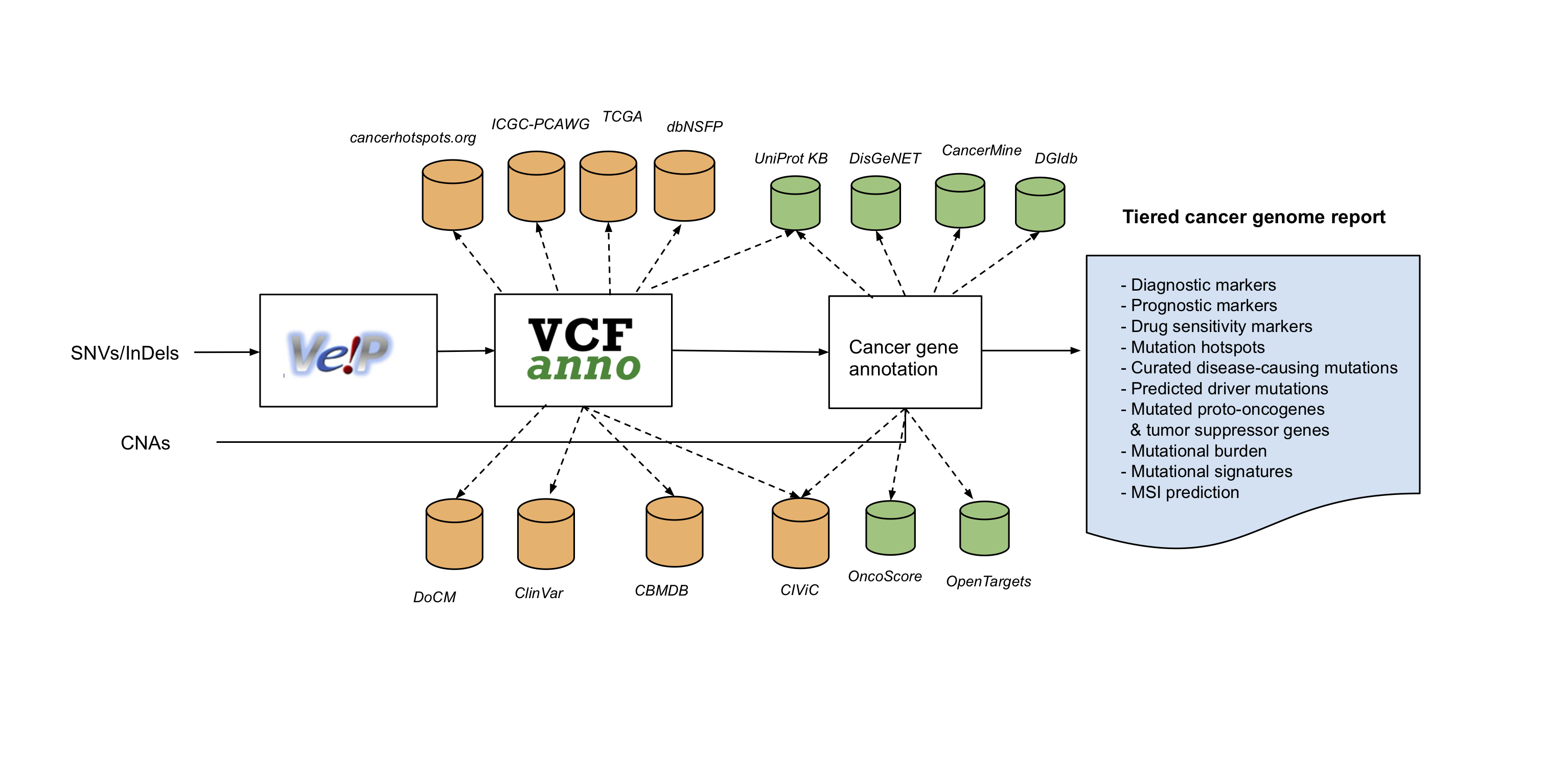
The Personal Cancer Genome Reporter (PCGR) is a stand-alone software package for functional annotation and translation of individual cancer genomes for precision oncology. Currently, it interprets both somatic SNVs/InDels and copy number aberrations. The software extends basic gene and variant annotations from the Ensembl’s Variant Effect Predictor (VEP) with oncology-relevant, up-to-date annotations retrieved flexibly through vcfanno, and produces interactive HTML reports intended for clinical interpretation.
- May 22nd 2019: 0.8.1 release
- Added Cancer_NOS.toml for unspecified tumor types
- Minor bugfixing
- May 20th 2019: 0.8.0 release
- Bundle update (VEP, CIViC, UniProt, CancerMine, dbNSFP, OpenTargets, DisGeNET, TCGA, ICGC-PCAWG)
- New functionality
- Ranking of variants in tiers 3-4/noncoding according to association scores from Open Targets Platform,(Carvalho-Silva et. al, NAR, 2019)
- Mutational burden in the context of TCGA distributions
- More extensive variant filtering options for tumor-only runs
- Possibility to feed a panel-of-normals VCF to PCGR for filtering purposes
- Possibility to add somatic CNA plot to report (provided as image file)
- Pre-made configuration files pr. tumor type
- Change pick order for primary transcript (VEP)
- Massive upgrade of the Cancer Predisposition Sequencing Reporter
- Choice between > 30 different virtual cancer predisposition gene panels
- Improved variant classification according to ACMG criteria
- Simplified report structure - organized according to pathogenicity levels
- Nov 27th 2018: 0.7.0 release
- Bundle update and bug fixing (see CHANGELOG )
- Reporting germline variants for cancer predisposition? Check out github.com/sigven/cpsr
- May 14th 2018: 0.6.2.1 release
- May 9th 2018: 0.6.2 release
- Fixed various bugs reported by users (see CHANGELOG)
- Data bundle update (ClinVar, KEGG, CIViC, UniProt, DiseaseOntology)
- May 2nd 2018: 0.6.1 release
- Fixed bugs in tier assignment
- April 25th 2018: 0.6.0 release
- Updated data sources
- Enabling specification of tumor type of input sample
- New tier system for classification of variants (ACMG-like)
- VCF validation can be turned off
- Tumor DP/AF presets
- JSON dump of report content
- GRCh38 support
- Runs under Python3
- November 29th 2017: 0.5.3 release
- Fixed bug with propagation of default options
- November 23rd 2017: 0.5.2 release
- November 15th 2017: 0.5.1 pre-release
- Bug fixing (VCF validation)
- November 14th 2017: 0.5.0 pre-release
- Updated version of VEP (v90)
- Updated versions of ClinVar, Uniprot KB, CIViC, CBMDB
- Removal of ExAC (replaced by gnomAD), removal of COSMIC due to licensing restrictions
- Users can analyze samples run without matching control (i.e. tumor-only)
- PCGR pipeline is now configured through a TOML-based configuration file
- Bug fixes / general speed improvements
- Work in progress: Export of report data through JSON
IMPORTANT: If you use PCGR, please cite the publication:
Sigve Nakken, Ghislain Fournous, Daniel Vodák, Lars Birger Aaasheim, Ola Myklebost, and Eivind Hovig. Personal Cancer Genome Reporter: variant interpretation report for precision oncology (2017). Bioinformatics. 34(10):1778–1780. doi:10.1093/bioinformatics/btx817
- VEP - Variant Effect Predictor v96 (GENCODE v30/v19 as the gene reference dataset)
- CIViC - Clinical interpretations of variants in cancer (May 18th 2019)
- ClinVar - Database of variants with clinical significance (May 2019)
- DoCM - Database of curated mutations (v3.2, Apr 2016)
- CBMDB - Cancer Biomarkers database (Jan 17th 2018)
- DisGeNET - Database of gene-tumor type associations (v6.0, Jan 2019)
- Cancer Hotspots - Resource for statistically significant mutations in cancer (v2 - 2017)
- dBNSFP - Database of non-synonymous functional predictions (v4.0, May 2019)
- TCGA - somatic mutations discovered across 33 tumor type cohorts (The Cancer Genome Atlas, release 16, Mar 2019)
- UniProt/SwissProt KnowledgeBase - Resource on protein sequence and functional information (2019_04, Apr 2019)
- Pfam - Database of protein families and domains (v32, Sep 2018)
- DGIdb - Database of targeted cancer drugs (v3.0.2, Jan 2018)
- ChEMBL - Manually curated database of bioactive molecules (v25.1, Mar 2019)
- CancerMine - Literature-derived database of tumor suppressor genes/proto-oncogenes (v12, May 2019)
An installation of Python (version 3.6) is required to run PCGR. Check that Python is installed by typing python --version in your terminal window. In addition, a Python library for parsing configuration files encoded with TOML is needed. To install, simply run the following command:
pip install toml
- Install the Docker engine on your preferred platform
- installing Docker on Linux
- installing Docker on Mac OS
- NOTE: We have not yet been able to perform enough testing on the Windows platform, and we have received feedback that particular versions of Docker/Windows do not work with PCGR (an example being mounting of data volumes)
- Test that Docker is running, e.g. by typing
docker psordocker imagesin the terminal window - Adjust the computing resources dedicated to the Docker, i.e.:
- Memory: minimum 5GB
- CPUs: minimum 4
- How to - Mac OS X
a. Clone the PCGR GitHub repository (includes run script and folder with configuration files pr tumor type): git clone https://github.com/sigven/pcgr.git
b. Download and unpack the latest data bundles in the PCGR directory
- grch37 data bundle - 20190519 (approx 15Gb)
- grch38 data bundle - 20190519 (approx 16Gb)
- Unpacking:
gzip -dc pcgr.databundle.grch37.YYYYMMDD.tgz | tar xvf -
c. Pull the PCGR Docker image (dev) from DockerHub (approx 5.1Gb):
docker pull sigven/pcgr:dev(PCGR annotation engine)
a. Download and unpack the latest software release (0.8.1)
b. Download and unpack the assembly-specific data bundle in the PCGR directory
-
grch37 data bundle - 20190519 (approx 15Gb)
-
grch38 data bundle - 20190519 (approx 16Gb)
- Unpacking:
gzip -dc pcgr.databundle.grch37.YYYYMMDD.tgz | tar xvf -
A data/ folder within the pcgr-X.X software folder should now have been produced
- Unpacking:
c. Pull the PCGR Docker image (0.8.1) from DockerHub (approx 5.2Gb):
docker pull sigven/pcgr:0.8.1(PCGR annotation engine)
The PCGR workflow accepts two types of input files:
- An unannotated, single-sample VCF file (>= v4.2) with called somatic variants (SNVs/InDels)
- A copy number segment file
PCGR can be run with either or both of the two input files present.
- We strongly recommend that the input VCF is compressed and indexed using bgzip and tabix
- If the input VCF contains multi-allelic sites, these will be subject to decomposition
- Variants used for reporting should be designated as 'PASS' in the VCF FILTER column
The tab-separated values file with copy number aberrations MUST contain the following four columns:
- Chromosome
- Start
- End
- Segment_Mean
Here, Chromosome, Start, and End denote the chromosomal segment, and Segment_Mean denotes the log(2) ratio for a particular segment, which is a common output of somatic copy number alteration callers. Note that coordinates must be one-based (i.e. chromosomes start at 1, not 0). Below shows the initial part of a copy number segment file that is formatted correctly according to PCGR's requirements:
Chromosome Start End Segment_Mean
1 3218329 3550598 0.0024
1 3552451 4593614 0.1995
1 4593663 6433129 -1.0277
There are pre-made configuration files pr. tumor type in the conf folder, formatted using TOML. In the configuration file, the user may configure a number of options in the PCGR workflow, related to the following:
- Sequencing depth/allelic support thresholds
- MSI prediction
- Mutational signatures analysis
- Mutational burden analysis (e.g. target size of region subject to sequencing)
- VCF to MAF conversion
- Tumor-only analysis options
- tick on/off various filtering schemes for exclusion of germline variants
- VEP/vcfanno options
- Log-ratio thresholds for gains/losses in CNA analysis
See here for more details about the exact usage of the configuration options.
A tumor sample report is generated by calling the Python script pcgr.py, which takes the following arguments and options:
usage: pcgr.py [options] <PCGR_DIR> <OUTPUT_DIR> <GENOME_ASSEMBLY> <CONFIG_FILE> <SAMPLE_ID>
Personal Cancer Genome Reporter (PCGR) workflow for clinical interpretation of
somatic nucleotide variants and copy number aberration segments
positional arguments:
pcgr_dir PCGR base directory with accompanying data directory,
e.g. ~/pcgr-0.8.1
output_dir Output directory
{grch37,grch38} Genome assembly build: grch37 or grch38
configuration_file PCGR configuration file (TOML format, in conf/ folder)
sample_id Tumor sample/cancer genome identifier - prefix for
output files
optional arguments:
-h, --help show this help message and exit
--input_vcf INPUT_VCF
VCF input file with somatic query variants
(SNVs/InDels). (default: None)
--input_cna INPUT_CNA
Somatic copy number alteration segments (tab-separated
values) (default: None)
--input_cna_plot INPUT_CNA_PLOT
Somatic copy number alteration plot (default: None)
--pon_vcf PON_VCF VCF file with germline calls from Panel of Normals
(PON) - i.e. blacklist variants (default: None)
--tumor_purity TUMOR_PURITY
Estimated tumor purity (between 0 and 1) (default:
None)
--tumor_ploidy TUMOR_PLOIDY
Estimated tumor ploidy (default: None)
--force_overwrite By default, the script will fail with an error if any
output file already exists. You can force the
overwrite of existing result files by using this flag
(default: False)
--version show program's version number and exit
--basic Run functional variant annotation on VCF through
VEP/vcfanno, omit other analyses (i.e. CNA, MSI,
report generation etc. (STEP 4) (default: False)
--no_vcf_validate Skip validation of input VCF with Ensembl's vcf-
validator (default: False)
--docker-uid DOCKER_USER_ID
Docker user ID. Default is the host system user ID. If
you are experiencing permission errors, try setting
this up to root (`--docker-uid root`) (default: None)
--no-docker Run the PCGR workflow in a non-Docker mode (see
install_no_docker/ folder for instructions (default:
False)
The examples folder contain input files from two tumor samples sequenced within TCGA (GRCh37 only). It also contains PCGR configuration files customized for these cases. A report for a colorectal tumor case can be generated by running the following command in your terminal window:
python pcgr.py --input_vcf ~/pcgr-0.8.1/examples/tumor_sample.COAD.vcf.gz
--input_cna ~/pcgr-0.8.1/examples/tumor_sample.COAD.cna.tsv --tumor_purity 0.9 --tumor_ploidy 2.0
~/pcgr-0.8.1 ~/pcgr-0.8.1/examples grch37 ~/pcgr-0.8.1/examples/examples_COAD.toml tumor_sample.COAD
This command will run the Docker-based PCGR workflow and produce the following output files in the examples folder:
- tumor_sample.COAD.pcgr_acmg.grch37.html - An interactive HTML report for clinical interpretation
- tumor_sample.COAD.pcgr_acmg.grch37.pass.vcf.gz - Bgzipped VCF file with rich set of annotations for precision oncology
- tumor_sample.COAD.pcgr_acmg.grch37.pass.tsv.gz - Compressed vcf2tsv-converted file with rich set of annotations for precision oncology
- tumor_sample.COAD.pcgr_acmg.grch37.snvs_indels.tiers.tsv - Tab-separated values file with variants organized according to tiers of functional relevance
- tumor_sample.COAD.pcgr_acmg.grch37.json.gz - Compressed JSON dump of HTML report content
- tumor_sample.COAD.pcgr_acmg.grch37.cna_segments.tsv.gz - Compressed tab-separated values file with annotations of gene transcripts that overlap with somatic copy number aberrations
sigven AT ifi.uio.no