English | Chinese
This is the official repository for IEPile: Unearthing Large-Scale Schema-Based Information Extraction Corpus
Datasets | Paper | Usage | Limitations | Statement & License | Citation
Please note that our IEPile may undergo updates (we will inform you upon their release). It is recommended to utilize the most current version.
- IEPile: A Large-Scale Information Extraction Corpus
- [2024/05] The paper IEPile: Unearthing Large-Scale Schema-Based Information Extraction Corpus is accepted by ACL 2024 main conference.
- [2024/04] We release a new bilingual (Chinese and English) schema-based information extraction model called OneKE based on Chinese-Alpaca-2-13B.
- [2024/02] We release a large-scale (0.32B tokens) high-quality bilingual (Chinese and English) Information Extraction (IE) instruction dataset named IEPile, along with two models trained with
IEPile, baichuan2-13b-iepile-lora and llama2-13b-iepile-lora. - [2023/10] We released a new bilingual (Chinese and English) theme-based Information Extraction (IE) instruction dataset named InstructIE with paper.
- [2023/08] We introduced a dedicated 13B model for Information Extraction (IE), named knowlm-13b-ie.
- [2023/05] We initiated an instruction-based Information Extraction project.
IEPile dataset download links: Google Drive | Hugging Face | WiseModel | ModelScpoe
Please be aware that the data contained in the dataset links provided above has already excluded any part related to the ACE2005 dataset. Should you require access to the unfiltered, complete dataset and have successfully obtained the necessary permissions, please do not hesitate to contact us via email at [email protected] or [email protected]. We will provide the complete dataset resources for your use.
Model download links for LLaMA2-IEPile | Baichuan2-IEPile | OneKE: zjunlp/llama2-13b-iepile-lora | zjunlp/baichuan2-13b-iepile-lora | zjunlp/OneKE
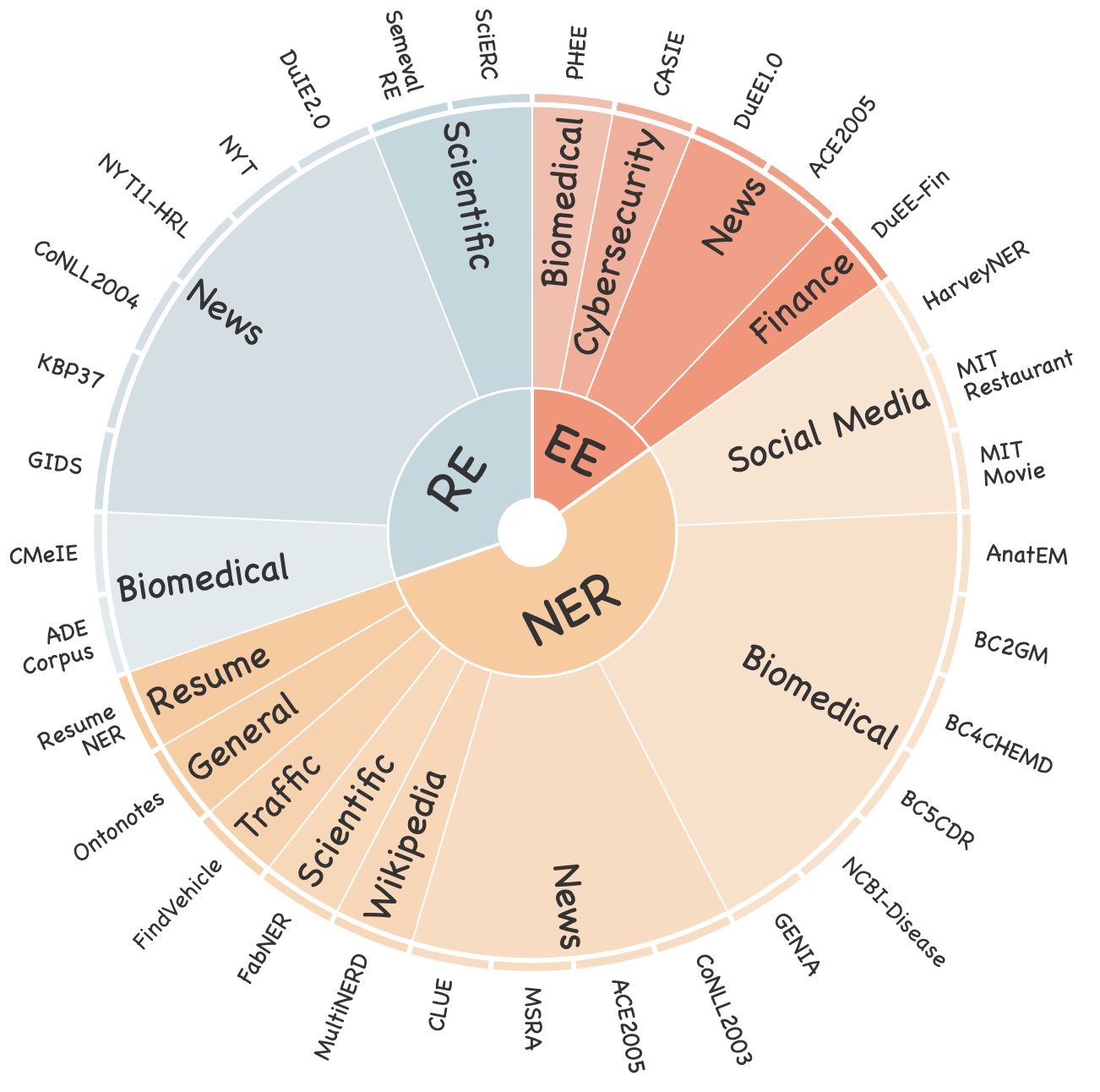
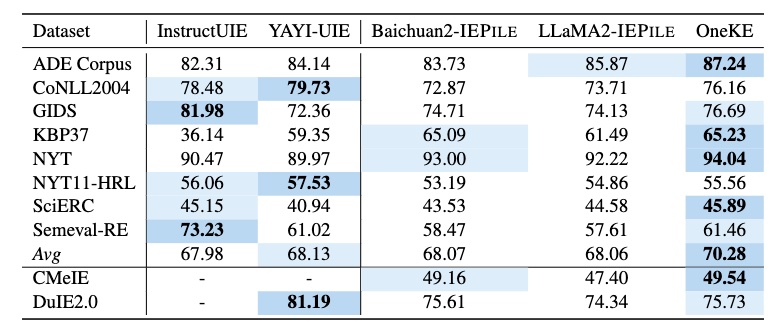
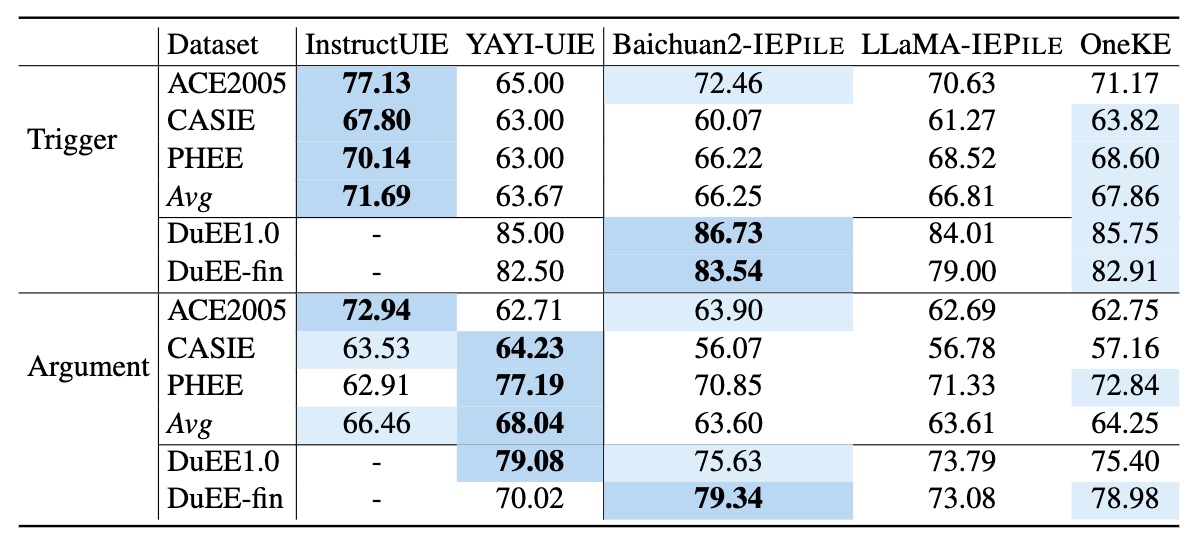
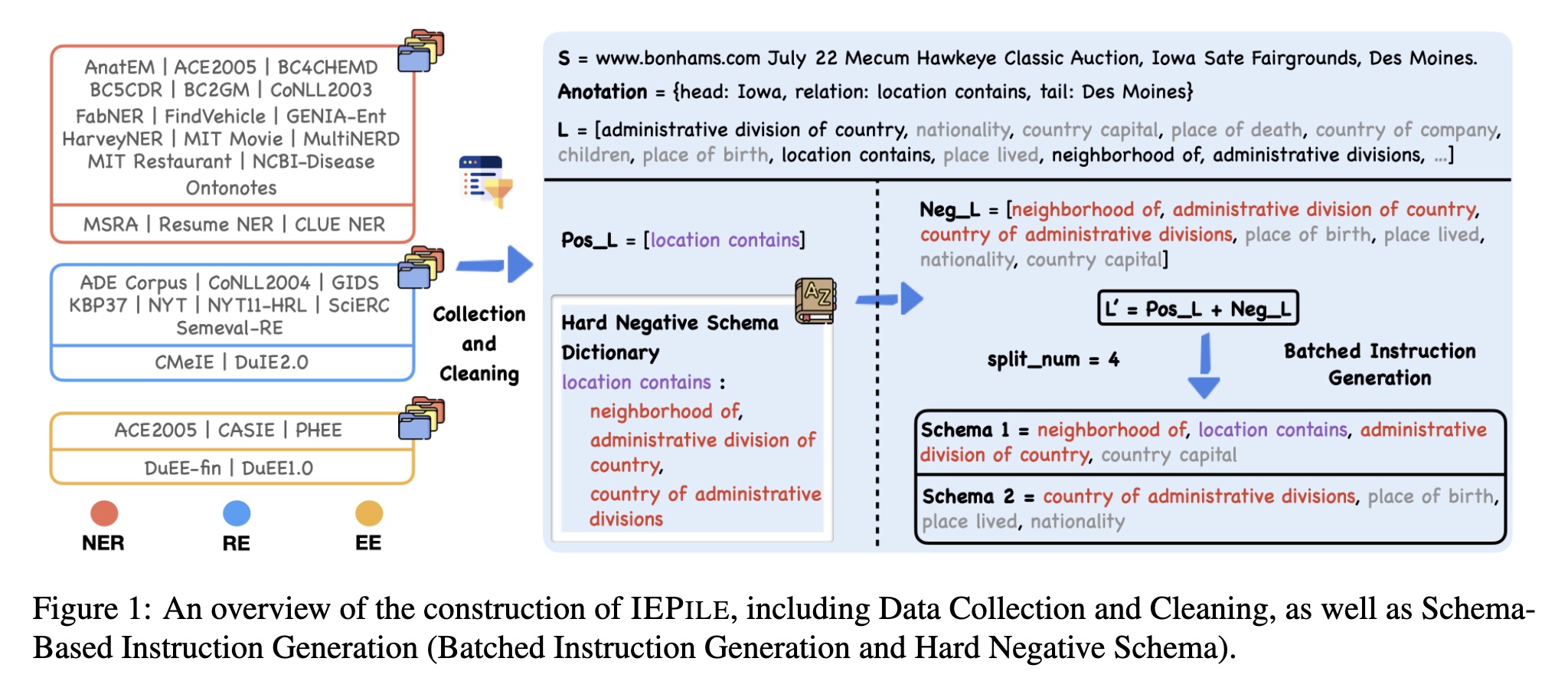
We have collected and cleaned existing Information Extraction (IE) datasets, integrating a total of 26 English IE datasets and 7 Chinese IE datasets. As shown in the Figure, these datasets cover multiple domains including general, medical, financial, and others.
In this study, we adopted the proposed "schema-based batched instruction generation strategy" to create a large-scale, high-quality, bilingual (Chinese and English) IE instruction tuning dataset named IEPile, containing approximately 0.32B tokens.
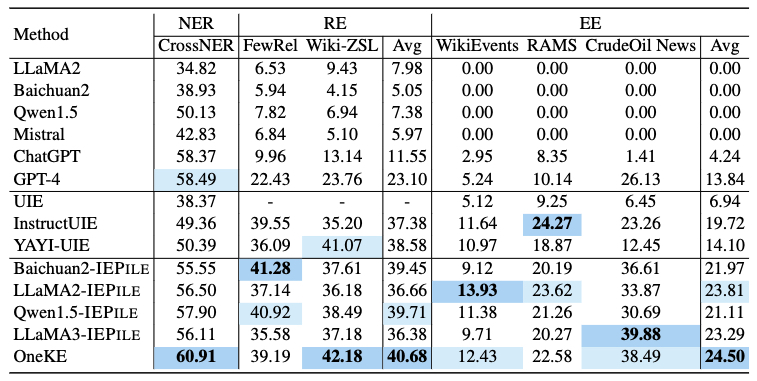
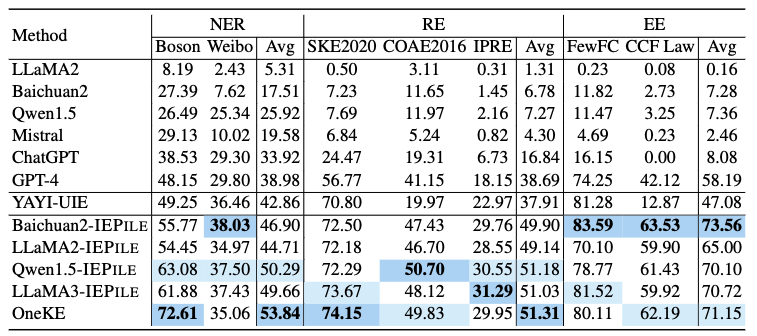
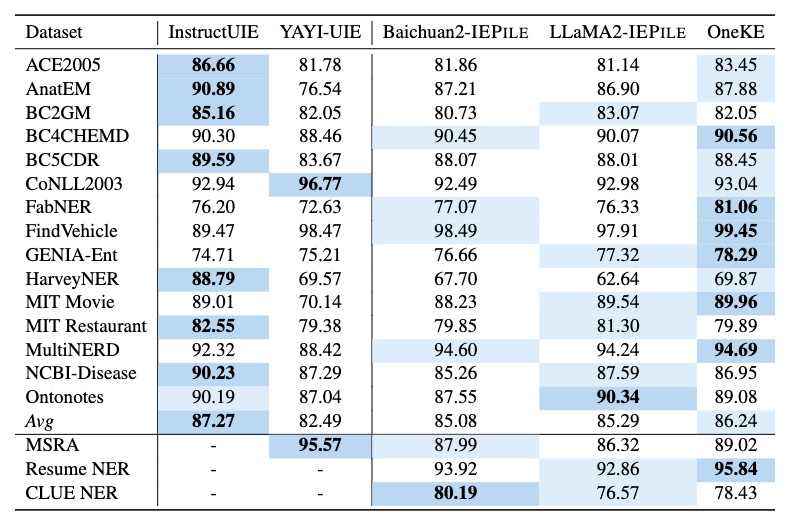
Based on IEPile, we fine-tuned the Baichuan2-13B-Chat and LLaMA2-13B-Chat models using the Lora technique. Experiments have demonstrated that the fine-tuned Baichuan2-IEPile and LLaMA2-IEPile models perform remarkably on fully supervised training sets and have achieved improvements in zero-shot information extraction tasks.
We concentrate on instruction-based IE, thus the construction of schema within the instructions is crucial. This is because they reflect the specific extraction requirements and are dynamically variable. Previous approaches with existing IE datasets often employ a rather extensive schema processing strategy when constructing instructions, utilizing all schemas within a label set for instruction building, raising two potential issues:
- Inconsistency in the number of schema queries within instruction between training and evaluation. For example, the model's performance will decrease if it is trained on about 20 schema queries but tested with either 10 or 30, even if the training and evaluation schemas are similar in content.
- Inadequate differentiation among schemas in the instructions. For example, semantically similar schemas like "layoffs", "depart" and "dismissals", may present co-occurrence ambiguities that could confuse the LLMs. Such schemas should co-occur more frequently within the instruction.
Therefore, we introduce the following solutions: 1)Hard Negative Schema; and 2) Batched Instruction Generation.
Hard Negative Schema
Assuming that dataset
Batched Instruction Generation
Subsequently, we obtain the final schema set
Each instance in IEPile contains four fields: task, source, instruction, and output.
Below is a data example:
{
"task": "NER",
"source": "CoNLL2003",
"instruction": "{\"instruction\": \"You are an expert in named entity recognition. Please extract entities that match the schema definition from the input. Return an empty list if the entity type does not exist. Please respond in the format of a JSON string.\", \"schema\": [\"person\", \"organization\", \"else\", \"location\"], \"input\": \"284 Robert Allenby ( Australia ) 69 71 71 73 , Miguel Angel Martin ( Spain ) 75 70 71 68 ( Allenby won at first play-off hole )\"}",
"output": "{\"person\": [\"Robert Allenby\", \"Allenby\", \"Miguel Angel Martin\"], \"organization\": [], \"else\": [], \"location\": [\"Australia\", \"Spain\"]}"
}The data instance belongs to the NER task, is part of the CoNLL2003 dataset, the schema list to be extracted includes ["person", "organization", "else", "location"], and the text to be extracted from is "284 Robert Allenby ( Australia ) 69 71 71 73 , Miguel Angel Martin ( Spain ) 75 70 71 68 ( Allenby won at first play-off hole )". The output is {"person": ["Robert Allenby", "Allenby", "Miguel Angel Martin"], "organization": [], "else": [], "location": ["Australia", "Spain"]}.
Note that the order of schemas in the output is consistent with the order in the instruction.
More Tasks Instance
{
"task": "EE",
"source": "PHEE",
"instruction": "{\"instruction\": \"You are an expert in event extraction. Please extract events from the input that conform to the schema definition. Return an empty list for events that do not exist, and return NAN for arguments that do not exist. If an argument has multiple values, please return a list. Respond in the format of a JSON string.\", \"schema\": [{\"event_type\": \"potential therapeutic event\", \"trigger\": true, \"arguments\": [\"Treatment.Time_elapsed\", \"Treatment.Route\", \"Treatment.Freq\", \"Treatment\", \"Subject.Race\", \"Treatment.Disorder\", \"Effect\", \"Subject.Age\", \"Combination.Drug\", \"Treatment.Duration\", \"Subject.Population\", \"Subject.Disorder\", \"Treatment.Dosage\", \"Treatment.Drug\"]}, {\"event_type\": \"adverse event\", \"trigger\": true, \"arguments\": [\"Subject.Population\", \"Subject.Age\", \"Effect\", \"Treatment.Drug\", \"Treatment.Dosage\", \"Treatment.Freq\", \"Subject.Gender\", \"Treatment.Disorder\", \"Subject\", \"Treatment\", \"Treatment.Time_elapsed\", \"Treatment.Duration\", \"Subject.Disorder\", \"Subject.Race\", \"Combination.Drug\"]}], \"input\": \"Our findings reveal that even in patients without a history of seizures, pregabalin can cause a cortical negative myoclonus.\"}",
"output": "{\"potential therapeutic event\": [], \"adverse event\": [{\"trigger\": \"cause \", \"arguments\": {\"Subject.Population\": \"NAN\", \"Subject.Age\": \"NAN\", \"Effect\": \"cortical negative myoclonus\", \"Treatment.Drug\": \"pregabalin\", \"Treatment.Dosage\": \"NAN\", \"Treatment.Freq\": \"NAN\", \"Subject.Gender\": \"NAN\", \"Treatment.Disorder\": \"NAN\", \"Subject\": \"patients without a history of seizures\", \"Treatment\": \"pregabalin\", \"Treatment.Time_elapsed\": \"NAN\", \"Treatment.Duration\": \"NAN\", \"Subject.Disorder\": \"NAN\", \"Subject.Race\": \"NAN\", \"Combination.Drug\": \"NAN\"}}]}"
}
{
"task": "RE",
"source": "NYT11",
"instruction": "{\"instruction\": \"You are an expert in relationship extraction. Please extract relationship triples that match the schema definition from the input. Return an empty list for relationships that do not exist. Please respond in the format of a JSON string.\", \"schema\": [\"neighborhood of\", \"nationality\", \"children\", \"place of death\"], \"input\": \" In the way New Jersey students know that Thomas Edison 's laboratory is in West Orange , the people of Colma know that Wyatt Earp 's ashes are buried at Hills of Eternity , a Jewish cemetery he was n't ; his wife was , and that Joe DiMaggio is at Holy Cross Cemetery , where visitors often lean bats against his gravestone . \"}",
"output": "{\"neighborhood of\": [], \"nationality\": [], \"children\": [], \"place of death\": [{\"subject\": \"Thomas Edison\", \"object\": \"West Orange\"}]}"
}Below are the explanations for each field:
| Field | Description |
|---|---|
| task | The task to which the instance belongs, one of the five types (NER, RE, EE, EET, EEA). |
| source | The dataset to which the instance belongs. |
| instruction | The instruction for inputting into the model, processed into a JSON string via json.dumps, including three parts: "instruction", "schema", and "input". |
| output | The output in the format of a dictionary's JSON string, where the key is the schema, and the value is the extracted content. |
In IEPile, the instruction format of IEPile adopts a JSON-like string structure, which is essentially a dictionary-type string composed of the following three main components:
(1) 'instruction': Task description, which outlines the task to be performed by the instruction (one of NER, RE, EE, EET, EEA).
(2) 'schema': A list of schemas to be extracted (entity types, relation types, event types).
(3) 'input': The text from which information is to be extracted.
The file instruction.py provides instructions for various tasks.
Before you begin, make sure to create an appropriate virtual environment following the instructions below:
conda create -n IEPile python=3.9 # Create a virtual environment
conda activate IEPile # Activate the environment
pip install -r requirements.txt # Install dependenciesIEPile dataset download links: Google Drive | Hugging Face
IEPile
├── train.json # Training set
└── dev.json # Validation setHere are some of the models supported by the code in this repository: [llama, alpaca, vicuna, zhixi, falcon, baichuan, chatglm, qwen, moss, openba]
mkdir data # Put data here
mkdir models # Put base models here
mkdir results # Put prediction results here
mkdir lora # Put LoRA fine-tuning results hereData should be placed in the ./data directory.
Important Note: All the commands below should be executed within the
IEPiledirectory. For example, if you want to run the fine-tuning script, you should use the following command:bash ft_scripts/fine_llama.bash. Please ensure your current working directory is correct. Please make sure that each entry in the training/validation files includes theinstruction,outputfields.
output_dir='lora/llama2-13b-chat-v1'
mkdir -p ${output_dir}
CUDA_VISIBLE_DEVICES="0,1,2,3" torchrun --nproc_per_node=4 --master_port=1287 src/test_finetune.py \
--do_train --do_eval \
--overwrite_output_dir \
--model_name_or_path 'models/llama2-13b-chat' \
--stage 'sft' \
--model_name 'llama' \
--template 'llama2' \
--train_file 'data/train.json' \
--valid_file 'data/dev.json' \
--output_dir=${output_dir} \
--per_device_train_batch_size 2 \
--per_device_eval_batch_size 2 \
--gradient_accumulation_steps 4 \
--preprocessing_num_workers 16 \
--num_train_epochs 10 \
--learning_rate 5e-5 \
--max_grad_norm 0.5 \
--optim "adamw_torch" \
--max_source_length 400 \
--cutoff_len 700 \
--max_target_length 300 \
--evaluation_strategy "epoch" \
--save_strategy "epoch" \
--save_total_limit 10 \
--lora_r 16 \
--lora_alpha 32 \
--lora_dropout 0.05 \
--bf16 CUDA_VISIBLE_DEVICES="0,1,2,3": used to specify which GPUs are available for the current training task. In this case, "0,1,2,3" means that the four GPUs with IDs 0, 1, 2, and 3 are being utilized. If your machine is equipped with more than four GPUs, this setting allows you to select any four of them for use.--nproc_per_node=4: specifies the number of processes to be launched on each node. Since four GPUs have been specified in this example, it is necessary to start four separate processes, with each process corresponding to one GPU.- For training tasks that use only a single GPU, the command
CUDA_VISIBLE_DEVICES=0 python src/finetune.pycan be used to initiate the training. Here, CUDA_VISIBLE_DEVICES=0 designates GPU number 0 for this training task. model_name: Specifies the name of the model architecture you want to use (7B, 13B, Base, Chat belong to the same model architecture). Currently supported models include: ["llama", "alpaca", "vicuna", "zhixi", "falcon", "baichuan", "chatglm", "qwen", "moss", "openba"]. Please note, this parameter should be distinguished from--model_name_or_path.model_name_or_path: Model path, please download the corresponding model from HuggingFace.template: The name of the template used, including:alpaca,baichuan,baichuan2,chatglm3, etc. Refer to src/datamodule/template.py to see all supported template names. The default is thealpacatemplate. ForChatversions of models, it is recommended to use the matching template, whileBaseversion models can default to usingalpaca.train_file,valid_file (optional): The file paths for the training set and the validation set, respectively. Note: Only JSON format files are currently supported.⚠️ Ifvalid_fileis not specified, a subset ofval_set_sizeentries will be automatically allocated fromtrain_fileto serve as the validation set.output_dir: The path to save the weight parameters after LoRA fine-tuning.val_set_size: The number of samples in the validation set, default is 1000.per_device_train_batch_size,per_device_eval_batch_size: Thebatch_sizeon each GPU device, adjust according to the size of the memory. For RTX3090, it is recommended to set between 2 and 4.max_source_length,max_target_length,cutoff_len: The maximum input and output lengths, and the cutoff length, which can simply be considered as the maximum input length + maximum output length. Set appropriate values according to specific needs and memory size.- If running out of GPU memory occurs when saving the model after the evaluation phase, please set
evaluation_strategytono.
Quantization can be performed by setting bits to 4; it is recommended for the RTX3090.
To learn more about parameter configuration, please refer to the src/utils/args.
The specific script for fine-tuning the LLaMA2-13B-Chat model can be found in ft_scripts/fine_llama.bash.
The specific script for fine-tuning the Baichuan2-13B-Chat model can be found in ft_scripts/fine_baichuan.bash.bash.
Although the Baichuan2-IEPile and LLaMA2-IEPile models have undergone extensive instruction fine-tuning on multiple general datasets and thus possess a degree of general information extraction capability, they may still exhibit certain limitations when processing data in specific domains (such as law, education, science, telecommunications). To address this challenge, it is recommended to conduct secondary training of these models on datasets specific to these domains. This will help the models better adapt to the semantic and structural characteristics of the specific domains, enhancing their information extraction capability within those domains.
Firstly, it's necessary to format the data to include instruction and output fields. For this purpose, we provide a script convert_func.py, which can batch convert data into a format that can be directly used by the model.
Before using the convert_func.py script, please make sure to refer to the data directory. This directory provides detailed instructions on the data format required for each task. Refer to
sample.jsonto understand the format of the data before conversion,schema.jsonto see the organization of the schema, andtrain.jsonto describe the data format after conversion.
Additionally, you can directly use the bilingual (Chinese and English) information extraction dataset zjunlp/InstructIE, which includes 12 themes such as characters, vehicles, works of art, natural science, man-made objects, astronomical objects, etc.
python ie2instruction/convert_func.py \
--src_path data/NER/sample.json \
--tgt_path data/NER/train.json \
--schema_path data/NER/schema.json \
--language zh \
--task NER \
--split_num 6 \
--random_sort \
--split trainlanguage: Supports two languages,zh(Chinese) anden(English), with different instruction templates used for each language.task: Currently supports five types of tasks: ['RE', 'NER', 'EE', 'EET', 'EEA'].split_num: Defines the maximum number of schemas that can be included in a single instruction. The default value is 4, and setting it to -1 means no splitting is done. The recommended number of task splits varies by task: 6 for NER, and 4 for RE, EE, EET, EEA.random_sort: Whether to randomize the order of schemas in the instructions. The default is False, which means schemas are sorted alphabetically.split: Specifies the type of dataset, with optionstrainortest.
The converted training data will contain four fields: task, source, instruction, output.
Generation of Hard Negative Samples: Promote co-occurrence of semantically close and easily confused schemas, reducing the amount of training samples.
python ie2instruction/convert_func.py \
--src_path data/SPO/sample.json \
--tgt_path data/SPO/train.json \
--schema_path data/SPO/schema.json \
--cluster_mode \
--hard_negative_path data/hard_negative/SPO_DuIE2.0.json \
--language zh \
--task SPO \
--split_num 4 \
--random_sort \
--split trainThe addition of the --cluster_mode and --hard_negative_path data/hard_negative/SPO_DuIE2.0.json parameters, where --hard_negative_path corresponds to the dictionary of difficult negative samples. The hard_dict.json contains dictionaries of hard negative samples for all datasets involved in IEPILE.
Model download links for LLaMA2-IEPile | Baichuan2-IEPile | LLaMA3-IEPile | Qwen1.5-IEPile | OneKE: zjunlp/llama2-13b-iepile-lora | zjunlp/baichuan2-13b-iepile-lora | zjunlp/llama3-8b-iepile-lora | zjunlp/qwen1.5-14b-iepile-lora | zjunlp/OneKE
| checkpoint_dir | model_name_or_path | moadel_name | fp16/bf16 | template |
|---|---|---|---|---|
| llama2-13b-iepile-lora | LLaMA2-13B-Chat | llama | bf16 | llama2 |
| baichuan2-13b-iepile-lora | BaiChuan2-13B-Chat | baichuan | bf16 | baichuan2 |
| llama3-8b-iepile-lora | LLaMA3-8B-Instruct | llama | bf16 | alpaca |
| qwen1.5-14b-iepile-lora | Qwen1.5-14B-Chat | qwen2 | bf16 | qwen |
| OneKE | OneKE | llama | bf16 | llama2_zh |
output_dir='lora/llama2-13b-chat-v1-continue'
mkdir -p ${output_dir}
CUDA_VISIBLE_DEVICES="0,1,2,3" torchrun --nproc_per_node=4 --master_port=1287 src/test_finetune.py \
--do_train --do_eval \
--overwrite_output_dir \
--model_name_or_path 'models/llama2-13B-Chat' \
--checkpoint_dir 'zjunlp/llama2-13b-iepile-lora' \
--stage 'sft' \
--model_name 'llama' \
--template 'llama2' \
--train_file 'data/train.json' \
--valid_file 'data/dev.json' \
--output_dir=${output_dir} \
--per_device_train_batch_size 2 \
--per_device_eval_batch_size 2 \
--gradient_accumulation_steps 4 \
--preprocessing_num_workers 16 \
--num_train_epochs 10 \
--learning_rate 5e-5 \
--max_grad_norm 0.5 \
--optim "adamw_torch" \
--max_source_length 400 \
--cutoff_len 700 \
--max_target_length 300 \
--evaluation_strategy "epoch" \
--save_strategy "epoch" \
--save_total_limit 10 \
--lora_r 64 \
--lora_alpha 64 \
--lora_dropout 0.05 \
--bf16 - Please refer to the 3.3LoRA Fine-tuning for further parameter description.
- To continue training based on the fine-tuned LoRA weights, simply point the
--checkpoint_dirparameter to the path of the LoRA weights, for example by setting it to'zjunlp/llama2-13b-iepile-lora'.
Quantization can be performed by setting bits to 4; it is recommended for the RTX3090.
Please note that when using
LLaMA2-IEPileorBaichuan2-IEPile, keep both lora_r and lora_alpha at 64. We do not provide recommended settings for these parameters.
- To continue training based on the fine-tuned model weights, just set the
--model_name_or_pathparameter to the path of the weights, such as'zjunlp/KnowLM-IE-v2', without setting--checkpoint_dir.
The script can be found at ft_scripts/fine_continue.bash.
output_dir='lora/OneKE-continue'
mkdir -p ${output_dir}
CUDA_VISIBLE_DEVICES="0,1,2,3" torchrun --nproc_per_node=4 --master_port=1287 src/test_finetune.py \
--do_train --do_eval \
--overwrite_output_dir \
--model_name_or_path 'models/OneKE' \
--stage 'sft' \
--model_name 'llama' \
--template 'llama2_zh' \
--train_file 'data/train.json' \
--valid_file 'data/dev.json' \
--output_dir=${output_dir} \
--per_device_train_batch_size 2 \
--per_device_eval_batch_size 2 \
--gradient_accumulation_steps 4 \
--preprocessing_num_workers 16 \
--num_train_epochs 10 \
--learning_rate 5e-5 \
--max_grad_norm 0.5 \
--optim "adamw_torch" \
--max_source_length 400 \
--cutoff_len 700 \
--max_target_length 300 \
--evaluation_strategy "epoch" \
--save_strategy "epoch" \
--save_total_limit 10 \
--bf16 output_dir='lora/OneKE-continue-lora'
mkdir -p ${output_dir}
CUDA_VISIBLE_DEVICES="0,1,2,3" torchrun --nproc_per_node=4 --master_port=1287 src/test_finetune.py \
--do_train --do_eval \
--overwrite_output_dir \
--model_name_or_path 'models/OneKE' \
--stage 'sft' \
--model_name 'llama' \
--template 'llama2_zh' \
--train_file 'data/train.json' \
--valid_file 'data/dev.json' \
--output_dir=${output_dir} \
--per_device_train_batch_size 2 \
--per_device_eval_batch_size 2 \
--gradient_accumulation_steps 4 \
--preprocessing_num_workers 16 \
--num_train_epochs 10 \
--learning_rate 5e-5 \
--max_grad_norm 0.5 \
--optim "adamw_torch" \
--max_source_length 400 \
--cutoff_len 700 \
--max_target_length 300 \
--evaluation_strategy "epoch" \
--save_strategy "epoch" \
--save_total_limit 10 \
--lora_r 64 \
--lora_alpha 64 \
--lora_dropout 0.05 \
--bf16 Before preparing the test data conversion, please visit the data directory to understand the data structure required for each task: 1) For the input data format, see sample.json. 2) For the schema format, please refer to schema.json. 3) For the format of the transformed data, refer to train.json. Unlike training data, test data input does not need to include annotation fields (entity, relation, event).
python ie2instruction/convert_func.py \
--src_path data/NER/sample.json \
--tgt_path data/NER/test.json \
--schema_path data/NER/schema.json \
--language zh \
--task NER \
--split_num 6 \
--split testWhen setting split to test, select the appropriate number of schemas according to the task type: 6 is recommended for NER, while 4 is recommended for RE, EE, EET, EEA. The transformed test data will contain five fields: id, task, source, instruction, label.
The label field will be used for subsequent evaluation. If the input data lacks the annotation fields (entity, relation, event), the transformed test data will not contain the label field, which is suitable for scenarios where no original annotated data is available.
Download the IEPile dataset from Google Drive | Hugging Face | WiseModel | ModelScpoe
The file tree is shown as follows:
IEPile
├── train.json # Training Set
├── dev.json # Validation Set
├── IE-en # English Unified Format Data
│ ├── NER
│ │ ├── CoNLL2003
│ │ │ ├── train.json
│ │ │ ├── dev.json
│ │ │ ├── schema.json # schema information file
│ │ │ └── test.json
│ │ ├── ...
│ ├── RE
│ ├── EE
│ ├── EET
│ ├── EEA
├── IE-zh # Chinese Unified Format Data
│ ├── NER
│ ├── RE
│ ├── EE
│ ├── EET
│ ├── EEABatch test instruction data can be obtained through the following script:
bash ie2instruction/eval_data_convert.bashYou need to set the dir_path in the first line of the script to the actual absolute path of the IEPile dataset. Note: Due to the possible inconsistency in the order of labels in the converted schema sequence, there may be slight deviations in the evaluation results.
Model download links for LLaMA2-IEPile | Baichuan2-IEPile : zjunlp/llama2-13b-iepile-lora | zjunlp/baichuan2-13b-iepile-lora
| checkpoint_dir | model_name_or_path | moadel_name | fp16/bf16 | template |
|---|---|---|---|---|
| llama2-13b-iepile-lora | LLaMA2-13B-Chat | llama | bf16 | llama2 |
| baichuan2-13b-iepile-lora | BaiChuan2-13B-Chat | baichuan | bf16 | baichuan2 |
| llama3-8b-iepile-lora | LLaMA3-8B-Instruct | llama | bf16 | alpaca |
| qwen1.5-14b-iepile-lora | Qwen1.5-14B-Chat | qwen2 | bf16 | qwen |
baichuan2-13b-iepile-lora (specified with --checkpoint_dir), you must also download BaiChuan2-13B-Chat (specified with --model_name_or_path). 🚫You cannot merely set --model_name_or_path lora/baichuan2-13b-iepile-lora.
CUDA_VISIBLE_DEVICES=0 python src/inference.py \
--stage sft \
--model_name_or_path 'models/llama2-13B-Chat' \
--checkpoint_dir 'lora/llama2-13b-IEPile-lora' \
--model_name 'llama' \
--template 'llama2' \
--do_predict \
--input_file 'data/NER/test.json' \
--output_file 'results/llama2-13b-IEPile-lora_output.json' \
--finetuning_type lora \
--output_dir 'lora/test' \
--predict_with_generate \
--cutoff_len 512 \
--bf16 \
--max_new_tokens 300 \
--bits 4- During inference,
model_name,template, andbf16must be the same as the settings used during training. model_name_or_path: Specify the path to the base model being used, which must match the corresponding LoRA model.checkpoint_dir: The path to the LoRA weight files.output_dir: This parameter does not take effect during inference and any path can be specified.input_file,output_file: Specify the input path for the test file and the output path for the prediction results, respectively.cutoff_len,max_new_tokens: Set the maximum input length and the number of new tokens to be generated, adjusting according to device performance.
Quantization can be performed by setting bits to 4; it is recommended for the RTX3090.
| checkpoint_dir | model_name_or_path | moadel_name | fp16/bf16 | template |
|---|---|---|---|---|
| OneKE | OneKE | llama | bf16 | llama2_zh |
Model download links for OneKE(based on chinese-alpaca2): zjunlp/OneKE
CUDA_VISIBLE_DEVICES=0 python src/inference.py \
--stage sft \
--model_name_or_path 'models/OneKE' \
--model_name 'llama' \
--template 'llama2_zh' \
--do_predict \
--input_file 'data/NER/test.json' \
--output_file 'results/OneKE_output.json' \
--output_dir 'lora/test' \
--predict_with_generate \
--cutoff_len 512 \
--bf16 \
--max_new_tokens 300 \
--bits 4model_name_or_path: The path to the weights of the model specialized for Information Extraction (IE).
We provide scripts for evaluating the F1 scores for various tasks.
python ie2instruction/eval_func.py \
--path1 data/NER/processed.json \
--task NER task: Currently supports five types of tasks: ['RE', 'NER', 'EE', 'EET', 'EEA'].- You can set
sort_bytosourceto calculate the F1 scores on each dataset separately.
We believe that annotated data contains the wisdom of humanity, and its existence is to promote the benefit of all humankind and help enhance our quality of life. We strongly urge all users not to use our corpus for any actions that may harm national or public security or violate legal regulations. We have done our best to ensure the quality and legality of the data provided. However, we also recognize that despite our efforts, there may still be some unforeseen issues, such as concerns about data protection and risks and problems caused by data misuse. We will not be responsible for these potential problems. For original data that is subject to usage permissions stricter than the CC BY-NC-SA 4.0 agreement, IEPile will adhere to those stricter terms. In all other cases, our operations will be based on the CC BY-NC-SA 4.0 license agreement.
From the data perspective, our study primarily focuses on schema-based IE, which limits our ability to generalize to human instructions that do not follow our specific format requirements. Additionally, we do not explore the field of Open Information Extraction (Open IE); however, if we remove schema constraints, our dataset would be suitable for Open IE scenarios. Besides, IEPile is confined to data in English and Chinese, and in the future, we hope to include data in more languages.
From the model perspective, due to computational resource limitations, our research only assessed two models: Baichuan and LLaMA, along with some baseline models. Our dataset can be applied to any other large language models (LLMs), such as Qwen, ChatGLM, Gemma.
If you use the IEPile or the code, please cite the paper:
@article{DBLP:journals/corr/abs-2402-14710,
author = {Honghao Gui and
Lin Yuan and
Hongbin Ye and
Ningyu Zhang and
Mengshu Sun and
Lei Liang and
Huajun Chen},
title = {IEPile: Unearthing Large-Scale Schema-Based Information Extraction
Corpus},
journal = {CoRR},
volume = {abs/2402.14710},
year = {2024},
url = {https://doi.org/10.48550/arXiv.2402.14710},
doi = {10.48550/ARXIV.2402.14710},
eprinttype = {arXiv},
eprint = {2402.14710},
timestamp = {Tue, 09 Apr 2024 07:32:43 +0200},
biburl = {https://dblp.org/rec/journals/corr/abs-2402-14710.bib},
bibsource = {dblp computer science bibliography, https://dblp.org}
}We are very grateful for the inspiration provided by the MathPile and KnowledgePile projects. Special thanks are due to the builders and maintainers of the following datasets: AnatEM、BC2GM、BC4CHEMD、NCBI-Disease、BC5CDR、HarveyNER、CoNLL2003、GENIA、ACE2005、MIT Restaurant、MIT Movie、FabNER、MultiNERD、Ontonotes、FindVehicle、CrossNER、MSRA NER、Resume NER、CLUE NER、Weibo NER、Boson、ADE Corpus、GIDS、CoNLL2004、SciERC、Semeval-RE、NYT11-HRL、KBP37、NYT、Wiki-ZSL、FewRel、CMeIE、DuIE、COAE2016、IPRE、SKE2020、CASIE、PHEE、CrudeOilNews、RAMS、WikiEvents、DuEE、DuEE-Fin、FewFC、CCF law, and more. These datasets have significantly contributed to the advancement of this research. We are also grateful for the valuable contributions in the field of information extraction made by InstructUIE and YAYI-UIE, both in terms of data and model innovation. Our research results have benefitted from their creativity and hard work as well. Additionally, our heartfelt thanks go to hiyouga/LLaMA-Factory; our fine-tuning code implementation owes much to their work. The assistance provided by these academic resources has been instrumental in the completion of our research, and for this, we are deeply appreciative.