two nucleic acids, the sequences are aligned for optimal comparison purposes (e.g., gaps can be introduced in the sequence of a first amino acid or nucleic acid sequence for optimal alignment with a second amino or nucleic acid sequence). The amino acid residues or nucleotides at corresponding amino acid positions or nucleotide positions are then compared. When a position in the first sequence is occupied by the same amino acid residue or nucleotide as the corresponding position in the second sequence, then the molecules are homologous at that position (i.e., as used herein amino acid or nucleic acid "homology" is equivalent to amino acid or nucleic acid "identity").
The nucleic acid sequence homology may be determined as the degree of identity between two sequences. The homology may be determined using computer programs known in the art, such as GAP software provided in the GCG program package. See, Needleman and Wunsch, 1970. JMol Biol 48: 443-453. Using GCG GAP software with the following settings for nucleic acid sequence comparison: GAP creation penalty of 5.0 and GAP extension penalty of 0.3, the coding region of the analogous nucleic acid sequences referred to above exhibits a degree of identity preferably of at least 70%, 75%, 80%, 85%, 90%, 95%, 98%, or 99%, with the CDS (encoding) part of the DNA sequence shown in SEQ ID NOS: 1, 3, 5, 7, 9, 11, 13, 15,
17, 19, 21, 23, 25, 27, 29, 31, 33, 35, 37, 39, 41 and 43.
The term "sequence identity" refers to the degree to which two polynucleotide or polypeptide sequences are identical on a residue-by-residue basis over a particular region of comparison. The term "percentage of sequence identity" is calculated by comparing two optimally aligned sequences over that region of comparison, determining the number of positions at which the identical nucleic acid base (e.g., A, T, C, G, U, or I, in the case of nucleic acids) occurs in both sequences to yield the number of matched positions, dividing the number of matched positions by the total number of positions in the region of comparison (i.e., the window size), and multiplying the result by 100 to yield the percentage of sequence identity. The term "substantial identity" as used herein denotes a characteristic of a polynucleotide sequence, wherein the polynucleotide comprises a sequence that has at least 80 percent sequence identity, preferably at least 85 percent identity and often 90 to 95 percent sequence identity, more usually at least 99 percent sequence identity as compared to a reference sequence over a comparison region.
Chimeric and Fusion Proteins
The invention also provides GPCRX chimeric or fusion proteins. As used herein, an GPCRX "chimeric protein" or "fusion protein" comprises an GPCRX polypeptide operatively- linked to a non-GPCRX polypeptide. An "GPCRX polypeptide" refers to a polypeptide having an amino acid sequence corresponding to an GPCRX protein (SEQ ID NOS:2, 4, 6, 8, 10, 12, 14, 16, 18, 20, 22, 24, 26, 28, 30, 32, 34, 36, 38, 40, 42 and 44), whereas a "non-GPCRX polypeptide" refers to a polypeptide having an amino acid sequence corresponding to a protein that is not substantially homologous to the GPCRX protein, e.g., a protein that is different from the GPCRX protein and that is derived from the same or a different organism. Within an GPCRX fusion protein the GPCRX polypeptide can correspond to all or a portion of an GPCRX protein, hi one embodiment, an GPCRX fusion protein comprises at least one biologically-active portion of an GPCRX protein. In another embodiment, an GPCRX fusion protein comprises at least two biologically-active portions of an GPCRX protein, hi yet another embodiment, an GPCRX fusion protein comprises at least three biologically-active portions of an GPCRX protein. Within the fusion protein, the term "operatively-linked" is intended to indicate that the GPCRX polypeptide and the non-GPCRX polypeptide are fused in-frame with one another. The non-GPCRX polypeptide can be fused to the N-terminus or C-terminus of the
GPCRX polypeptide.
In one embodiment, the fusion protein is a GST-GPCRX fusion protein in which the
GPCRX sequences are fused to the C-terminus of the GST (glutathione S-transferase)
sequences. Such fusion proteins can facilitate the purification of recombinant GPCRX polypeptides.
In another embodiment, the fusion protein is an GPCRX protein containing a heterologous signal sequence at its N-terminus. In certain host cells (e.g., mammalian host cells), expression and/or secretion of GPCRX can be increased through use of a heterologous signal sequence.
In yet another embodiment, the fusion protein is an GPCRX-immunoglobulin fusion protein in which the GPCRX sequences are fused to sequences derived from a member of the immunoglobulin protein family. The GPCRX-immunoglobulin fusion proteins of the invention can be incorporated into pharmaceutical compositions and administered to a subject to inhibit an interaction between an GPCRX ligand and an GPCRX protein on the surface of a cell, to thereby suppress GPCRX-mediated signal transduction in vivo. The GPCRX-immunoglobulin fusion proteins can be used to affect the bioavailability of an GPCRX cognate ligand. Inhibition of the GPCRX ligand/GPCRX interaction may be useful therapeutically for both the treatment of proliferative and differentiative disorders, as well as modulating (e.g. promoting or inhibiting) cell survival. Moreover, the GPCRX-immunoglobulin fusion proteins of the invention can be used as immunogens to produce anti-GPCRX antibodies in a subject, to purify GPCRX ligands, and in screening assays to identify molecules that inhibit the interaction of GPCRX with an GPCRX ligand. An GPCRX chimeric or fusion protein of the invention can be produced by standard recombinant DNA techniques. For example, DNA fragments coding for the different polypeptide sequences are ligated together in-frame in accordance with conventional techniques, e.g., by employing blunt-ended or stagger-ended termini for ligation, restriction enzyme digestion to provide for appropriate termini, filling-in of cohesive ends as appropriate, alkaline phosphatase treatment to avoid undesirable joining, and enzymatic ligation. In another embodiment, the fusion gene can be synthesized by conventional techniques including automated DNA synthesizers. Alternatively, PCR amplification of gene fragments can be carried out using anchor primers that give rise to complementary overhangs between two consecutive gene fragments that can subsequently be annealed and reamplified to generate a chimeric gene sequence (see, e.g., Ausubel, et al (eds.) CURRENT PROTOCOLS IN MOLECULAR
BIOLOGY, John Wiley & Sons, 1992). Moreover, many expression vectors are commercially available that already encode a fusion moiety (e.g., a GST polypeptide). An GPCRX-encoding nucleic acid can be cloned into such an expression vector such that the fusion moiety is linked in-frame to the GPCRX protein.
GPCRX Agonists and Antagonists
The invention also pertains to variants of the GPCRX proteins that function as either GPCRX agonists (i.e., mimetics) or as GPCRX antagonists. Variants of the GPCRX protein can be generated by mutagenesis (e.g., discrete point mutation or truncation of the GPCRX protein). An agonist of the GPCRX protein can retain substantially the same, or a subset of, the biological activities of the naturally occurring form of the GPCRX protein. An antagonist of the GPCRX protein can inhibit one or more of the activities of the naturally occurring form of the GPCRX protein by, for example, competitively binding to a downstream or upstream member of a cellular signaling cascade which includes the GPCRX protein. Thus, specific biological effects can be elicited by treatment with a variant of limited function. In one embodiment, treatment of a subject with a variant having a subset of the biological activities of the naturally occurring form of the protein has fewer side effects in a subject relative to treatment with the naturally occurring form of the GPCRX proteins. Variants of the GPCRX proteins that function as either GPCRX agonists (i.e., mimetics) or as GPCRX antagonists can be identified by screening combinatorial libraries of mutants (e.g., truncation mutants) of the GPCRX proteins for GPCRX protein agonist or antagonist activity. In one embodiment, a variegated library of GPCRX variants is generated by combinatorial mutagenesis at the nucleic acid level and is encoded by a variegated gene library. A variegated library of GPCRX variants can be produced by, for example, enzymatically ligating a mixture of synthetic oligonucleotides into gene sequences such that a degenerate set of potential GPCRX sequences is expressible as individual polypeptides, or alternatively, as a set of larger fusion proteins (e.g., for phage display) containing the set of GPCRX sequences therein. There are a variety of methods which can be used to produce libraries of potential GPCRX variants from a degenerate oligonucleotide sequence. Chemical synthesis of a degenerate gene sequence can be performed in an automatic DNA synthesizer, and the synthetic gene then ligated into an appropriate expression vector. Use of a degenerate set of genes allows for the provision, in one mixture, of all of the sequences encoding the desired set of potential GPCRX sequences. Methods for synthesizing degenerate oligonucleotides are well-known within the art. See, e.g., Narang, 1983. Tetrahedron 39: 3; Itakura, et al, 1984. Annu. Rev. Biochem. 53: 323; Itakura, et al, 1984. Science 198: 1056; Ike, et al, 1983. Nucl. Acids Res. 11: 477.
Polypeptide Libraries
In addition, libraries of fragments of the GPCRX protein coding sequences can be used to generate a variegated population of GPCRX fragments for screening and subsequent selection of variants of an GPCRX protein, hi one embodiment, a library of coding sequence fragments can be generated by treating a double stranded PCR fragment of an GPCRX coding sequence with a nuclease under conditions wherein nicking occurs only about once per molecule, denaturing the double stranded DNA, renaturing the DNA to form double-stranded DNA that can include sense/antisense pairs from different nicked products, removing single stranded portions from reformed duplexes by treatment with Si nuclease, and ligating the resulting fragment library into an expression vector. By this method, expression libraries can be derived which encodes N-terminal and internal fragments of various sizes of the GPCRX proteins.
Various techniques are known in the art for screening gene products of combinatorial libraries made by point mutations or truncation, and for screening cDNA libraries for gene products having a selected property. Such techniques are adaptable for rapid screening of the gene libraries generated by the combinatorial mutagenesis of GPCRX proteins. The most widely used techniques, which are amenable to high throughput analysis, for screening large gene libraries typically include cloning the gene library into replicable expression vectors, transforming appropriate cells with the resulting library of vectors, and expressing the combinatorial genes under conditions in which detection of a desired activity facilitates isolation of the vector encoding the gene whose product was detected. Recursive ensemble mutagenesis (REM), a new technique that enhances the frequency of functional mutants in the libraries, can be used in combination with the screening assays to identify GPCRX variants. See, e.g., Arkin and Yourvan, 1992. Proc. Natl. Acad. Sci. USA 89: 7811-7815; Delgrave, et al, 1993. Protein Engineering 6:327-331.
Anti-GPCRX Antibodies
The invention encompasses antibodies and antibody fragments, such as Fa or (Fab)2, that bind immunospecifically to any of the GPCRX polypeptides of said invention. An isolated GPCRX protein, or a portion or fragment thereof, can be used as an immunogen to generate antibodies that bind to GPCRX polypeptides using standard techniques for polyclonal and monoclonal antibody preparation. The full-length GPCRX proteins can be used or, alternatively, the invention provides antigenic peptide fragments of GPCRX proteins
for use as immunogens. The antigenic GPCRX peptides comprises at least 4 amino acid residues of the amino acid sequence shown in SEQ ID NOS:2, 4, 6, 8, 10, 12, 14, 16, 18, 20, 22, 24, 26, 28, 30, 32, 34, 36, 38, 40, 42 and 44 and encompasses an epitope of GPCRX such that an antibody raised against the peptide forms a specific immune complex with GPCRX. Preferably, the antigemc peptide comprises at least 6, 8, 10, 15, 20, or 30 amino acid residues. Longer antigenic peptides are sometimes preferable over shorter antigenic peptides, depending on use and according to methods well known to someone skilled in the art. hi certain embodiments of the invention, at least one epitope encompassed by the antigenic peptide is a region of GPCRX that is located on the surface of the protein (e.g., a hydrophilic region). As a means for targeting antibody production, hydropathy plots showing regions of hydrophilicity and hydrophobicity may be generated by any method well known in the art, including, for example, the Kyte Doolittle or the Hopp Woods methods, either with or without Fourier transformation (see, e.g., Hopp and Woods, 1981. Proc. Nat. Acad. Sci. USA 78: 3824-3828; Kyte and Doolittle, 1982. J. Mol. Biol. 157: 105-142, each incoφorated herein by reference in their entirety) .
As disclosed herein, GPCRX protein sequences of SEQ ID NOS:2, 4, 6, 8, 10, 12, 14, 16, 18, 20, 22, 24, 26, 28, 30, 32, 34, 36, 38, 40, 42 and 44, or derivatives, fragments, analogs or homologs thereof, may be utilized as immunogens in the generation of antibodies that immunospecifically-bind these protein components. The term "antibody" as used herein refers to immunoglobulin molecules and immunologically-active portions of immunoglobulin molecules, i.e., molecules that contain an antigen binding site that specifically-binds (immunoreacts with) an antigen, such as GPCRX. Such antibodies include, but are not limited to, polyclonal, monoclonal, chimeric, single chain, Fa and F(a ')2 fragments, and an Fa expression library, hi a specific embodiment, antibodies to human GPCRX proteins are disclosed. Various procedures known within the art may be used for the production of polyclonal or monoclonal antibodies to an GPCRX protein sequence of SEQ ID NOS:2, 4, 6, 8, 10, 12, 14, 16, 18, 20, 22, 24, 26, 28, 30, 32, 34, 36, 38, 40, 42 and 44, or a derivative, fragment, analog or homolog thereof. Some of these proteins are discussed below.
For the production of polyclonal antibodies, various suitable host animals (e.g., rabbit, goat, mouse or other mammal) may be immunized by injection with the native protein, or a synthetic variant thereof, or a derivative of the foregoing. An appropriate immunogenic preparation can contain, for example, recombinantly-expressed GPCRX protein or a chemically-synthesized GPCRX polypeptide. The preparation can further include an adjuvant.
Various adjuvants used to increase the immunological response include, but are not limited to,
Freund's (complete and incomplete), mineral gels (e.g., aluminum hydroxide), surface active substances (e.g., lysolecithin, pluronic polyols, polyanions, peptides, oil emulsions, dinitrophenol, etc.), human adjuvants such as Bacille Calmette-Guerin and Corynebacterium parvum, or similar immunostimulatory agents. If desired, the antibody molecules directed against GPCRX can be isolated from the mammal (e.g. , from the blood) and further purified by well known techniques, such as protein A cliromatography to obtain the IgG fraction.
The term "monoclonal antibody" or "monoclonal antibody composition", as used herein, refers to a population of antibody molecules that contain only one species of an antigen binding site capable of immunoreacting with a particular epitope of GPCRX. A monoclonal antibody composition thus typically displays a single binding affinity for a particular GPCRX protein with which it immunoreacts. For preparation of monoclonal antibodies directed towards a particular GPCRX protein, or derivatives, fragments, analogs or homologs thereof, any technique that provides for the production of antibody molecules by continuous cell line culture may be utilized. Such techniques include, but are not limited to, the hybridoma technique (see, e.g., Kohler & Milstein, 1975. Nature 256: 495-497); the trioma technique; the human B-cell hybridoma technique (see, e.g., Kozbor, et al, 1983. Immunol Today 4: 72) and the EBV hybridoma technique to produce human monoclonal antibodies (see, e.g., Cole, et al, 1985. In: MONOCLONAL ANTIBODIES AND CANCER THERAPY, Alan R. Liss, Inc., pp. 77-96). Human monoclonal antibodies may be utilized in the practice of the invention and may be produced by using human hybridomas (see, e.g., Cote, et al, 1983. Proc Natl Acad Sci USA 80: 2026-2030) or by transforming human B-cells with Epstein Barr Virus in vitro (see, e.g., Cole, et al, 1985. hi: MONOCLONAL ANTIBODIES AND CANCER THERAPY, Alan R. Liss, Inc., pp. 77-96). Each of the above citations is incoφorated herein by reference in their entirety.
According to the invention, techniques can be adapted for the production of single-chain antibodies specific to an GPCRX protein (see, e.g., U.S. Patent No. 4,946,778). In addition, methods can be adapted for the construction of Fa expression libraries (see, e.g., Huse, et al, 1989. Science 246: 1275-1281) to allow rapid and effective identification of monoclonal Fab fragments with the desired specificity for an GPCRX protein or derivatives, fragments, analogs or homologs thereof. Non-human antibodies can be "humanized" by techniques well known in the art. See, e.g., U.S. Patent No. 5,225,539. Antibody fragments that contain the idiotypes to an GPCRX protein may be produced by techniques known in the art including, but not limited to: (i) an F(ab') fragment produced by pepsin digestion of an antibody molecule; (ii) an Fa fragment generated by reducing the disulfide bridges of an F ay)2 fragment; (Hi) an Fab fragment
generated by the treatment of the antibody molecule with papain and a reducing agent; and (iv) Fv fragments.
Additionally, recombinant anti-GPCRX antibodies, such as chimeric and humanized monoclonal antibodies, comprising both human and non-human portions, which can be made using standard recombinant DNA techniques, are within the scope of the invention. Such chimeric and humanized monoclonal antibodies can be produced by recombinant DNA techniques known in the art, for example using methods described in International Application No. PCT/US86/02269; European Patent Application No. 184,187; European Patent Application No. 171,496; European Patent Application No. 173,494; PCT International Publication No. WO 86/01533; U.S. Patent No. 4,816,567; U.S. Pat. No. 5,225,539; European Patent Application No. 125,023; Better, et al, 1988. Science 240: 1041-1043; Liu, et al, 1987. Proc. Natl. Acad. Sci. USA 84: 3439-3443; Liu, et al, 1987. J. Immunol. 139: 3521-3526; Sun, et al, 1987. Proc. Natl. Acad. Sci. USA 84: 214-218; Nishimura, et al, 1987. Cancer Res. 47: 999-1005; Wood, et al, 1985. Nature 314 :446-449; Shaw, et al, 1988. J. Natl. Cancer Inst. 80: 1553-1559); Morrison(1985) Science 229:1202-1207; Oi, et al. (1986) BioTechniques 4:214; Jones, et al, 1986. Nature 321: 552-525; Verhoeyan, et al, 1988. Science 239: 1534; and Beidler, et al, 1988. J. Immunol. 141: 4053-4060. Each of the above citations are incoφorated herein by reference in their entirety.
In one embodiment, methods for the screening of antibodies that possess the desired specificity include, but are not limited to, enzyme- linked immunosorbent assay (ELISA) and other immunologically-mediated techniques known within the art. In a specific embodiment, selection of antibodies that are specific to a particular domain of an GPCRX protein is facilitated by generation of hybridomas that bind to the fragment of an GPCRX protein possessing such a domain. Thus, antibodies that are specific for a desired domain within an GPCRX protein, or derivatives, fragments, analogs or homologs thereof, are also provided herein.
Anti-GPCRX antibodies may be used in methods known within the art relating to the localization and/or quantitation of an GPCRX protein (e.g., for use in measuring levels of the GPCRX protein within appropriate physiological samples, for use in diagnostic methods, for use in imaging the protein, and the like), i a given embodiment, antibodies for GPCRX proteins, or derivatives, fragments, analogs or homologs thereof, that contain the antibody derived binding domain, are utilized as pharmacologically-active compounds (hereinafter "Therapeutics").
An anti-GPCRX antibody (e.g., monoclonal antibody) can be used to isolate an GPCRX polypeptide by standard techniques, such as affinity chromatography or immunoprecipitation. An anti-GPCRX antibody can facilitate the purification of natural GPCRX polypeptide from cells and of recombinantly-produced GPCRX polypeptide expressed in host cells. Moreover, an anti-GPCRX antibody can be used to detect GPCRX protein (e.g., in a cellular lysate or cell supernatant) in order to evaluate the abundance and pattern of expression of the GPCRX protein. Anti-GPCRX antibodies can be used diagnostically to monitor protein levels in tissue as part of a clinical testing procedure, e.g., to, for example, determine the efficacy of a given treatment regimen. Detection can be facilitated by coupling (i.e., physically linking) the antibody to a detectable substance. Examples of detectable substances include various enzymes, prosthetic groups, fluorescent materials, luminescent materials, bioluminescent materials, and radioactive materials. Examples of suitable enzymes include horseradish peroxidase, alkaline phosphatase, β-galactosidase, or acetylcholinesterase; examples of suitable prosthetic group complexes include streptavidin/biotin and avidin/biotin; examples of suitable fluorescent materials include umbelliferone, fluorescein, fluorescein isothiocyanate, rhodamine, dichlorotriazinylamine fluorescein, dansyl chloride or phycoerythrin; an example of a luminescent material includes luminol; examples of bioluminescent materials include luciferase, luciferin, and aequorin, and examples of suitable radioactive material include 125I, 131I, 35S or 3H.
GPCRX Recombinant Expression Vectors and Host Cells
Another aspect of the invention pertains to vectors, preferably expression vectors, containing a nucleic acid encoding an GPCRX protein, or derivatives, fragments, analogs or homologs thereof. As used herein, the term "vector" refers to a nucleic acid molecule capable of transporting another nucleic acid to which it has been linked. One type of vector is a
"plasmid", which refers to a circular double stranded DNA loop into which additional DNA segments can be ligated. Another type of vector is a viral vector, wherein additional DNA segments can be ligated into the viral genome. Certain vectors are capable of autonomous replication in a host cell into which they are introduced (e.g., bacterial vectors having a bacterial origin of replication and episomal mammalian vectors). Other vectors (e.g., non-episomal mammalian vectors) are integrated into the genome of a host cell upon introduction into the host cell, and thereby are replicated along with the host genome.
Moreover, certain vectors are capable of directing the expression of genes to which they are operatively-linked. Such vectors are referred to herein as "expression vectors". In general,
expression vectors of utility in recombinant DNA techniques are often in the form of plasmids. In the present specification, "plasmid" and "vector" can be used interchangeably as the plasmid is the most commonly used form of vector. However, the invention is intended to include such other forms of expression vectors, such as viral vectors (e.g., replication defective retro viruses, adenoviruses and adeno-associated viruses), which serve equivalent functions.
The recombinant expression vectors of the invention comprise a nucleic acid of the invention in a form suitable for expression of the nucleic acid in a host cell, which means that the recombinant expression vectors include one or more regulatory sequences, selected on the basis of the host cells to be used for expression, that is operatively-linked to the nucleic acid sequence to be expressed. Within a recombinant expression vector, "operably-linked" is intended to mean that the nucleotide sequence of interest is linked to the regulatory sequence(s) in a manner that allows for expression of the nucleotide sequence (e.g., in an in vitro transcription/translation system or in a host cell when the vector is introduced into the host cell). The term "regulatory sequence" is intended to includes promoters, enhancers and other expression control elements (e.g., polyadenylation signals). Such regulatory sequences are described, for example, in Goeddel, GENE EXPRESSION TECHNOLOGY: METHODS IN ENZYMOLOGY 185, Academic Press, San Diego, Calif. (1990). Regulatory sequences include those that direct constitutive expression of a nucleotide sequence in many types of host cell and those that direct expression of the nucleotide sequence only in certain host cells (e.g., tissue-specific regulatory sequences). It will be appreciated by those skilled in the art that the design of the expression vector can depend on such factors as the choice of the host cell to be transformed, the level of expression of protein desired, etc. The expression vectors of the invention can be introduced into host cells to thereby produce proteins or peptides, including fusion proteins or peptides, encoded by nucleic acids as described herein (e.g., GPCRX proteins, mutant forms of GPCRX proteins, fusion proteins, etc.).
The recombinant expression vectors of the invention can be designed for expression of GPCRX proteins in prokaryotic or eukaryotic cells. For example, GPCRX proteins can be expressed in bacterial cells such as Escherichia coli, insect cells (using baculovirus expression vectors) yeast cells or mammalian cells. Suitable host cells are discussed further in Goeddel, GENE EXPRESSION TECHNOLOGY: METHODS IN ENZYMOLOGY 185, Academic Press, San Diego, Calif. (1990). Alternatively, the recombinant expression vector can be transcribed and translated in vitro, for example using T7 promoter regulatory sequences and T7 polymerase.
Expression of proteins in prokaryotes is most often carried out in Escherichia coli with vectors containing constitutive or inducible promoters directing the expression of either fusion or non-fusion proteins. Fusion vectors add a number of amino acids to a protein encoded therein, usually to the amino terminus of the recombinant protein. Such fusion vectors typically serve three puφoses: (i) to increase expression of recombinant protein; (ii) to increase the solubility of the recombinant protein; and (Hi) to aid in the purification of the recombinant protein by acting as a ligand in affinity purification. Often, in fusion expression vectors, a proteolytic cleavage site is introduced at the junction of the fusion moiety and the recombinant protein to enable separation of the recombinant protein from the fusion moiety subsequent to purification of the fusion protein. Such enzymes, and their cognate recognition sequences, include Factor Xa, thrombin and enterokinase. Typical fusion expression vectors include pGEX (Pharmacia Biotech Inc; Smith and Johnson, 1988. Gene 61: 31-40), pMAL (New England Biolabs, Beverly, Mass.) and pRIT5 (Pharmacia, Piscataway, N. J.) that fuse glutathione S-transferase (GST), maltose E binding protein, or protein A, respectively, to the target recombinant protein.
Examples of suitable inducible non-fusion E. coli expression vectors include pTrc (Amrann et al, (1988) Gene 69:301-315) and pET lid (Srudier et al, GENE EXPRESSION TECHNOLOGY: METHODS IN ENZYMOLOGY 185, Academic Press, San Diego, Calif. (1990) 60-89). One strategy to maximize recombinant protein expression in E. coli is to express the protein in a host bacteria with an impaired capacity to proteolytically cleave the recombinant protein. See, e.g., Gottesman, GENE EXPRESSION TECHNOLOGY: METHODS IN ENZYMOLOGY 185, Academic Press, San Diego, Calif. (1990) 119-128. Another strategy is to alter the nucleic acid sequence of the nucleic acid to be inserted into an expression vector so that the individual codons for each amino acid are those preferentially utilized in E. coli (see, e.g., Wada, et al, 1992. Nucl. Acids Res. 20: 2111-2118). Such alteration of nucleic acid sequences of the invention can be carried out by standard DNA synthesis techniques.
In another embodiment, the GPCRX expression vector is a yeast expression vector. Examples of vectors for expression in yeast Saccharomyces cerivisae include pYepSecl (Baldari, et al, 1987. EMBO J. 6: 229-234), pMFa (Kurjan and Herskowitz, 1982. Cell 30:
933-943), ρJRY88 (Schultz et al, 1987. Gene 54: 113-123), ρYES2 (hivitrogen Coφoration,
San Diego, Calif.), and picZ (InVitrogen Corp, San Diego, Calif).
Alternatively, GPCRX can be expressed in insect cells using baculovirus expression vectors. Baculovirus vectors available for expression of proteins in cultured insect cells (e.g.,
SF9 cells) include the pAc series (Smith, et al, 1983. Mol. Cell. Biol. 3: 2156-2165) and the pVL series (Lucklow and Summers, 1989. Virology 170: 31-39).
In yet another embodiment, a nucleic acid of the invention is expressed in mammalian cells using a mammalian expression vector. Examples of mammalian expression vectors include pCDM8 (Seed, 1987. Nature 329: 840) and pMT2PC (Kaufman, et al, 1987. EMBO J. 6: 187-195). When used in mammalian cells, the expression vector's control functions are often provided by viral regulatory elements. For example, commonly used promoters are derived from polyoma, adenovirus 2, cytomegalovirus, and simian virus 40. For other suitable expression systems for both prokaryotic and eukaryotic cells see, e.g., Chapters 16 and 17 of Sambrook, et al, MOLECULAR CLONING: A LABORATORY MANUAL. 2nd ed., Cold Spring Harbor Laboratory, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y., 1989. In another embodiment, the recombinant mammalian expression vector is capable of directing expression of the nucleic acid preferentially in a particular cell type (e.g., tissue-specific regulatory elements are used to express the nucleic acid). Tissue-specific regulatory elements are known in the art. Non-limiting examples of suitable tissue-specific promoters include the albumin promoter (liver-specific; Pinkert, et al, 1987. Genes Dev. 1: 268-277), lymphoid-specific promoters (Calame and Eaton, 1988. Adv. Immunol. 43: 235-275), in particular promoters of T cell receptors (Winoto and Baltimore, 1989. EMBO J. 8: 729-733) and immunoglobulins (Banerji, et al, 1983. Cell 33: 729-740; Queen and Baltimore, 1983. Cell 33: 741-748), neuron-specific promoters (e.g., the neurofilament promoter; Byrne and Ruddle, 1989. Proc. Natl. Acad. Sci. USA 86: 5473-5477), pancreas-specific promoters (Edlund, et al, 1985. Science 230: 912-916), and mammary gland-specific promoters (e.g., milk whey promoter; U.S. Pat. No. 4,873,316 and European Application Publication No. 264,166). Developmentally-regulated promoters are also encompassed, e.g., the murine hox promoters (Kessel and Gruss, 1990. Science 249: 374-379) and the α-fetoprotein promoter (Campes and Tilghman, 1989. Genes Dev. 3: 537-546).
The invention further provides a recombinant expression vector comprising a DNA molecule of the invention cloned into the expression vector in an antisense orientation. That is, the DNA molecule is operatively-linked to a regulatory sequence in a manner that allows for expression (by transcription of the DNA molecule) of an RNA molecule that is antisense to
GPCRX mRNA. Regulatory sequences operatively linked to a nucleic acid cloned in the antisense orientation can be chosen that direct the continuous expression of the antisense RNA molecule in a variety of cell types, for instance viral promoters and/or enhancers, or regulatory sequences can be chosen that direct constitutive, tissue specific or cell type specific expression
of antisense RNA. The antisense expression vector can be in the form of a recombinant plasmid, phagemid or attenuated virus in which antisense nucleic acids are produced under the control of a high efficiency regulatory region, the activity of which can be determined by the cell type into which the vector is introduced. For a discussion of the regulation of gene expression using antisense genes see, e.g., Weintraub, et al, "Antisense RNA as a molecular tool for genetic analysis," Reviews-Trends in Genetics, Vol. 1(1) 1986.
Another aspect of the invention pertains to host cells into which a recombinant expression vector of the invention has been introduced. The terms "host cell" and "recombinant host cell" are used interchangeably herein. It is understood that such terms refer not only to the particular subj ect cell but also to the progeny or potential progeny of such a cell. Because certain modifications may occur in succeeding generations due to either mutation or environmental influences, such progeny may not, in fact, be identical to the parent cell, but are still included within the scope of the term as used herein.
A host cell can be any prokaryotic or eukaryotic cell. For example, GPCRX protein can be expressed in bacterial cells such as E. coli, insect cells, yeast or mammalian cells (such as Chinese hamster ovary cells (CHO) or COS cells). Other suitable host cells are known to those skilled in the art.
Vector DNA can be introduced into prokaryotic or eukaryotic cells via conventional transformation or transfection techniques. As used herein, the terms "transformation" and "transfection" are intended to refer to a variety of art-recognized techniques for introducing foreign nucleic acid (e.g., DNA) into a host cell, including calcium phosphate or calcium chloride co-precipitation, DEAE-dextran-mediated transfection, lipofection, or electroporation. Suitable methods for transforming or transfecting host cells can be found in Sambrook, et al. (MOLECULAR CLONING: A LABORATORY MANUAL. 2nd ed., Cold Spring Harbor Laboratory, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y., 1989), and other laboratory manuals.
For stable transfection of mammalian cells, it is known that, depending upon the expression vector and transfection technique used, only a small fraction of cells may integrate the foreign DNA into their genome, hi order to identify and select these integrants, a gene that encodes a selectable marker (e.g., resistance to antibiotics) is generally introduced into the host cells along with the gene of interest. Various selectable markers include those that confer resistance to drugs, such as G418, hygromycin and methotrexate. Nucleic acid encoding a selectable marker can be introduced into a host cell on the same vector as that encoding
GPCRX or can be introduced on a separate vector. Cells stably transfected with the introduced
nucleic acid can be identified by drug selection (e.g., cells that have incoφorated the selectable marker gene will survive, while the other cells die).
A host cell of the invention, such as a prokaryotic or eukaryotic host cell in culture, can be used to produce (i.e., express) GPCRX protein. Accordingly, the invention further provides methods for producing GPCRX protein using the host cells of the invention, hi one embodiment, the method comprises culturing the host cell of invention (into which a recombinant expression vector encoding GPCRX protein has been introduced) in a suitable medium such that GPCRX protein is produced. In another embodiment, the method further comprises isolating GPCRX protein from the medium or the host cell.
Transgenic GPCRX Animals
The host cells of the invention can also be used to produce non-human transgenic animals. For example, in one embodiment, a host cell of the invention is a fertilized oocyte or an embryonic stem cell into which GPCRX protein-coding sequences have been introduced. Such host cells can then be used to create non-human transgenic animals in which exogenous GPCRX sequences have been introduced into their genome or homologous recombinant animals in which endogenous GPCRX sequences have been altered. Such animals are useful for studying the function and/or activity of GPCRX protein and for identifying and/or evaluating modulators of GPCRX protein activity. As used herein, a "transgenic animal" is a non-human animal, preferably a mammal, more preferably a rodent such as a rat or mouse, in which one or more of the cells of the animal includes a transgene. Other examples of transgenic animals include non-human primates, sheep, dogs, cows, goats, chickens, amphibians, etc. A transgene is exogenous DNA that is integrated into the genome of a cell from which a transgenic animal develops and that remains in the genome of the mature animal, thereby directing the expression of an encoded gene product in one or more cell types or tissues of the transgenic animal. As used herein, a "homologous recombinant animal" is a non-human animal, preferably a mammal, more preferably a mouse, in which an endogenous GPCRX gene has been altered by homologous recombination between the endogenous gene and an exogenous DNA molecule introduced into a cell of the animal, e.g., an embryonic cell of the animal, prior to development of the animal. A transgenic animal of the invention can be created by introducing GPCRX-encoding nucleic acid into the male pronuclei of a fertilized oocyte (e.g., by microinjection, retro viral infection) and allowing the oocyte to develop in a pseudopregnant female foster animal. The human GPCRX cDNA sequences of SEQ ID NOS:l, 3, 5, 7, 9, 11, 13, 15, 17, 19, 21, 23, 25,
27, 29, 31, 33, 35, 37, 39, 41 and 43 can be introduced as a transgene into the genome of a non-human animal. Alternatively, a non-human homologue of the human GPCRX gene, such as a mouse GPCRX gene, can be isolated based on hybridization to the human GPCRX cDNA (described further supra) and used as a transgene. Intronic sequences and polyadenylation signals can also be included in the transgene to increase the efficiency of expression of the transgene. A tissue-specific regulatory sequence(s) can be operably-linked to the GPCRX transgene to direct expression of GPCRX protein to particular cells. Methods for generating transgenic animals via embryo manipulation and microinjection, particularly animals such as mice, have become conventional in the art and are described, for example, in U.S. Patent Nos. 4,736,866; 4,870,009; and 4,873,191; and Hogan, 1986. hi: MANIPULATING THE MOUSE
EMBRYO, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y. Similar methods are used for production of other transgenic animals. A transgenic founder animal can be identified based upon the presence of the GPCRX transgene in its genome and/or expression of GPCRX mRNA in tissues or cells of the animals. A transgenic founder animal can then be used to breed additional animals carrying the transgene. Moreover, transgenic animals carrying a transgene- encoding GPCRX protein can further be bred to other transgenic animals carrying other transgenes.
To create a homologous recombinant animal, a vector is prepared which contains at least a portion of an GPCRX gene into which a deletion, addition or substitution has been introduced to thereby alter, e.g., functionally disrupt, the GPCRX gene. The GPCRX gene can be a human gene (e.g., the cDNA of SEQ ED NOS:l, 3, 5, 7, 9, 11, 13, 15, 17, 19, 21, 23, 25, 27, 29, 31, 33, 35, 37, 39, 41 and 43), but more preferably, is a non-human homologue of a human GPCRX gene. For example, a mouse homologue of human GPCRX gene of SEQ ID NOS:l, 3, 5, 7, 9, 11, 13, 15, 17, 19, 21, 23, 25, 27, 29, 31, 33, 35, 37, 39, 41 and 43 can be used to construct a homologous recombination vector suitable for altering an endogenous
GPCRX gene in the mouse genome. In one embodiment, the vector is designed such that, upon homologous recombination, the endogenous GPCRX gene is functionally disrupted (i.e., no longer encodes a functional protein; also referred to as a "knock out" vector).
Alternatively, the vector can be designed such that, upon homologous recombination, the endogenous GPCRX gene is mutated or otherwise altered but still encodes functional protein (e.g., the upstream regulatory region can be altered to thereby alter the expression of the endogenous GPCRX protein). In the homologous recombination vector, the altered portion of the GPCRX gene is flanked at its 5'- and 3'-termini by additional nucleic acid of the GPCRX gene to allow for homologous recombination to occur between the exogenous GPCRX gene
carried by the vector and an endogenous GPCRX gene in an embryonic stem cell. The additional flanking GPCRX nucleic acid is of sufficient length for successful homologous recombination with the endogenous gene. Typically, several kilobases of flanking DNA (both at the 5'- and 3'-termini) are included in the vector. See, e.g., Thomas, et al, 1987. Cell 51: 503 for a description of homologous recombination vectors. The vector is ten introduced into an embryonic stem cell line (e.g., by electroporation) and cells in which the introduced GPCRX gene has homologously-recombined with the endogenous GPCRX gene are selected. See, e.g., Li, et al, 1992. Cell 69: 915.
The selected cells are then injected into a blastocyst of an animal (e.g., a mouse) to form aggregation chimeras. See, e.g., Bradley, 1987. In: TERATOCARCINOMAS AND EMBRYONIC
STEM CELLS: A PRACTICAL APPROACH, Robertson, ed. IRL, Oxford, pp. 113-152. A chimeric embryo can then be implanted into a suitable pseudopregnant female foster animal and the embryo brought to term. Progeny harboring the homologously-recombined DNA in their germ cells can be used to breed animals in which all cells of the animal contain the homologously- recombined DNA by germline transmission of the transgene. Methods for constructing homologous recombination vectors and homologous recombinant animals are described further in Bradley, 1991. Curr. Opin. Biotechnol 2: 823-829; PCT International Publication Nos.: WO 90/11354; WO 91/01140; WO 92/0968; and WO 93/04169.
In another embodiment, transgenic non-humans animals can be produced that contain selected systems that allow for regulated expression of the transgene. One example of such a system is the cre/loxP recombinase system of bacteriophage PI. For a description of the cre/loxP recombinase system, See, e.g., Lakso, et al, 1992. Proc. Natl. Acad. Sci. USA 89: 6232-6236. Another example of a recombinase system is the FLP recombinase system of Saccharomyces cerevisiae. See, O'Gorman, et al, 1991. Science 251:1351-1355. If a cre/loxP recombinase system is used to regulate expression of the transgene, animals containing transgenes encoding both the Cre recombinase and a selected protein are required. Such animals can be provided through the construction of "double" transgenic animals, e.g., by mating two transgenic animals, one containing a transgene encoding a selected protein and the other containing a transgene encoding a recombinase. Clones of the non-human transgenic animals described herein can also be produced according to the methods described in Wilmut, et al, 1997. Nature 385: 810-813. h brief, a cell (e.g., a somatic cell) from the transgenic animal can be isolated and induced to exit the growth cycle and enter Go phase. The quiescent cell can then be fused, e.g., through the use of electrical pulses, to an enucleated oocyte from an animal of the same species from which the
quiescent cell is isolated. The reconstructed oocyte is then cultured such that it develops to morula or blastocyte and then transferred to pseudopregnant female foster animal. The offspring borne of this female foster animal will be a clone of the animal from which the cell (e.g., the somatic cell) is isolated.
Pharmaceutical Compositions
The GPCRX nucleic acid molecules, GPCRX proteins, and anti-GPCRX antibodies (also referred to herein as "active compounds") of the invention, and derivatives, fragments, analogs and homologs thereof, can be incoφorated into pharmaceutical compositions suitable for administration. Such compositions typically comprise the nucleic acid molecule, protein, or antibody and a pharmaceutically acceptable carrier. As used herein, "pharmaceutically acceptable carrier" is intended to include any and all solvents, dispersion media, coatings, antibacterial and antifungal agents, isotonic and absoφtion delaying agents, and the like, compatible with pharmaceutical administration. Suitable carriers are described in the most recent edition of Remington's Pharmaceutical Sciences, a standard reference text in the field, which is incoφorated herein by reference. Preferred examples of such carriers or diluents include, but are not limited to, water, saline, finger's solutions, dextrose solution, and 5% human serum albumin. Liposomes and non-aqueous vehicles such as fixed oils may also be used. The use of such media and agents for pharmaceutically active substances is well known in the art. Except insofar as any conventional media or agent is incompatible with the active compound, use thereof in the compositions is contemplated. Supplementary active compounds can also be incoφorated into the compositions.
A pharmaceutical composition of the invention is formulated to be compatible with its intended route of administration. Examples of routes of administration include parenteral, e.g., intravenous, intradermal, subcutaneous, oral (e.g., inhalation), transdermal (i.e., topical), transmucosal, and rectal administration. Solutions or suspensions used for parenteral, intradermal, or subcutaneous application can include the following components: a sterile diluent such as water for injection, saline solution, fixed oils, polyethylene glycols, glycerine, propylene glycol or other synthetic solvents; antibacterial agents such as benzyl alcohol or methyl parabens; antioxidants such as ascorbic acid or sodium bisulfite; chelating agents such as ethylenediaminetetraacetic acid (EDTA); buffers such as acetates, citrates or phosphates, and agents for the adjustment of tonicity such as sodium chloride or dextrose. The pH can be adjusted with acids or bases, such as hydrochloric acid or sodium hydroxide. The parenteral
preparation can be enclosed in ampoules, disposable syringes or multiple dose vials made of glass or plastic.
Pharmaceutical compositions suitable for injectable use include sterile aqueous solutions (where water soluble) or dispersions and sterile powders for the extemporaneous preparation of sterile injectable solutions or dispersion. For intravenous administration, suitable carriers include physiological saline, bacteriostatic water, Cremophor EL (BASF, Parsippany, N. J.) or phosphate buffered saline (PBS), hi all cases, the composition must be sterile and should be fluid to the extent that easy syringeability exists. It must be stable under the conditions of manufacture and storage and must be preserved against the contaminating action of microorganisms such as bacteria and fungi. The carrier can be a solvent or dispersion medium containing, for example, water, ethanol, polyol (for example, glycerol, propylene glycol, and liquid polyethylene glycol, and the like), and suitable mixtures thereof. The proper fluidity can be maintained, for example, by the use of a coating such as lecithin, by the maintenance of the required particle size in the case of dispersion and by the use of surfactants. Prevention of the action of microorganisms can be achieved by various antibacterial and antifungal agents, for example, parabens, chlorobutanol, phenol, ascorbic acid, thimerosal, and the like. In many cases, it will be preferable to include isotonic agents, for example, sugars, polyalcohols such as manitol, sorbitol, sodium chloride in the composition. Prolonged absoφtion of the injectable compositions can be brought about by including in the composition an agent which delays absoφtion, for example, aluminum monostearate and gelatin.
Sterile injectable solutions can be prepared by incoφorating the active compound (e.g., an GPCRX protein or anti-GPCRX antibody) in the required amount in an appropriate solvent with one or a combination of ingredients enumerated above, as required, followed by filtered sterilization. Generally, dispersions are prepared by incoφorating the active compound into a sterile vehicle that contains a basic dispersion medium and the required other ingredients from those enumerated above, hi the case of sterile powders for the preparation of sterile injectable solutions, methods of preparation are vacuum drying and freeze-drying that yields a powder of the active ingredient plus any additional desired ingredient from a previously sterile-filtered solution thereof. Oral compositions generally include an inert diluent or an edible carrier. They can be enclosed in gelatin capsules or compressed into tablets. For the pm ose of oral therapeutic administration, the active compound can be incoφorated with excipients and used in the form of tablets, troches, or capsules. Oral compositions can also be prepared using a fluid carrier for use as a mouthwash, wherein the compound in the fluid carrier is applied orally and swished
and expectorated or swallowed. Pharmaceutically compatible binding agents, and/or adjuvant materials can be included as part of the composition. The tablets, pills, capsules, troches and the like can contain any of the following ingredients, or compounds of a similar nature: a binder such as microcrystalline cellulose, gum tragacanth or gelatin; an excipient such as starch or lactose, a disintegrating agent such as alginic acid, Primogel, or corn starch; a lubricant such as magnesium stearate or Sterotes; a glidant such as colloidal silicon dioxide; a sweetening agent such as sucrose or saccharin; or a flavoring agent such as peppermint, methyl salicylate, or orange flavoring.
For administration by inhalation, the compounds are delivered in the form of an aerosol spray from pressured container or dispenser which contains a suitable propellant, e.g., a gas such as carbon dioxide, or a nebulizer.
Systemic administration can also be by transmucosal or transdermal means. For. transmucosal or transdermal administration, penetrants appropriate to the barrier to be permeated are used in the formulation. Such penetrants are generally known in the art, and include, for example, for transmucosal administration, detergents, bile salts, and fusidic acid derivatives. Transmucosal administration can be accomplished through the use of nasal sprays or suppositories. For transdennal administration, the active compounds are formulated into ointments, salves, gels, or creams as generally known in the art.
The compounds can also be prepared in the form of suppositories (e.g., with conventional suppository bases such as cocoa butter and other glycerides) or retention enemas for rectal delivery.
In one embodiment, the active compounds are prepared with carriers that will protect the compound against rapid elimination from the body, such as a controlled release formulation, including implants and microencapsulated delivery systems. Biodegradable, biocompatible polymers can be used, such as ethylene vinyl acetate, polyanhydrides, polyglycolic acid, collagen, polyorthoesters, and polylactic acid. Methods for preparation of such formulations will be apparent to those skilled in the art. The materials can also be obtained commercially from Alza Coφoration and Nova Pharmaceuticals, Inc. Liposomal suspensions (including liposomes targeted to infected cells with monoclonal antibodies to viral antigens) can also be used as phannaceutically acceptable carriers. These can be prepared according to methods known to those skilled in the art, for example, as described in U.S. Patent No. 4,522,811.
It is especially advantageous to formulate oral or parenteral compositions in dosage unit form for ease of administration and uniformity of dosage. Dosage unit form as used herein refers to physically discrete units suited as unitary dosages for the subject to be treated; each
unit containing a predetermined quantity of active compound calculated to produce the desired therapeutic effect in association with the required pharmaceutical carrier. The specification for the dosage unit forms of the invention are dictated by and directly dependent on the unique characteristics of the active compound and the particular therapeutic effect to be achieved, and the limitations inherent in the art of compounding such an active compound for the treatment of individuals.
The nucleic acid molecules of the invention can be inserted into vectors and used as gene therapy vectors. Gene therapy vectors can be delivered to a subject by, for example, intravenous injection, local administration (see, e.g., U.S. Patent No. 5,328,470) or by stereotactic injection (see, e.g., Chen, et al, 1994. Proc. Natl. Acad. Sci. USA 91: 3054-3057). The pharmaceutical preparation of the gene therapy vector can include the gene therapy vector in an acceptable diluent, or can comprise a slow release matrix in which the gene delivery vehicle is imbedded. Alternatively, where the complete gene delivery vector can be produced intact from recombinant cells, e.g., retro viral vectors, the pharmaceutical preparation can include one or more cells that produce the gene delivery system.
The pharmaceutical compositions can be included in a container, pack, or dispenser together with instructions for administration.
Screening and Detection Methods
The isolated nucleic acid molecules of the invention can be used to express GPCRX protein (e.g. , via a recombinant expression vector in a host cell in gene therapy applications), to detect GPCRX mRNA (e.g., in a biological sample) or a genetic lesion in an GPCRX gene, and to modulate GPCRX activity, as described further, below. In addition, the GPCRX proteins can be used to screen drugs or compounds that modulate the GPCRX protein activity or expression as well as to treat disorders characterized by insufficient or excessive production of GPCRX protein or production of GPCRX protein forms that have decreased or aberrant activity compared to GPCRX wild-type protein (e.g.; diabetes (regulates insulin release); obesity (binds and transport lipids); metabolic disturbances associated with obesity, the metabolic syndrome X as well as anorexia and wasting disorders associated with chronic diseases and various cancers, and infectious disease(possesses anti-microbial activity) and the various dyslipidemias. In addition, the anti-GPCRX antibodies of the invention can be used to detect and isolate GPCRX proteins and modulate GPCRX activity. In yet a further aspect, the invention can be used in methods to influence appetite, absoφtion of nutrients and the disposition of metabolic substrates in both a positive and negative fashion.
The invention further pertains to novel agents identified by the screening assays described herein and uses thereof for treatments as described, supra.
Screening Assays The invention provides a method (also referred to herein as a "screening assay") for identifying modulators, i.e., candidate or test compounds or agents (e.g., peptides, peptidomimetics, small molecules or other drugs) that bind to GPCRX proteins or have a stimulatory or inhibitory effect on, e.g., GPCRX protein expression or GPCRX protein activity. The invention also includes compounds identified in the screening assays described herein. In one embodiment, the invention provides assays for screening candidate or test compounds which bind to or modulate the activity of the membrane-bound form of an GPCRX protein or polypeptide or biologically-active portion thereof. The test compounds of the invention can be obtained using any of the numerous approaches in combinatorial library methods known in the art, including: biological libraries; spatially addressable parallel solid phase or solution phase libraries; synthetic library methods requiring deconvolution; the "one-bead one-compound" library method; and synthetic library methods using affinity chromatography selection. The biological library approach is limited to peptide libraries, while the other four approaches are applicable to peptide, non-peptide oligomer or small molecule libraries of compounds. See, e.g., Lam, 1997 '. Anticancer Drug Design 12: 145. A "small molecule" as used herein, is meant to refer to a composition that has a molecular weight of less than about 5 kD and most preferably less than about 4 kD. Small molecules can be, e.g., nucleic acids, peptides, polypeptides, peptidomimetics, carbohydrates, lipids or other organic or inorganic molecules. Libraries of chemical and/or biological mixtures, such as fungal, bacterial, or algal extracts, are known in the art and can be screened with any of the assays of the invention.
Examples of methods for the synthesis of molecular libraries can be found in the art, for example in: DeWitt, et al, 1993. Proc. Natl. Acad. Sci. U.S.A. 90: 6909; Erb, et al, 1994. Proc. Natl. Acad. Sci. U.S.A. 91: 11422; Zuckermann, et α/.,.1994. J. Med. Chem. 37: 2678; Cho, et al, 1993. Science 261: 1303; Carrell, et al, 1994. Angew. Chem. Int. Ed. Engl. 33: 2059; Carell, et al, 1994. Angew. Chem. Int. Ed. Engl. 33: 2061; and Gallop, et al, 1994. J. Med.
Chem. 37: 1233.
Libraries of compounds maybe presented in solution (e.g., Houghten, 1992.
Biotechniques 13: 412-421), or on beads (Lam, 1991. Nature 354: 82-84), on chips (Fodor,
1993. Nature 364: 555-556), bacteria (Ladner, U.S. Patent No. 5,223,409), spores (Ladner, U.S.
Patent 5,233,409), plasmids (Cull, et al, 1992. Proc. Natl. Acad. Sci. USA 89: 1865-1869) or on phage (Scott and Smith, 1990. Science 249: 386-390; Devlin, 1990. Science 249: 404-406; Cwirla, et al, 1990. Proc. Natl. Acad. Sci. U.S.A. 87: 6378-6382; Felici, 1991. J. Mol. Biol 222: 301-310; Ladner, U.S. Patent No. 5,233,409.). hi one embodiment, an assay is a cell-based assay in which a cell which expresses a membrane-bound form of GPCRX protein, or a biologically-active portion thereof, on the cell surface is contacted with a test compound and the ability of the test compound to bind to an GPCRX protein determined. The cell, for example, can of mammalian origin or a yeast cell. Determining the ability of the test compound to bind to the GPCRX protein can be accomplished, for example, by coupling the test compound with a radioisotope or enzymatic label such that binding of the test compound to the GPCRX protein or biologically-active portion thereof can be determined by detecting the labeled compound in a complex. For example, test compounds can be labeled with 1251, 35S, 14C, or 3H, either directly or indirectly, and the radioisotope detected by direct counting of radioemission or by scintillation counting. Alternatively, test compounds can be enzymatically-labeled with, for example, horseradish peroxidase, alkaline phosphatase, or luciferase, and the enzymatic label detected by determination of conversion of an appropriate substrate to product. In one embodiment, the assay comprises contacting a cell which expresses a membrane-bound form of GPCRX protein, or a biologically-active portion thereof, on the cell surface with a known compound which binds GPCRX to form an assay mixture, contacting the assay mixture with a test compound, and determining the ability of the test compound to interact with an GPCRX protein, wherein determining the ability of the test compound to interact with an GPCRX protein comprises determining the ability of the test compound to preferentially bind to GPCRX protein or a biologically-active portion thereof as compared to the known compound. hi another embodiment, an assay is a cell-based assay comprising contacting a cell expressing a membrane-bound form of GPCRX protein, or a biologically-active portion thereof, on the cell surface with a test compound and determining the ability of the test compound to modulate (e.g., stimulate or inhibit) the activity of the GPCRX protein or biologically- active portion thereof. Determining the ability of the test compound to modulate the activity of GPCRX or a biologically-active portion thereof can be accomplished, for example, by determining the ability of the GPCRX protein to bind to or interact with an GPCRX target molecule. As used herein, a "target molecule" is a molecule with which an GPCRX protein binds or interacts in nature, for example, a molecule on the surface of a cell which expresses an
GPCRX interacting protein, a molecule on the surface of a second cell, a molecule in the
extracellular milieu, a molecule associated with the internal surface of a cell membrane or a cytoplasmic molecule. An GPCRX target molecule can be a non-GPCRX molecule or an GPCRX protein or polypeptide of the invention. In one embodiment, an GPCRX target molecule is a component of a signal transduction pathway that facilitates transduction of an extracellular signal (e.g. a signal generated by binding of a compound to a membrane-bound GPCRX molecule) through the cell membrane and into the cell. The target, for example, can be a second intercellular protein that has catalytic activity or a protein that facilitates the association of downstream signaling molecules with GPCRX.
Determining the ability of the GPCRX protein to bind to or interact with an GPCRX target molecule can be accomplished by one of the methods described above for determining direct binding. In one embodiment, determining the ability of the GPCRX protein to bind to or interact with an GPCRX target molecule can be accomplished by determining the activity of the target molecule. For example, the activity of the target molecule can be determined by detecting induction of a cellular second messenger of the target (i.e. intracellular Ca2+, diacylglycerol, IP3, etc.), detecting catalytic/enzymatic activity of the target an appropriate substrate, detecting the induction of a reporter gene (comprising an GPCRX-responsive regulatory element operatively linked to a nucleic acid encoding a detectable marker, e.g., luciferase), or detecting a cellular response, for example, cell survival, cellular differentiation, or cell proliferation. In yet another embodiment, an assay of the invention is a cell-free assay comprising contacting an GPCRX protein or biologically-active portion thereof with a test compound and determining the ability of the test compound to bind to the GPCRX protein or biologically- active portion thereof. Binding of the test compound to the GPCRX protein can be determined either directly or indirectly as described above. In one such embodiment, the assay comprises contacting the GPCRX protein or biologically-active portion thereof with a known compound which binds GPCRX to form an assay mixture, contacting the assay mixture with a test compound, and determining the ability of the test compound to interact with an GPCRX protein, wherein determining the ability of the test compound to interact with an GPCRX protein comprises determining the ability of the test compound to preferentially bind to GPCRX or biologically-active portion thereof as compared to the known compound.
In still another embodiment, an assay is a cell-free assay comprising contacting GPCRX protein or biologically-active portion thereof with a test compound and determining the ability of the test compound to modulate (e.g. stimulate or inhibit) the activity of the GPCRX protein or biologically-active portion thereof. Determining the ability of the test compound to
modulate the activity of GPCRX can be accomplished, for example, by determining the ability of the GPCRX protein to bind to an GPCRX target molecule by one of the methods described above for determining direct binding. In an alternative embodiment, determining the ability of the test compound to modulate the activity of GPCRX protein can be accomplished by determining the ability of the GPCRX protein further modulate an GPCRX target molecule. For example, the catalytic/enzymatic activity of the target molecule on an appropriate substrate can be detennined as described, supra.
In yet another embodiment, the cell-free assay comprises contacting the GPCRX protein or biologically- active portion thereof with a known compound which binds GPCRX protein to form an assay mixture, contacting the assay mixture with a test compound, and determining the ability of the test compound to interact with an GPCRX protein, wherein determining the ability of the test compound to interact with an GPCRX protein comprises determining the ability of the GPCRX protein to preferentially bind to or modulate the activity of an GPCRX target molecule. The cell-free assays of the invention are amenable to use of both the soluble form or the membrane-bound form of GPCRX protein. In the case of cell-free assays comprising the membrane-bound form of GPCRX protein, it may be desirable to utilize a solubilizing agent such that the membrane-bound form of GPCRX protein is maintained in solution. Examples of such solubilizing agents include non-ionic detergents such as n-octylglucoside, n-dodecylglucoside, n-dodecylmaltoside, octanoyl-N-methylglucamide, decanoyl-N-methylglucamide, Triton® X-100, Triton® X-114, Thesit®, Isotridecypoly(ethylene glycol ether)n, N-dodecyl~N,N-dimethyl-3-ammonio-l -propane sulfonate, 3-(3-cholamidopropyl) dimethylamminiol-1 -propane sulfonate (CHAPS), or 3-(3-cholamidopropyl)dimethylaιmniniol-2-hydroxy- 1 -propane sulfonate (CHAPSO). hi more than one embodiment of the above assay methods of the invention, it may be desirable to immobilize either GPCRX protein or its target molecule to facilitate separation of complexed from uncomplexed fonns of one or both of the proteins, as well as to accommodate automation of the assay. Binding of a test compound to GPCRX protein, or interaction of GPCRX protein with a target molecule in the presence and absence of a candidate compound, can be accomplished in any vessel suitable for containing the reactants. Examples of such vessels include microtiter plates, test tubes, and micro-centrifuge tubes. In one embodiment, a fusion protein can be provided that adds a domain that allows one or both of the proteins to be bound to a matrix. For example, GST-GPCRX fusion proteins or GST-target fusion proteins can be adsorbed onto glutathione sepharose beads (Sigma Chemical, St. Louis, MO) or
glutathione derivatized microtiter plates, that are then combined with the test compound or the test compound and either the non-adsorbed target protein or GPCRX protein, and the mixture is incubated under conditions conducive to complex formation (e.g., at physiological conditions for salt and pH). Following incubation, the beads or microtiter plate wells are washed to remove any unbound components, the matrix immobilized in the case of beads, complex detennined either directly or indirectly, for example, as described, supra. Alternatively, the complexes can be dissociated from the matrix, and the level of GPCRX protein binding or activity determined using standard techniques.
Other techniques for immobilizing proteins on matrices can also be used in the screening assays of the invention. For example, either the GPCRX protein or its target molecule can be immobilized utilizing conjugation of biotin and streptavidin. Biotinylated GPCRX protein or target molecules can be prepared from biotin-NHS (N-hydroxy-succinimide) using techniques well-known within the art (e.g., biotinylation kit, Pierce Chemicals, Rockford, 111.), and immobilized in the wells of streptavidin-coated 96 well plates (Pierce Chemical). Alternatively, antibodies reactive with GPCRX protein or target molecules, but which do not interfere with binding of the GPCRX protein to its target molecule, can be derivatized to the wells of the plate, and unbound target or GPCRX protein trapped in the wells by antibody conjugation. Methods for detecting such complexes, in addition to those described above for the GST-immobilized complexes, include immunodetection of complexes using antibodies reactive with the GPCRX protein or target molecule, as well as enzyme-linked assays that rely on detecting an enzymatic activity associated with the GPCRX protein or target molecule.
In another embodiment, modulators of GPCRX protein expression are identified in a method wherein a cell is contacted with a candidate compound and the expression of GPCRX mRNA or protein in the cell is determined. The level of expression of GPCRX mRNA or protein in the presence of the candidate compound is compared to the level of expression of
GPCRX mRNA or protein in the absence of the candidate compound. The candidate compound can then be identified as a modulator of GPCRX mRNA or protein expression based upon this comparison. For example, when expression of GPCRX mRNA or protein is greater (i.e., statistically significantly greater) in the presence of the candidate compound than in its absence, the candidate compound is identified as a stimulator of GPCRX mRNA or protein expression. Alternatively, when expression of GPCRX mRNA or protein is less (statistically significantly less) in the presence of the candidate compound than in its absence, the candidate compound is identified as an inhibitor of GPCRX mRNA or protein expression. The level of GPCRX
mRNA or protein expression in the cells can be determined by methods described herein for detecting GPCRX mRNA or protein.
In yet another aspect of the invention, the GPCRX proteins can be used as "bait proteins" in a two-hybrid assay or three hybrid assay (see, e.g., U.S. Patent No. 5,283,317; Zervos, et al, 1993. Cell 72: 223-232; Madura, et al, 1993. J. Biol. Chem. 268: 12046-12054; Bartel, et al, 1993. Biotechniques 14: 920-924; Iwabuchi, et al, 1993. Oncogene 8: 1693-1696; and Brent WO 94/10300), to identify other proteins that bind to or interact with GPCRX ("GPCRX-binding proteins" or "GPCRX-bp") and modulate GPCRX activity. Such GPCRX-binding proteins are also likely to be involved in the propagation of signals by the GPCRX proteins as, for example, upstream or downstream elements of the GPCRX pathway. The two-hybrid system is based on the modular nature of most transcription factors, which consist of separable DNA-binding and activation domains. Briefly, the assay utilizes two different DNA constructs. In one construct, the gene that codes for GPCRX is fused to a gene encoding the DNA binding domain of a known transcription factor (e.g. , GAL-4). In the other construct, a DNA sequence, from a library of DNA sequences, that encodes an unidentified protein ("prey" or "sample") is fused to a gene that codes for the activation domain of the known transcription factor. If the "bait" and the "prey" proteins are able to interact, in vivo, forming an GPCRX-dependent complex, the DNA-binding and activation domains of the transcription factor are brought into close proximity. This proximity allows transcription of a reporter gene (e.g., LacZ) that is operably linked to a transcriptional regulatory site responsive to the transcription factor. Expression of the reporter gene can be detected and cell colonies containing the functional transcription factor can be isolated and used to obtain the cloned gene that encodes the protein which interacts with GPCRX.
The invention further pertains to novel agents identified by the aforementioned screening assays and uses thereof for treatments as described herein.
Detection Assays
Portions or fragments of the cDNA sequences identified herein (and the corresponding complete gene sequences) can be used in numerous ways as polynucleotide reagents. By way of example, and not of limitation, these sequences can be used to: (i) map their respective genes on a chromosome; and, thus, locate gene regions associated with genetic disease; (ii) identify an individual from a minute biological sample (tissue typing); and (Hi) aid in forensic identification of a biological sample. Some of these applications are described in the subsections, below.
Chromosome Mapping
Once the sequence (or a portion of the sequence) of a gene has been isolated, this sequence can be used to map the location of the gene on a chromosome. This process is called chromosome mapping. Accordingly, portions or fragments of the GPCRX sequences, SEQ ID NOS:l, 3, 5, 7, 9, 11, 13, 15, 17, 19, 21, 23, 25, 27, 29, 31, 33, 35, 37, 39, 41 and 43, or fragments or derivatives thereof, can be used to map the location of the GPCRX genes, respectively, on a chromosome. The mapping of the GPCRX sequences to chromosomes is an important first step in correlating these sequences with genes associated with disease. Briefly, GPCRX genes can be mapped to chromosomes by preparing PCR primers
(preferably 15-25 bp in length) from the GPCRX sequences. Computer analysis of the GPCRX, sequences can be used to rapidly select primers that do not span more than one exon in the genomic DNA, thus complicating the amplification process. These primers can then be used for PCR screening of somatic cell hybrids containing individual human chromosomes. Only those hybrids containing the human gene coreesponding to the GPCRX sequences will yield an amplified fragment.
Somatic cell hybrids are prepared by fusing somatic cells from different mammals (e.g., human and mouse cells). As hybrids of human and mouse cells grow and divide, they gradually lose human chromosomes in random order, but retain the mouse chromosomes. By using media in which mouse cells cannot grow, because they lack a particular enzyme, but in which human cells can, the one human chromosome that contains the gene encoding the needed enzyme will be retained. By using various media, panels of hybrid cell lines can be established. Each cell line in a panel contains either a single human chromosome or a small number of human chromosomes, and a full set of mouse chromosomes, allowing easy mapping of individual genes to specific human chromosomes. See, e.g., D'Eustachio, et al, 1983. Science 220: 919-924. Somatic cell hybrids containing only fragments of human chromosomes can also be produced by using human chromosomes with translocations and deletions.
PCR mapping of somatic cell hybrids is a rapid procedure for assigning a particular sequence to a particular chromosome. Three or more sequences can be assigned per day using a single thermal cycler. Using the GPCRX sequences to design oligonucleotide primers, sub- localization can be achieved with panels of fragments from specific chromosomes.
Fluorescence in situ hybridization (FISH) of a DNA sequence to a metaphase chromosomal spread can further be used to provide a precise chromosomal location in one step.
Chromosome spreads can be made using cells whose division has been blocked in metaphase
by a chemical like colcemid that disrupts the mitotic spindle. The chromosomes can be treated briefly with trypsin, and then stained with Giemsa. A pattern of light and dark bands develops on each chromosome, so that the chromosomes can be identified individually. The FISH technique can be used with a DNA sequence as short as 500 or 600 bases. However, clones larger than 1,000 bases have a higher likelihood of binding to a unique chromosomal location with sufficient signal intensity for simple detection. Preferably 1,000 bases, and more preferably 2,000 bases, will suffice to get good results at a reasonable amount of time. For a review of this technique, see, Verma, et al, HUMAN CHROMOSOMES: A MANUAL OF BASIC TECHNIQUES (Pergamon Press, New York 1988). Reagents for chromosome mapping can be used individually to mark a single chromosome or a single site on that chromosome, or panels of reagents can be used for marking multiple sites and or multiple chromosomes. Reagents corresponding to noncoding regions of the genes actually are preferred for mapping puφoses. Coding sequences are more likely to be conserved within gene families, thus increasing the chance of cross hybridizations during chromosomal mapping.
Once a sequence has been mapped to a precise chromosomal location, the physical position of the sequence on the chromosome can be correlated with genetic map data. Such data are found, e.g., in McKusick, MENDELIAN INHERITANCE IN MAN, available on-line through Johns Hopkins University Welch Medical Library). The relationship between genes and disease, mapped to the same chromosomal region, can then be identified through linkage analysis (co-inheritance of physically adjacent genes), described in, e.g., Egeland, et al, 1987. Nature, 325: 783-787.
Moreover, differences in the DNA sequences between individuals affected and unaffected with a disease associated with the GPCRX gene, can be determined. If a mutation is observed in some or all of the affected individuals but not in any unaffected individuals, then the mutation is likely to be the causative agent of the particular disease. Comparison of affected and unaffected individuals generally involves first looking for structural alterations in the chromosomes, such as deletions or translocations that are visible from chromosome spreads or detectable using PCR based on that DNA sequence. Ultimately, complete sequencing of genes from several individuals can be performed to confirm the presence of a mutation and to distinguish mutations from polymoφhisms.
Tissue Typing
The GPCRX sequences of the invention can also be used to identify individuals from minute biological samples. In this technique, an individual's genomic DNA is digested with one or more restriction enzymes, and probed on a Southern blot to yield unique bands for identification. The sequences of the invention are useful as additional DNA markers for RFLP ("restriction fragment length polymoφhisms," described in U.S. Patent No. 5,272,057).
Furthermore, the sequences of the invention can be used to provide an alternative technique that determines the actual base-by-base DNA sequence of selected portions of an individual's genome. Thus, the GPCRX sequences described herein can be used to prepare two PCR primers from the 5'- and 3'-termini of the sequences. These primers can then be used to amplify an individual's DNA and subsequently sequence it.
Panels of corresponding DNA sequences from individuals, prepared in this manner, can provide unique individual identifications, as each individual will have a unique set of such DNA sequences due to allelic differences. The sequences of the invention can be used to obtain such identification sequences from individuals and from tissue. The GPCRX sequences of the invention uniquely represent portions of the human genome. Allelic variation occurs to some degree in the coding regions of these sequences, and to a greater degree in the noncoding regions. It is estimated that allelic variation between individual humans occurs with a frequency of about once per each 500 bases. Much of the allelic variation is due to single nucleotide polymoφhisms (SNPs), which include restriction fragment length polymoφhisms (RFLPs).
Each of the sequences described herein can, to some degree, be used as a standard against which DNA from an individual can be compared for identification puφoses. Because greater numbers of polymoφhisms occur in the noncoding regions, fewer sequences are necessary to differentiate individuals. The noncoding sequences can comfortably provide positive individual identification with a panel of perhaps 10 to 1,000 primers that each yield a noncoding amplified sequence of 100 bases. If predicted coding sequences, such as those in SEQ ID NOS:l, 3, 5, 7, 9, 11, 13, 15, 17, 19, 21, 23, 25, 27, 29, 31, 33, 35, 37, 39, 41 and 43 are used, a more appropriate number of primers for positive individual identification would be 500-2,000.
Predictive Medicine
The invention also pertains to the field of predictive medicine in which diagnostic assays, prognostic assays, pharmacogenomics, and monitoring clinical trials are used for prognostic (predictive) p uposes to thereby treat an individual prophylactically. Accordingly, one aspect of the invention relates to diagnostic assays for determining GPCRX protein and/or nucleic acid expression as well as GPCRX activity, in the context of a biological sample (e.g., blood, serum, cells, tissue) to thereby determine whether an individual is afflicted with a disease or disorder, or is at risk of developing a disorder, associated with aberrant GPCRX expression or activity. The disorders include metabolic disorders, diabetes, obesity, infectious disease, anorexia, cancer- associated cachexia, cancer, neurodegenerative disorders, Alzheimer's Disease, Parkinson's Disorder, immune disorders, and hematopoietic disorders, and the various dyslipidemias, metabolic disturbances associated with obesity, the metabolic syndrome X and wasting disorders associated with chronic diseases and various cancers. The invention also provides for prognostic (or predictive) assays for determining whether an individual is at risk of developing a disorder associated with GPCRX protein, nucleic acid expression or activity. For example, mutations in an GPCRX gene can be assayed in a biological sample. Such assays can be used for prognostic or predictive puφose to thereby prophylactically treat an individual prior to the onset of a disorder characterized by or associated with GPCRX protein, nucleic acid expression, or biological activity.
Another aspect of the invention provides methods for determining GPCRX protein, nucleic acid expression or activity in an individual to thereby select appropriate therapeutic or prophylactic agents for that individual (referred to herein as "pharmacogenomics"). Pharmacogenomics allows for the selection of agents (e.g., drugs) for therapeutic or prophylactic treatment of an individual based on the genotype of the individual (e.g., the genotype of the individual examined to determine the ability of the individual to respond to a particular agent.)
Yet another aspect of the invention pertains to monitoring the influence of agents (e.g., drugs, compounds) on the expression or activity of GPCRX in clinical trials. These and other agents are described in further detail in the following sections.
Diagnostic Assays
An exemplary method for detecting the presence or absence of GPCRX in a biological sample involves obtaining a biological sample from a test subject and contacting the biological
sample with a compound or an agent capable of detecting GPCRX protein or nucleic acid (e.g., mRNA, genomic DNA) that encodes GPCRX protein such that the presence of GPCRX is detected in the biological sample. An agent for detecting GPCRX mRNA or genomic DNA is a labeled nucleic acid probe capable of hybridizing to GPCRX mRNA or genomic DNA. The nucleic acid probe can be, for example, a full-length GPCRX nucleic acid, such as the nucleic acid of SEQ ID NOS:l, 3, 5, 7, 9, 11, 13, 15, 17, 19, 21, 23, 25, 27, 29, 31, 33, 35, 37, 39, 41 and 43, or a portion thereof, such as an oligonucleotide of at least 15, 30, 50, 100, 250 or 500 nucleotides in length and sufficient to specifically hybridize under stringent conditions to GPCRX mRNA or genomic DNA. Other suitable probes for use in the diagnostic assays of the invention are described herein.
An agent for detecting GPCRX protein is an antibody capable of binding to GPCRX protein, preferably an antibody with a detectable label. Antibodies can be polyclonal, or more preferably, monoclonal. An intact antibody, or a fragment thereof (e.g., Fab or F(ab') ) can be "used. The term "labeled", with regard to the probe or antibody, is intended to encompass direct labeling of the probe or antibody by coupling (z. e. , physically linking) a detectable substance to the probe or antibody, as well as indirect labeling of the probe or antibody by reactivity with another reagent that is directly labeled. Examples of indirect labeling include detection of a primary antibody using a fluorescently-labeled secondary antibody and end-labeling of a DNA probe with biotin such that it can be detected with fluorescently-labeled streptavidin. The term "biological sample" is intended to include tissues, cells and biological fluids isolated from a subject, as well as tissues, cells and fluids present within a subject. That is, the detection method of the invention can be used to detect GPCRX mRNA, protein, or genomic DNA in a biological sample in vitro as well as in vivo. For example, in vitro techniques for detection of GPCRX mRNA include Northern hybridizations and in situ hybridizations. In vitro techniques for detection of GPCRX protein include enzyme linked immunosorbent assays (ELIS As), Western blots, immunoprecipitations, and immunofluorescence. In vitro techniques for detection of GPCRX genomic DNA include Southern hybridizations. Furthermore, in vivo techniques for detection of GPCRX protein include introducing into a subject a labeled anti-GPCRX antibody. For example, the antibody can be labeled with a radioactive marker whose presence and location in a subject can be detected by standard imaging techniques.
In one embodiment, the biological sample contains protein molecules from the test subject. Alternatively, the biological sample can contain mRNA molecules from the test subject or genomic DNA molecules from the test subject. A prefened biological sample is a peripheral blood leukocyte sample isolated by conventional means from a subject.
In another embodiment, the methods further involve obtaining a control biological sample from a control subject, contacting the control sample with a compound or agent capable of detecting GPCRX protein, mRNA, or genomic DNA, such that the presence of GPCRX protein, mRNA or genomic DNA is detected in the biological sample, and comparing the presence of GPCRX protein, mRNA or genomic DNA in the control sample with the presence of GPCRX protein, mRNA or genomic DNA in the test sample.
The invention also encompasses kits for detecting the presence of GPCRX in a biological sample. For example, the kit can comprise: a labeled compound or agent capable of detecting GPCRX protein or mRNA in a biological sample; means for determining the amount of GPCRX in the sample; and means for comparing the amount of GPCRX in the sample with a standard. The compound or agent can be packaged in a suitable container. The kit can further comprise instructions for using the kit to detect GPCRX protein or nucleic acid.
Prognostic Assays The diagnostic methods described herein can furthermore be utilized to identify subj ects having or at risk of developing a disease or disorder associated with abereant GPCRX expression or activity. For example, the assays described herein, such as the preceding diagnostic assays or the following assays, can be utilized to identify a subject having or at risk of developing a disorder associated with GPCRX protein, nucleic acid expression or activity. Alternatively, the prognostic assays can be utilized to identify a subject having or at risk for developing a disease or disorder. Thus, the invention provides a method for identifying a disease or disorder associated with aberrant GPCRX expression or activity in which a test sample is obtained from a subject and GPCRX protein or nucleic acid (e.g., mRNA, genomic DNA) is detected, wherein the presence of GPCRX protein or nucleic acid is diagnostic for a subject having or at risk of developing a disease or disorder associated with aberrant GPCRX expression or activity. As used herein, a "test sample" refers to a biological sample obtained from a subject of interest. For example, a test sample can be a biological fluid (e.g., serum), cell sample, or tissue.
Furthermore, the prognostic assays described herein can be used to determine whether a subject can be administered an agent (e.g., an agonist, antagonist, peptidomimetic, protein, peptide, nucleic acid, small molecule, or other drug candidate) to treat a disease or disorder associated with aberrant GPCRX expression or activity. For example, such methods can be used to determine whether a subject can be effectively treated with an agent for a disorder.
Thus, the invention provides methods for determining whether a subject can be effectively
treated with an agent for a disorder associated with aberrant GPCRX expression or activity in which a test sample is obtained and GPCRX protein or nucleic acid is detected (e.g., wherein the presence of GPCRX protein or nucleic acid is diagnostic for a subject that can be administered the agent to treat a disorder associated with aberrant GPCRX expression or activity).
The methods of the invention can also be used to detect genetic lesions in an GPCRX gene, thereby determining if a subject with the lesioned gene is at risk for a disorder characterized by aberrant cell proliferation and/or differentiation. In various embodiments, the methods include detecting, in a sample of cells from the subject, the presence or absence of a genetic lesion characterized by at least one of an alteration affecting the integrity of a gene encoding an GPCRX-protein, or the misexpression of the GPCRX gene. For example, such genetic lesions can be detected by ascertaining the existence of at least one of: (i) a deletion of one or more nucleotides from an GPCRX gene; (ii) an addition of one or more nucleotides to an GPCRX gene; (Hi) a substitution of one or more nucleotides of an GPCRX gene, (iv) a chromosomal rearrangement of an GPCRX gene; (v) an alteration in the level of a messenger RNA transcript of an GPCRX gene, (vi) abereant modification of an GPCRX gene, such as of the methylation pattern of the genomic DNA, (vii) the presence of a non- wild-type splicing pattern of a messenger RNA transcript of an GPCRX gene, (viii) a non- wild-type level of an GPCRX protein, (ix) allelic loss of an GPCRX gene, and (x) inappropriate post-translational modification of an GPCRX protein. As described herein, there are a large number of assay techniques known in the art which can be used for detecting lesions in an GPCRX gene. A preferred biological sample is a peripheral blood leukocyte sample isolated by conventional means from a subject. However, any biological sample containing nucleated cells may be used, including, for example, buccal mucosal cells. h certain embodiments, detection of the lesion involves the use of a probe/primer in a polymerase chain reaction (PCR) (.see, e.g., U.S. Patent Nos. 4,683,195 and 4,683,202), such as anchor PCR or RACE PCR, or, alternatively, in a ligation chain reaction (LCR) (see, e.g., Landegran, et αl., 1988. Science 241: 1077-1080; and akazawa, et αl, 1994. Proc. Nαtl Acαd. Sci. USA 91 : 360-364), the latter of which can be particularly useful for detecting point mutations in the GPCRX-gene (see, Abravaya, et αl, 1995. Nucl. Acids Res. 23: 675-682). This method can include the steps of collecting a sample of cells from a patient, isolating nucleic acid (e.g., genomic, mRNA or both) from the cells of the sample, contacting the nucleic acid sample with one or more primers that specifically hybridize to an GPCRX gene under conditions such that hybridization and amplification of the GPCRX gene (if present) occurs,
and detecting the presence or absence of an amplification product, or detecting the size of the amplification product and comparing the length to a control sample. It is anticipated that PCR and/or LCR may be desirable to use as a preliminary amplification step in conjunction with any of the techniques used for detecting mutations described herein. Alternative amplification methods include: self sustained sequence replication (see,
Guatelli, et al, 1990. Proc. Natl. Acad. Sci. USA 87: 1874-1878), transcriptional amplification system (see, Kwoh, et al, 1989. Proc. Natl. Acad. Sci. USA 86: 1173-1177); Qβ Replicase (see, Lizardi, et al, 1988. BioTechnology 6: 1197), or any other nucleic acid amplification method, followed by the detection of the amplified molecules using techniques well known to those of skill in the art. These detection schemes are especially useful for the detection of nucleic acid molecules if such molecules are present in very low numbers.
In an alternative embodiment, mutations in an GPCRX gene from a sample cell can be identified by alterations in restriction enzyme cleavage patterns. For example, sample and control DNA is isolated, amplified (optionally), digested with one or more restriction endonucleases, and fragment length sizes are determined by gel electrophoresis and compared. Differences in fragment length sizes between sample and control DNA indicates mutations in the sample DNA. Moreover, the use of sequence specific ribozymes (see, e.g., U.S. Patent No. 5,493,531) can be used to score for the presence of specific mutations by development or loss of a ribozyme cleavage site. In other embodiments, genetic mutations in GPCRX can be identified by hybridizing a sample and control nucleic acids, e.g., DNA or RNA, to high-density arrays containing hundreds or thousands of oligonucleotides probes. See, e.g., Cronin, et al, 1996. Human Mutation 1: 244-255; Kozal, et al, 1996. Nat. Med. 2: 753-759. For example, genetic mutations in GPCRX can be identified in two dimensional arrays containing light-generated DNA probes as described in Cronin, et al, supra. Briefly, a first hybridization array of probes can be used to scan through long stretches of DNA in a sample and control to identify base changes between the sequences by making linear arrays of sequential overlapping probes. This step allows the identification of point mutations. This is followed by a second hybridization array that allows the characterization of specific mutations by using smaller, specialized probe arrays complementary to all variants or mutations detected. Each mutation array is composed of parallel probe sets, one complementary to the wild-type gene and the other complementary to the mutant gene.
In yet another embodiment, any of a variety of sequencing reactions known in the art can be used to directly sequence the GPCRX gene and detect mutations by comparing the
sequence of the sample GPCRX with the corresponding wild-type (control) sequence. Examples of sequencing reactions include those based on techniques developed by Maxim and Gilbert, 1977. Proc. Natl. Acad. Sci. USA 74: 560 or Sanger, 1977. Proc. Natl. Acad. Sci. USA 74: 5463. It is also contemplated that any of a variety of automated sequencing procedures can be utilized when perfonning the diagnostic assays (see, e.g., Naeve, et al, 1995. Biotechniques 19: 448), including sequencing by mass spectromerry (see, e.g., PCT International Publication No. WO 94/16101; Cohen, et al, 1996. Adv. Chromatography 36: 127-162; and Griffin, et al, 1993. Appl. Biochem. Biotechnol 38: 147-159).
Other methods for detecting mutations in the GPCRX gene include methods in which protection from cleavage agents is used to detect mismatched bases in RNA/RNA or
RNA/DNA heteroduplexes. See, e.g., Myers, et al, 1985. Science 230: 1242. In general, the art technique of "mismatch cleavage" starts by providing heteroduplexes of formed by hybridizing (labeled) RNA or DNA containing the wild-type GPCRX sequence with potentially mutant RNA or DNA obtained from a tissue sample. The double-stranded duplexes are treated with an agent that cleaves single-stranded regions of the duplex such as which will exist due to basepair mismatches between the control and sample strands. For instance, RNA/DNA duplexes can be treated with RNase and DNA/DNA hybrids treated with Si nuclease to enzymatically digesting the mismatched regions. In other embodiments, either DNA/DNA or RNA/DNA duplexes can be treated with hydroxylamine or osmium tetroxide and with piperidine in order to digest mismatched regions. After digestion of the mismatched regions, the resulting material is then separated by size on denaturing polyacrylamide gels to determine the site of mutation. See, e.g., Cotton, et al, 1988. Proc. Natl Acad. Sci. USA 85: 4397; Saleeba, et al, 1992. Methods Enzymol 217: 286-295. In an embodiment, the control DNA or RNA can be labeled for detection. In still another embodiment, the mismatch cleavage reaction employs one or more proteins that recognize mismatched base pairs in double-stranded DNA (so called "DNA mismatch repair" enzymes) in defined systems for detecting and mapping point mutations in GPCRX cDNAs obtained from samples of cells. For example, the mutY enzyme of E. coli cleaves A at G/A mismatches and the thymidine DNA glycosylase from HeLa cells cleaves T at G/T mismatches. See, e.g., Hsu, et al, 1994. Carcinogenesis 15: 1657-1662. According to an exemplary embodiment, a probe based on an GPCRX sequence, e.g., a wild- type GPCRX sequence, is hybridized to a cDNA or other DNA product from a test cell(s). The duplex is treated with a DNA mismatch repair enzyme, and the cleavage products, if any, can be detected from electrophoresis protocols or the like. See, e.g., U.S. Patent No. 5,459,039.
In other embodiments, alterations in electrophoretic mobility will be used to identify mutations in GPCRX genes. For example, single strand conformation polymoφhism (SSCP) may be used to detect differences in electrophoretic mobility between mutant and wild type nucleic acids. See, e.g., Orita, et al, 1989. Proc. Natl. Acad. Sci. USA: 86: 2766; Cotton, 1993. Mutat. Res. 285: 125-144; Hayashi, 1992. Genet. Anal. Tech. Appl. 9: 73-79. Single-stranded DNA fragments of sample and control GPCRX nucleic acids will be denatured and allowed to renature. The secondary structure of single-stranded nucleic acids varies according to sequence, the resulting alteration in electrophoretic mobility enables the detection of even a single base change. The DNA fragments may be labeled or detected with labeled probes. The sensitivity of the assay may be enhanced by using RNA (rather than DNA), in which the secondary structure is more sensitive to a change in sequence, hi one embodiment, the subject method utilizes heteroduplex analysis to separate double stranded heteroduplex molecules on the basis of changes in electrophoretic mobility. See, e.g., Keen, et al, 1991. Trends Genet. 1: 5. hi yet another embodiment, the movement of mutant or wild-type fragments in polyacrylamide gels containing a gradient of denaturant is assayed using denaturing gradient gel electrophoresis (DGGE). See, e.g., Myers, et al, 1985. Nature 313: 495. When DGGE is used as the method of analysis, DNA will be modified to insure that it does not completely denature, for example by adding a GC clamp of approximately 40 bp of high-melting GC-rich DNA by PCR. hi a further embodiment, a temperature gradient is used in place of a denaturing gradient to identify differences in the mobility of control and sample DNA. See, e.g., Rosenbaum and Reissner, 1987. Biophys. Chem. 265: 12753.
Examples of other techniques for detecting point mutations include, but are not limited to, selective oligonucleotide hybridization, selective amplification, or selective primer extension. For example, oligonucleotide primers may be prepared in which the known mutation is placed centrally and then hybridized to target DNA under conditions that permit hybridization only if a perfect match is found. See, e.g., Saiki, et al, 1986. Nature 324: 163; Saiki, et al, 1989. Proc. Natl. Acad. Sci. USA 86: 6230. Such allele specific oligonucleotides are hybridized to PCR amplified target DNA or a number of different mutations when the oligonucleotides are attached to the hybridizing membrane and hybridized with labeled target
DNA.
Alternatively, allele specific amplification technology that depends on selective PCR amplification may be used in conjunction with the instant invention. Oligonucleotides used as primers for specific amplification may carry the mutation of interest in the center of the
molecule (so that amplification depends on differential hybridization; see, e.g., Gibbs, et al, 1989. Nucl. Acids Res. 17: 2437-2448) or at the extreme 3'-terminus of one primer where, under appropriate conditions, mismatch can prevent, or reduce polymerase extension (see, e.g., Prossner, 1993. Tibtech. 11 : 238). In addition it may be desirable to introduce a novel restriction site in the region of the mutation to create cleavage-based detection. See, e.g., Gasparini, et al, 1992. Mol. Cell Probes 6: 1. It is anticipated that in certain embodiments amplification may also be performed using Taq ligase for amplification. See, e.g., Barany, 1991. Proc. Natl. Acad. Sci. USA 88: 189. In such cases, ligation will occur only if there is a perfect match at the 3 '-terminus of the 5' sequence, making it possible to detect the presence of a known mutation at a specific site by looking for the presence or absence of amplification. The methods described herein may be perfonned, for example, by utilizing pre-packaged diagnostic kits comprising at least one probe nucleic acid or antibody reagent described herein, which may be conveniently used, e.g., in clinical settings to diagnose patients exhibiting symptoms or family history of a disease or illness involving an GPCRX gene. Furthermore, any cell type or tissue, preferably peripheral blood leukocytes, in which
GPCRX is expressed may be utilized in the prognostic assays described herein. However, any biological sample containing nucleated cells may be used, including, for example, buccal mucosal cells.
Pharmacogenomics
Agents, or modulators that have a stimulatory or inhibitory effect on GPCRX activity (e.g., GPCRX gene expression), as identified by a screening assay described herein can be administered to individuals to treat (prophylactically or therapeutically) disorders (The disorders include metabolic disorders, diabetes, obesity, infectious disease, anorexia, cancer- associated cachexia, cancer, neurodegenerative disorders, Alzheimer's Disease, Parkinson's Disorder, immune disorders, and hematopoietic disorders, and the various dyslipidemias, metabolic disturbances associated with obesity, the metabolic syndrome X and wasting disorders associated with chronic diseases and various cancers.) In conjunction with such treatment, the pharmacogenomics (i.e., the study of the relationship between an individual's genotype and that individual's response to a foreign compound or drug) of the individual may be considered. Differences in metabolism of therapeutics can lead to severe toxicity or therapeutic failure by altering the relation between dose and blood concentration of the pharmacologically active drug. Thus, the pharmacogenomics of the individual permits the selection of effective agents (e.g., drugs) for prophylactic or therapeutic treatments based on a
consideration of the individual's genotype. Such pharmacogenomics can further be used to determine appropriate dosages and therapeutic regimens. Accordingly, the activity of GPCRX protein, expression of GPCRX nucleic acid, or mutation content of GPCRX genes in an individual can be determined to thereby select appropriate agent(s) for therapeutic or prophylactic treatment of the individual.
Pharmacogenomics deals with clinically significant hereditary variations in the response to drugs due to altered drug disposition and abnormal action in affected persons. See e.g., Eichelbaum, 1996. Clin. Exp. Pharmacol. Physiol, 23: 983-985; Linder, 1997. Clin. Chem., 43: 254-266. In general, two types of pharmaco genetic conditions can be differentiated. Genetic conditions transmitted as a single factor altering the way drugs act on the body (altered drug action) or genetic conditions transmitted as single factors altering the way the body acts on drugs (altered drug metabolism). These pharmacogenetic conditions can occur either as rare defects or as polymoφhisms. For example, glucose-6-phosphate dehydrogenase (G6PD) deficiency is a common inherited enzymopathy in which the main clinical complication is hemolysis after ingestion of oxidant drugs (anti-malarials, sulfonamides, analgesics, nitrofurans) and consumption of fava beans.
As an illustrative embodiment, the activity of drug metabolizing enzymes is a major determinant of both the intensity and duration of drug action. The discovery of genetic polymoφhisms of drug metabolizing enzymes (e.g., N-acetyltransferase 2 (NAT 2) and cytochrome P450 enzymes CYP2D6 and CYP2C19) has provided an explanation as to why some patients do not obtain the expected drug effects or show exaggerated drug response and serious toxicity after taking the standard and safe dose of a drug. These polymoφhisms are expressed in two phenotypes in the population, the extensive metabolizer (EM) and poor metabolizer (PM). The prevalence of PM is different among different populations. For example, the gene coding for CYP2D6 is highly polymoφhic and several mutations have been identified in PM, which all lead to the absence of functional CYP2D6. Poor metabolizers of CYP2D6 and CYP2C19 quite frequently experience exaggerated drug response and side effects when they receive standard doses. If a metabolite is the active therapeutic moiety, PM show no therapeutic response, as demonstrated for the analgesic effect of codeine mediated by its CYP2D6-formed metabolite moφhine. At the other extreme are the so called ultra-rapid metabolizers who do not respond to standard doses. Recently, the molecular basis of ultra-rapid metabolism has been identified to be due to CYP2D6 gene amplification.
Thus, the activity of GPCRX protein, expression of GPCRX nucleic acid, or mutation content of GPCRX genes in an individual can be determined to thereby select appropriate
agent(s) for therapeutic or prophylactic treatment of the individual, hi addition, pharmacogenetic studies can be used to apply genotyping of polymoφhic alleles encoding drug-metabolizing enzymes to the identification of an individual's drug responsiveness phenotype. This knowledge, when applied to dosing or drug selection, can avoid adverse reactions or therapeutic failure and thus enhance therapeutic or prophylactic efficiency when treating a subject with an GPCRX modulator, such as a modulator identified by one of the exemplary screening assays described herein.
Monitoring of Effects During Clinical Trials Monitoring the influence of agents (e.g., drugs, compounds) on the expression or activity of GPCRX (e.g., the ability to modulate abereant cell proliferation and/or differentiation) can be applied not only in basic drug screening, but also in clinical trials. For example, the effectiveness of an agent determined by a screening assay as described herein to increase GPCRX gene expression, protein levels, or upregulate GPCRX activity, can be monitored in clinical trails of subjects exhibiting decreased GPCRX gene expression, protein levels, or downregulated GPCRX activity. Alternatively, the effectiveness of an agent determined by a screening assay to decrease GPCRX gene expression, protein levels, or downregulate GPCRX activity, can be monitored in clinical trails of subjects exhibiting increased GPCRX gene expression, protein levels, or upregulated GPCRX activity. In such clinical trials, the expression or activity of GPCRX and, preferably, other genes that have been implicated in, for example, a cellular proliferation or immune disorder can be used as a "read out" or markers of the immune responsiveness of a particular cell.
By way of example, and not of limitation, genes, including GPCRX, that are modulated in cells by treatment with an agent (e.g. , compound, drug or small molecule) that modulates GPCRX activity (e.g., identified in a screening assay as described herein) can be identified.
Thus, to study the effect of agents on cellular proliferation disorders, for example, in a clinical trial, cells can be isolated and RNA prepared and analyzed for the levels of expression of GPCRX and other genes implicated in the disorder. The levels of gene expression (i.e., a gene expression pattern) can be quantified by Northern blot analysis or RT-PCR, as described herein, or alternatively by measuring the amount of protein produced, by one of the methods as described herein, or by measuring the levels of activity of GPCRX or other genes. In this manner, the gene expression pattern can serve as a marker, indicative of the physiological response of the cells to the agent. Accordingly, this response state may be determined before, and at various points during, treatment of the individual with the agent.
In one embodiment, the invention provides a method for monitoring the effectiveness of treatment of a subject with an agent (e.g., an agonist, antagonist, protein, peptide, peptidomimetic, nucleic acid, small molecule, or other drug candidate identified by the screening assays described herein) comprising the steps of (i) obtaining a pre-administration sample from a subject prior to administration of the agent; (ii) detecting the level of expression of an GPCRX protein, mRNA, or genomic DNA in the preadministration sample; (Hi) obtaining one or more post-administration samples from the subject; (iv) detecting the level of expression or activity of the GPCRX protein, mRNA, or genomic DNA in the post-administration samples; (v) comparing the level of expression or activity of the GPCRX protein, mRNA, or genomic DNA in the pre-administration sample with the GPCRX protein, mRNA, or genomic DNA in the post administration sample or samples; and (vi) altering the administration of the agent to the subject accordingly. For example, increased administration of the agent may be desirable to increase the expression or activity of GPCRX to higher levels than detected, i.e., to increase the effectiveness of the agent. Alternatively, decreased administration of the agent may be desirable to decrease expression or activity of GPCRX to lower levels than detected, i.e., to decrease the effectiveness of the agent.
Methods of Treatment
The invention provides for both prophylactic and therapeutic methods of treating a subject at risk of (or susceptible to) a disorder or having a disorder associated with abenant GPCRX expression or activity. The disorders include cardiomyopathy, atherosclerosis, hypertension, congenital heart defects, aortic stenosis, atrial septal defect (ASD), atrioventricular (A-V) canal defect, ductus arteriosus, pulmonary stenosis, subaortic stenosis, ventricular septal defect (VSD), valve diseases, tuberous sclerosis, scleroderma, obesity, transplantation, adrenoleukodystrophy, congenital adrenal hypeφlasia, prostate cancer, neoplasm; adenocarcinoma, lymphoma, uterus cancer, fertility, hemophilia, hypercoagulation, idiopathic thrombocytopenic puφura, immunodeficiencies, graft versus host disease, ADDS, bronchial asthma, Crohn's disease; multiple sclerosis, treatment of Albright Hereditary Ostoeodystrophy, and other diseases, disorders and conditions of the like. These methods of treatment will be discussed more fully, below.
Disease and Disorders
Diseases and disorders that are characterized by increased (relative to a subject not suffering from the disease or disorder) levels or biological activity may be treated with
Therapeutics that antagonize (i.e., reduce or inhibit) activity. Therapeutics that antagonize activity may be administered in a therapeutic or prophylactic manner. Therapeutics that may be utilized include, but are not limited to: (i) an aforementioned peptide, or analogs, derivatives, fragments or homologs thereof; (ii) antibodies to an aforementioned peptide; (Hi) nucleic acids encoding an aforementioned peptide; (zv) administration of antisense nucleic acid and nucleic acids that are "dysfunctional" (z'.e., due to a heterologous insertion within the coding sequences of coding sequences to an aforementioned peptide) that are utilized to "knockout" endoggenous function of an aforementioned peptide by homologous recombination (see, e.g., Capecchi, 1989. Science 244: 1288-1292); or (v) modulators ( i.e., inhibitors, agonists and antagonists, including additional peptide mimetic of the invention or antibodies specific to a peptide of the invention) that alter the interaction between an aforementioned peptide and its binding partner.
Diseases and disorders that are characterized by decreased (relative to a subject not suffering from the disease or disorder) levels or biological activity may be treated with Therapeutics that increase (i.e., are agonists to) activity. Therapeutics that upregulate activity may be administered in a therapeutic or prophylactic manner. Therapeutics that may be utilized include, but are not limited to, an aforementioned peptide, or analogs, derivatives, fragments or homologs thereof; or an agonist that increases bioavailability.
Increased or decreased levels can be readily detected by quantifying peptide and/or RNA, by obtaining a patient tissue sample (e.g., from biopsy tissue) and assaying it in vitro for RNA or peptide levels, structure and or activity of the expressed peptides (or niRNAs of an aforementioned peptide). Methods that are well-known within the art include, but are not limited to, immunoassays (e.g., by Western blot analysis, immunoprecipitation followed by sodium dodecyl sulfate (SDS) polyacrylamide gel electrophoresis, irnmunocytochemistry, etc.) and/or hybridization assays to detect expression of lnRNAs (e.g., Northern assays, dot blots, in situ hybridization, and the like).
Prophylactic Methods
In one aspect, the invention provides a method for preventing, in a subject, a disease or condition associated with an aberrant GPCRX expression or activity, by administering to the subject an agent that modulates GPCRX expression or at least one GPCRX activity. Subjects at risk for a disease that is caused or contributed to by aberrant GPCRX expression or activity can be identified by, for example, any or a combination of diagnostic or prognostic assays as described herein. Administration of a prophylactic agent can occur prior to the manifestation of symptoms characteristic of the GPCRX aberrancy, such that a disease or disorder is prevented
or, alternatively, delayed in its progression. Depending upon the type of GPCRX aberrancy, for example, an GPCRX agonist or GPCRX antagonist agent can be used for treating the subject. The appropriate agent can be determined based on screening assays described herein. The prophylactic methods of the invention are further discussed in the following subsections.
Therapeutic Methods
Another aspect of the invention pertains to methods of modulating GPCRX expression or activity for therapeutic puφoses. The modulatory method of the invention involves contacting a cell with an agent that modulates one or more of the activities of GPCRX protein activity associated with the cell. An agent that modulates GPCRX protein activity can be an agent as described herein, such as a nucleic acid or a protein, a naturally-occurring cognate ligand of an GPCRX protein, a peptide, an GPCRX peptidomimetic, or other small molecule, hi one embodiment, the agent stimulates one or more GPCRX protein activity. Examples of such stimulatory agents include active GPCRX protein and a nucleic acid molecule encoding GPCRX that has been introduced into the cell. In another embodiment, the agent inhibits one or more GPCRX protein activity. Examples of such inhibitory agents include antisense GPCRX nucleic acid molecules and anti-GPCRX antibodies. These modulatory methods can be performed in vitro (e.g., by culturing the cell with the agent) or, alternatively, in vivo (e.g., by administering the agent to a subject). As such, the invention provides methods of treating an individual afflicted with a disease or disorder characterized by aberrant expression or activity of an GPCRX protein or nucleic acid molecule, hi one embodiment, the method involves administering an agent (e.g., an agent identified by a screening assay described herein), or combination of agents that modulates (e.g., up-regulates or down-regulates) GPCRX expression or activity. In another embodiment, the method involves administering an GPCRX protein or nucleic acid molecule as therapy to compensate for reduced or aberrant GPCRX expression or activity.
Stimulation of GPCRX activity is desirable in situations in which GPCRX is abnormally downregulated and/or in which increased GPCRX activity is likely to have a beneficial effect. One example of such a situation is where a subject has a disorder characterized by aberrant cell proliferation and/or differentiation (e.g., cancer or immune associated disorders). Another example of such a situation is where the subject has a gestational disease (e.g., preclampsia).
Determination of the Biological Effect of the Therapeutic
In various embodiments of the invention, suitable in vitro or in vivo assays are perfonned to determine the effect of a specific Therapeutic and whether its administration is indicated for treatment of the affected tissue. In various specific embodiments, in vitro assays may be performed with representative cells of the type(s) involved in the patient's disorder, to determine if a given Therapeutic exerts the desired effect upon the cell type(s). Compounds for use in therapy may be tested in suitable animal model systems including, but not limited to rats, mice, chicken, cows, monkeys, rabbits, and the like, prior to testing in human subjects. Similarly, for in vivo testing, any of the animal model system known in the art may be used prior to administration to human subjects.
Prophylactic and Therapeutic Uses of the Compositions of the Invention
The GPCRX nucleic acids and proteins of the invention are useful in potential prophylactic and therapeutic applications implicated in a variety of disorders including, but not limited to: metabolic disorders, diabetes, obesity, infectious disease, anorexia, cancer-associated cancer, neurodegenerative disorders, Alzheimer's Disease, Parkinson's Disorder, immune disorders, hematopoietic disorders, and the various dyslipidemias, metabolic disturbances associated with obesity, the metabolic syndrome X and wasting disorders associated with chronic diseases and various cancers.
As an example, a cDNA encoding the GPCRX protein of the invention may be useful in gene therapy, and the protein may be useful when administered to a subject in need thereof. By way of non-limiting example, the compositions of the invention will have efficacy for treatment of patients suffering from: metabolic disorders, diabetes, obesity, infectious disease, anorexia, cancer-associated cachexia, cancer, neurodegenerative disorders, Alzheimer's Disease, Parkinson's Disorder, immune disorders, hematopoietic disorders, and the various dyslipidemias.
Both the novel nucleic acid encoding the GPCRX protein, and the GPCRX protein of the invention, or fragments thereof, may also be useful in diagnostic applications, wherein the presence or amount of the nucleic acid or the protein are to be assessed. A further use could be as an anti-bacterial molecule (z'.e., some peptides have been found to possess anti-bacterial properties). These materials are further useful in the generation of antibodies which immunospecifically-bind to the novel substances of the invention for use in therapeutic or diagnostic methods.
Examples
Example 1. Quantitative expression analysis of clones in various cells and tissues
The quantitative expression of various clones was assessed using microtiter plates containing RNA samples from a variety of normal and pathology-derived cells, cell lines and tissues using real time quantitative PCR (RTQ PCR; TAQMAN®). RTQ PCR was performed on a Perkin-Elmer Biosystems ABI PRISM® 7700 Sequence Detection System. Various collections of samples are assembled on the plates, and referred to as Panel 1 (containing cells and cell lines from normal and cancer sources), Panel 2 (containing samples derived from tissues, in particular from surgical samples, from normal and cancer sources), Panel 3 (containing samples derived from a wide variety of cancer sources) and Panel 4 (containing cells and cell lines from normal cells and cells related to inflammatory conditions).
First, the RNA samples were normalized to constitutively expressed genes such as β- actin and GAPDH. RNA (~50 ng total or ~1 ng polyA+) was converted to cDNA using the TAQMAN® Reverse Transcription Reagents Kit (PE Biosystems, Foster City, CA; Catalog No. N808-0234) and random hexamers according to the manufacturer' s protocol. Reactions were performed in 20 ul and incubated for 30 min. at 48°C. cDNA (5 ul) was then transfened to a separate plate for the TAQMAN® reaction using β-actin and GAPDH TAQMAN® Assay Reagents (PE Biosystems; Catalog Nos. 4310881E and 4310884E, respectively) and TAQMAN® universal PCR Master Mix (PE Biosystems; Catalog No. 4304447) according to the manufacturer's protocol. Reactions were performed in 25 ul using the following parameters: 2 min. at 50°C; 10 min. at 95°C; 15 sec. at 95°C/1 min. at 60°C (40 cycles). Results were recorded as CT values (cycle at which a given sample crosses a threshold level of fluorescence) using a log scale, with the difference in RNA concentration between a given sample and the sample with the lowest CT value being represented as 2 to the power of delta CT. The percent relative expression is then obtained by taking the reciprocal of this RNA difference and multiplying by 100. The average CT values obtained for β-actin and GAPDH were used to normalize RNA samples. The RNA sample generating the highest CT value required no further diluting, while all other samples were diluted relative to this sample according to their β-actin /GAPDH average CT values. Normalized RNA (5 ul) was converted to cDNA and analyzed via TAQMAN® using
One Step RT-PCR Master Mix Reagents (PE Biosystems; Catalog No. 4309169) and gene- specific primers according to the manufacturer's instructions. Probes and primers were designed for each assay according to Perkin Elmer Biosystem's Primer Express Software
package (version I for Apple Computer's Macintosh Power PC) or a similar algorithm using the target sequence as input. Default settings were used for reaction conditions and the following parameters were set before selecting primers: primer concentration = 250 nM, primer melting temperature (Tm) range = 58°-60° C, primer optimal Tm = 59° C, maximum primer difference = 2° C, probe does not have 5"G, probe Tm must be 10° C greater than primer Tm, amplicon size 75 bp to 100 bp. The probes and primers selected (see below) were synthesized by Synthegen (Houston, TX, USA). Probes were double purified by HPLC to remove uncoupled dye and evaluated by mass spectroscopy to verify coupling of reporter and quencher dyes to the 5' and 3' ends of the probe, respectively. Their final concentrations were: forward and reverse primers, 900 nM each, and probe, 200nM.
PCR conditions: Normalized RNA from each tissue and each cell line was spotted in each well of a 96 well PCR plate (Perkin Elmer Biosystems). PCR cocktails including two probes (a probe specific for the target clone and another gene-specific probe multiplexed with the target probe) were set up using IX TaqMan™ PCR Master Mix for the PE Biosystems 7700, with 5 mM MgC12, dNTPs (dA, G, C, U at 1 : 1 : 1 :2 ratios), 0.25 U/ml AmpliTaq Gold™ (PE Biosystems), and 0.4 U/μl RNase inhibitor, and 0.25 U/μl reverse transcriptase. Reverse transcription was performed at 48° C for 30 minutes followed by amplification/PCR cycles as follows: 95° C 10 min, then 40 cycles of 95° C for 15 seconds, 60° C for 1 minute.
In the results for Panel 1, the following abbreviations are used: ca. = carcinoma,
* = established from metastasis, met = metastasis, s cell var= small cell variant, non-s = non-sm =non-small, squam = squamous, pi. eff = pi effusion = pleural effusion, glio = glioma, astro = astrocytoma, and neuro = neuroblastoma.
Panel 2
The plates for Panel 2 generally include 2 control wells and 94 test samples composed of RNA or cDNA isolated from human tissue procured by surgeons working in close cooperation with the National Cancer Institute's Cooperative Human Tissue Network (CHTN) or the National Disease Research Initiative (NDRI). The tissues are derived from human malignancies and in cases where indicated many malignant tissues have "matched margins" obtained from noncancerous tissue just adjacent to the tumor. These are termed normal adjacent tissues and are denoted "NAT" in the results below. The tumor tissue and the "matched margins" are evaluated by two independent pathologists (the surgical pathologists and again by a pathologists at NDRI or CHTN). This analysis provides a gross histopathological assessment of tumor differentiation grade. Moreover, most samples include the original surgical pathology report that provides information regarding the clinical stage of the patient. These matched margins are taken from the tissue surrounding (i.e. immediately proximal) to the zone of surgery (designated "NAT", for normal adjacent tissue, in Table RR). In addition, RNA and cDNA samples were obtained from various human tissues derived from autopsies performed on elderly people or sudden death victims (accidents, etc.). These tissue were ascertained to be free of disease and were purchased from various commercial sources such as Clontech (Palo Alto, CA), Research Genetics, and Invitrogen.
RNA integrity from all samples is controlled for quality by visual assessment of agarose gel electropherograms using 28S and 18S ribosomal RNA staining intensity ratio as a guide (2:1 to 2.5:1 28s: 18s) and the absence of low molecular weight RNAs that would be indicative of degradation products. Samples are controlled against genomic DNA contamination by RTQ PCR reactions run in the absence of reverse transcriptase using probe and primer sets designed to amplify across the span of a single exon.
Panel 4
Panel 4 includes samples on a 96 well plate (2 control wells, 94 test samples) composed of RNA (Panel 4r) or cDNA (Panel 4d) isolated from various human cell lines or tissues related to inflammatory conditions. Total RNA from control normal tissues such as colon and lung
(Stratagene ,La Jolla, CA) and thymus and kidney (Clontech) were employed. Total RNA from liver tissue from cirrhosis patients and kidney from lupus patients was obtained from
BioChain (Biochain Institute, ie, Hayward, CA). Intestinal tissue for RNA preparation from patients diagnosed as having Crohn's disease and ulcerative colitis was obtained from the National Disease Research Interchange (NDRI) (Philadelphia, PA).
Astrocytes, lung fibroblasts, dermal fibroblasts, coronary artery smooth muscle cells, small airway epithelium, bronchial epithelium, microvascular dermal endothelial cells, microvascular lung endothelial cells, human pulmonary aortic endothelial cells, human umbilical vein endothelial cells were all purchased from Clonetics (Walkersville, MD) and grown in the media supplied for these cell types by Clonetics. These primary cell types were activated with various cytokines or combinations of cytokines for 6 and/or 12-14 hours, as indicated. The following cytokines were used; IL-1 beta at approximately 1-5 ng/ml, TNF alpha at approximately 5-10 ng/ml, IFN gamma at approximately 20-50 ng/ml, IL-4 at approximately 5-10 ng/ml, IL-9 at approximately 5-10 ng/ml, IL-13 at approximately 5-10 ng/ml. Endothelial cells were sometimes starved for various times by culture in the basal media from Clonetics with 0.1% serum.
Mononuclear cells were prepared from blood of employees at CuraGen Coφoration, using Ficoll. LAK cells were prepared from these cells by culture in DMEM 5% FCS (Hyclone), 100 μM non essential amino acids (Gibco/Life Technologies, Rockville, MD), 1 mM sodium pyruvate (Gibco), mercaptoethanol 5.5 x 10"5 M (Gibco), and 10 mM Hepes
(Gibco) and Interleukin 2 for 4-6 days. Cells were then either activated with 10-20 ng/ml PMA and 1-2 μg/ml ionomycin, IL-12 at 5-10 ng/ml, IFN gamma at 20-50 ng/ml and E -18 at 5-10 ng/ml for 6 hours. In some cases, mononuclear cells were cultured for 4-5 days in DMEM 5% FCS (Hyclone), 100 μM non essential amino acids (Gibco), 1 mM sodium pyruvate (Gibco), mercaptoethanol 5.5 x 10"5 M (Gibco), and 10 mM Hepes (Gibco) with PHA
(phytohemagglutinin) or PWM (pokeweed mitogen) at approximately 5 μg/ml. Samples were taken at 24, 48 and 72 hours for RNA preparation. MLR (mixed lymphocyte reaction) samples were obtained by taking blood from two donors, isolating the mononuclear cells using Ficoll and mixing the isolated mononuclear cells 1:1 at a final concentration of approximately 2x 106 cells/ml in DMEM 5% FCS (Hyclone), 100 μM non essential amino acids (Gibco), 1 mM sodium pyruvate (Gibco), mercaptoethanol (5.5 x 10"5 M) (Gibco), and 10 mM Hepes (Gibco). The MLR was cultured and samples taken at various time points ranging from 1- 7 days for RNA preparation.
Monocytes were isolated from mononuclear cells using CD 14 Miltenyi Beads, +ve VS selection columns and a Vario Magnet according to the manufacturer's instructions. Monocytes were differentiated into dendritic cells by culture in DMEM 5% fetal calf serum (FCS) (Hyclone, Logan, UT), 100 μM non essential amino acids (Gibco), 1 mM sodium pyruvate (Gibco), mercaptoethanol 5.5 x 10"5 M (Gibco), and 10 mM Hepes (Gibco), 50 ng/ml GMCSF and 5 ng/ml IL-4 for 5-7 days. Macrophages were prepared by culture of monocytes for 5-7 days in DMEM 5% FCS (Hyclone), 100 μM non essential amino acids (Gibco), 1 mM sodium pyruvate (Gibco), mercaptoethanol 5.5 x 10"5 M (Gibco), 10 mM Hepes (Gibco) and 10% AB Human Serum or MCSF at approximately 50 ng/ml. Monocytes, macrophages and dendritic cells were stimulated for 6 and 12-14 hours with lipopolysaccharide (LPS) at 100 ng/ml. Dendritic cells were also stimulated with anti-CD40 monoclonal antibody (Pharmingen) at 10 μg/ml for 6 and 12-14 hours.
CD4 lymphocytes, CD8 lymphocytes and NK cells were also isolated from mononuclear cells using CD4, CD8 and CD56 Miltenyi beads, positive VS selection columns and a Vario Magnet according to the manufacturer's instructions. CD45RA and CD45RO CD4 lymphocytes were isolated by depleting mononuclear cells of CD8, CD56, CD14 and CD19 cells using CD8, CD56, CD14 and CD19 Miltenyi beads and positive selection. Then CD45RO beads were used to isolate the CD45RO CD4 lymphocytes with the remaining cells being CD45RA CD4 lymphocytes. CD45RA CD4, CD45RO CD4 and CD8 lymphocytes were placed in DMEM 5% FCS (Hyclone), 100 μM non essential amino acids (Gibco), 1 mM sodium pyruvate (Gibco), mercaptoethanol 5.5 x 10"5 M (Gibco), and 10 mM Hepes (Gibco) and plated at 106 cells/ml onto Falcon 6 well tissue culture plates that had been coated overnight with 0.5 μg/ml anti-CD28 (Pharmingen) and 3 ug/ml anti-CD3 (OKT3, ATCC) in PBS. After 6 and 24 hours, the cells were harvested for RNA preparation. To prepare chronically activated CD8 lymphocytes, we activated the isolated CD8 lymphocytes for 4 days on anti-CD28 and anti-CD3 coated plates and then harvested the cells and expanded them in DMEM 5% FCS (Hyclone), 100 μM non essential amino acids (Gibco), 1 mM sodium pyruvate (Gibco), mercaptoethanol 5.5 x 10"5 M (Gibco), and 10 mM Hepes (Gibco) and IL-2. The expanded CD8 cells were then activated again with plate bound anti-CD3 and anti-CD28 for 4 days and expanded as before. RNA was isolated 6 and 24 hours after the second activation and after 4 days of the second expansion culture. The isolated NK cells were cultured in DMEM 5% FCS
(Hyclone), 100 μM non essential amino acids (Gibco), 1 mM sodium pyruvate (Gibco), mercaptoethanol 5.5 x 10"5 M (Gibco), and 10 mM Hepes (Gibco) and IL-2 for 4-6 days before RNA was prepared.
To obtain B cells, tonsils were procured from NDRI. The tonsil was cut up with sterile dissecting scissors and then passed through a sieve. Tonsil cells were then spun down and resupended at 106 cells/ml in DMEM 5% FCS (Hyclone), 100 μM non essential amino acids (Gibco), 1 mM sodium pyruvate (Gibco), mercaptoethanol 5.5 x 10"5 M (Gibco), and 10 mM Hepes (Gibco). To activate the cells, we used PWM at 5 μg/ml or anti-CD40 (Pharmingen) at approximately 10 μg/ml and IL-4 at 5-10 ng/ml. Cells were harvested for RNA preparation at 24,48 and 72 hours.
To prepare the primary and secondary Thl/Th2 and Trl cells, six-well Falcon plates were coated overnight with 10 μg/ml anti-CD28 (Pharmingen) and 2 μg/ml OKT3 (ATCC), and then washed twice with PBS. Umbilical cord blood CD4 lymphocytes (Poietic Systems,
5 6
German Town, MD) were cultured at 10 -10 cells/ml in DMEM 5% FCS (Hyclone), 100 μM non essential amino acids (Gibco), 1 mM sodium pyruvate (Gibco), mercaptoethanol 5.5 x 10"5 M (Gibco), 10 mM Hepes (Gibco) and IL-2 (4 ng/ml). IL-12 (5 ng/ml) and anti-IL4 (1 Dg/ml) were used to direct to Thl, while IL-4 (5 ng/ml) and anti-IFN gamma (1 Dg/ml) were used to direct to Th2 and IX- 10 at 5 ng/ml was used to direct to Trl . After 4-5 days, the activated Thl, Th2 and Trl lymphocytes were washed once in DMEM and expanded for 4-7 days in DMEM 5% FCS (Hyclone), 100 μM non essential amino acids (Gibco), 1 mM sodium pyruvate (Gibco), mercaptoethanol 5.5 x 10"5 M (Gibco), 10 mM Hepes (Gibco) and IL-2 (1 ng/ml). Following this, the activated Thl, Th2 and Trl lymphocytes were re-stimulated for 5 days with anti-CD28/OKT3 and cytokines as described above, but with the addition of anti-CD95L (1
Dg/ml) to prevent apoptosis. After 4-5 days, the Thl, Th2 and Trl lymphocytes were washed and then expanded again with IL-2 for 4-7 days. Activated Thl and Th2 lymphocytes were maintained in this way for a maximum of three cycles. RNA was prepared from primary and secondary Thl, Th2 and Trl after 6 and 24 hours following the second and third activations with plate bound anti-CD3 and anti-CD28 mAbs and 4 days into the second and third expansion cultures in Interleukin 2.
The following leukocyte cells lines were obtained from the ATCC: Ramos, EOL-1, KU- 812. EOL cells were further differentiated by culture in 0.1 mM dbcAMP at 5 xlO5 cells/ml for 8 days, changing the media every 3 days and adjusting the cell concentration to 5 xlO5 cells/ml. For the culture of these cells, we used DMEM or RPMI (as recommended by the ATCC), with the addition of 5% FCS (Hyclone), 100 μM non essential amino acids (Gibco), 1 mM sodium pyruvate (Gibco), mercaptoethanol 5.5 x 10"5 M (Gibco), 10 mM Hepes (Gibco). RNA was either prepared from resting cells or cells activated with PMA at 10 ng/ml and ionomycin at 1 μg/ml for 6 and 14 hours. Keratinocyte line CCD106 and an airway epithelial tumor line NCI- H292 were also obtained from the ATCC. Both were cultured in DMEM 5% FCS (Hyclone), 100 μM non essential amino acids (Gibco), 1 mM sodium pyruvate (Gibco), mercaptoethanol 5.5 x 10"5 M (Gibco), and 10 mM Hepes (Gibco). CCD1106 cells were activated for 6 and 14 hours with approximately 5 ng/ml TNF alpha and 1 ng/ml IL-1 beta, while NCI-H292 cells were activated for 6 and 14 hours with the following cytokines: 5 ng/ml IL-4, 5 ng/ml IL-9, 5 ng/ml IL-13 and 25 ng/ml IFN gamma.
For these cell lines and blood cells, RNA was prepared by lysing approximately 107 cells/ml using Trizol (Gibco BRL). Briefly, 1/10 volume of bromochloropropane (Molecular Research Coφoration) was added to the RNA sample, vortexed and after 10 minutes at room temperature, the tubes were spun at 14,000 rpm in a Sorvall SS34 rotor. The aqueous phase was removed and placed in a 15 ml Falcon Tube. An equal volume of isopropanol was added and left at -20 degrees C overnight. The precipitated RNA was spun down at 9,000 rpm for 15 min in a Sorvall SS34 rotor and washed in 70% ethanol. The pellet was redissolved in 300 μl of RNAse-free water and 35 μl buffer (Promega) 5 μl DTT, 7 μl RNAsin and 8 μl DNAse were added. The tube was incubated at 37 degrees C for 30 minutes to remove contaminating genomic DNA, extracted once with phenol chloroform and re- precipitated with 1/10 volume of 3 M sodium acetate and 2 volumes of 100% ethanol. The RNA was spun down and placed in RNAse free water. RNA was stored at -80 degrees C.
GPCRla
Expression of gene GPCRla was assessed using the primer-probe set Agl 151, described in Table 10. Results of the RTQ-PCR runs are shown in Tables 11 and 12.
PAGE INTENTIONALLY LEFT BLANK
Table 10. Probe Name: Agll51
Table 11. Panel 1.3D
Table 12. Panel 4D
Panel 1.3D Summary: All samples show expression of gene AP001884_A at very low to undetectable levels (Ct values >35).
Panel 4D Summary: Of three replicate experiments, one run was not evaluated (and no data is shown) due to a poor amplification plot. The other two runs show low to undetectable levels of expression, except in the IBD colitis 1 sample, which is likely due to genomic DNA contamination.
GPCR2a
Expression of gene GPCR2a was assessed using the primer-probe set Agl 118, described in Table 13.
Table 13. Probe Name: Aglll8
Reverse 5'-CAGACGATGGACACATAAGACA-3* 58.7 22 714 71
Expression of gene GPCR2a was low/undetectable (Ct values >35) in panels 1.2 and
4D.
GPCR3a
Expression of gene GPCR3a was assessed using the primer-probe set Agl644, described in Table 14. Results of the RTQ-PCR runs are shown in Table 15.
Table 14. Probe Name: Agl 644
Table 15. Panels 4D and 4.1D
Panel 1.3D Summary: Expression of geneGPCR3a is low/undetectable (Ct calues >35) in this panel (data not shown).
Panel 4D Summary: Expression of gene GPCR3a seen in colitis 1 and 2 but not in normal colon. The protein encoded for by this transcript may be expressed by the normal tissue within the colon in response to inflammation or by the infiltrating leukocytes. It may be important in cell extravasation, activation or signal transduction. Protein therapeutics designed from this GPCR could reduce or inhibit inflammation due to colitis.
Panel 4. ID Summary: The only detectable expression in this panel is in the kidney (Ct = 33.1). These results do not correlate with expression data from either panel 4D or 1.3D and therefore require replication before any conclusions can be drawn.
GPCR4
Expression of gene GPCR4 was assessed using the primer-probe set Agl 152, described in Table 16. Results of the RTQ-PCR runs are shown in Table 17.
Table 16. Probe Name: Agll52
Table 17. Panels 1.2 and 4D
Panel 1.2 Summary: The expression of GPCR4 is highest in the adipose, probably as a result of genomic DNA contamination. Expression of GPCR4 is moderate in colorectal tissues and the cerebral cortex and low in fetal, kidney and the prostate tissues. Expression in disease tissue is high in certain samples of colon, lung and ovarian cancer. Therefore antibodies against this protein may be used as a therapeutic in these types of cancers.
Panel 4D Summary: Expression of GPCR4 is at detectable levels only in the IBD colitis 1 sample, probably due to genomic DNA contammation.
GPCR5b
Expression of gene GPCR5b was assessed using the primer-probe set Ag2370, described in Table 18. Results of the RTQ-PCR runs are shown in Table 19.
Table 18. Probe Name: Ag2370
Table 19. Panel 4D
Panel 4D Summary: The gene GPCR5b is highly expressed in dermal fibroblasts treated with IFN- gamma or IL-4 as against down regulation in IL-l/TNF treated fibroblasts. This result indicates that the expression of GPCR5b is stimulus-specific. Thus GPCR5b may play an important role in signal transduction, upregulation of adhesion molecules or induction of cytokine release during inflammation. Antibody, antisense or small molecule therapeutics designed against the antigen encoded for by this transcript could reduce or inhibit inflammation and may be important for the treatment of diseases such as psoriasis or delayed type hypersensitivity.
GPCR5c
Expression of gene GPCR5c was assessed using the primer-probe set Agl 645, described in Table 20. Results of the RTQ-PCR runs are shown in Tables 21 and 22.
Table 20. Probe Name: Agl 645
Table 21. Panel 1.3D
Table 22. Panel 4D
Panel 1.3D Summary: Expression levels of gene GPCR5c are highest in the spleen, with lower levels in the amygdala region of the brain. Expression in other samples was undetectable. This gene may be involved in B lymphocyte maturation and amygdalar functions such as appetite control and fear conditioning.
Panel 4D Summary: High expression of the GPCR5c gene is seen in colitis 1. The protein encoded for by this antigen may be important in the inflammatory process. Antagonistic antibodies or small molecule therapeutics may reduce or inhibit inflammation in the bowel due to inflammatory bowel disease.
GPCRόa
Expression of gene GPCR6a was assessed using the primer-probe set Agl 646, described in Table 23.
Table 23. Probe Name: Agl 646
There was no detectable expression using Agl 646 in panels 1.3D or 4D.
GPCRόd
Expression of gene GPCR6d was assessed using the primer-probe sets Ag2373 and Ag2498 (identical sequences), described in Table 24. Results of the RTQ-PCR runs are shown in Table 25.
Table 24. Probe Name: Ag2373/Ag2498
Table 25. Panel 4D
Panel 4D Summary: Runs with the two identical probe/primer pairs do not correlate well with one another. Expression of GPCRόd with probe Ag2373 shows undetectable expression in all samples with Ct values >35. Expression of GPCRόd with probe Ag 2498 is highly expressed in the thymus and in primary regulatory T cells (Trl). Thus the gene GPCRόd may encode a receptor involved in differentiation, activation or the regulatory activity of T cells. Antagonistic or agonistic antibodies, small molecule or protein therapeutics may be able to regulate immune responses and be important for organ transplant (antagonistic) and cancer therapeutics (agonistic).
Expression of gene GPCRόd was low to undetectable (Ct values >35) in panel 1.3D (data not shown).
GPCR7
Expression of gene GPCR7 was assessed using the primer-probe set Agl 647, described in Table 26. Results of the RTQ-PCR runs are shown in Table 27.
Table 26. Probe Name: Agl 647
Panel 4D Summary: Expression of gene GPCR7 is seen in three tissues - normal thymus, cirrhotic liver and peripheral blood monocytes (PBMCs) treated with LPS. This transcript is not expressed in normal liver tissue (panel 1.3D, data not shown) and antibodies or small molecule therapies may reduce inflammation and tissue destruction associated with cirrhosis.
Expression of GPCR7 was low to undetectable (Ct>35) on panel 1.3D.
GPCR8e
Expression of gene GPCR8e was assessed using the primer-probe set Agl 648, described in Table 28. Results of the RTQ-PCR runs are shown in Tables 29 and 30.
Table 28. Probe Name: Agl 648
Table 29. Panel 1.3D
Table 30. Panel 4D
Panel 1.3D Summary: Gene GPCR8e is highly expressed by spleen among normal tissues. In addition, it is seen in the ovarian cancer cell line SK-OV3. GPCR8e may be involved in B lymphocyte maturation. Small molecule therapeutics or antibody therapeutics designed to the protein encoded by this gene could be useful in certain kinds of cancer.
Panel 4D Summary: GPCR8e shows high expression of the transcript in colitis 1. The protein encoded for by this antigen may be important in the inflammatory process. Antagonistic antibodies or small molecule therapeutics may reduce or inhibit inflammation in the bowel resulting from inflammatory bowel disease.
EQUIVALENTS
Although particular embodiments have been disclosed herein in detail, this has been done by way of example for purposes of illustration only, and is not intended to be limiting with respect to the scope of the appended claims, which follow. In particular, it is contemplated by the inventors that various substitutions, alterations, and modifications may be made to the invention without departing from the spirit and scope of the invention as defined by the claims. The choice of nucleic acid starting material, clone of interest, or library type is believed to be a matter of routine for a person of ordinary skill in the art with knowledge of the embodiments described herein. Other aspects, advantages, and modifications considered to be within the scope of the following claims.