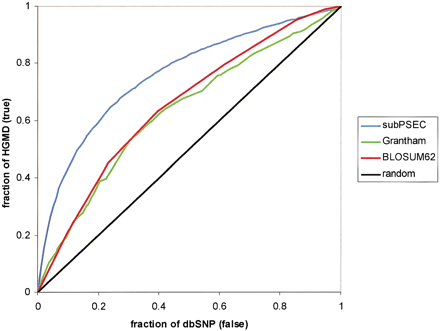
Predicting whether a missense SNP will have an effect on protein function: comparison between position-specific scores (subPEC) and “average” substitution scores. Position-specific scores from PANTHER HMMs (blue line) make a larger number of correct predictions (true positives shown on Y-axis) for a given number of errors (false positives shown on X-axis) than scores from the two most commonly referenced substitution scores: the Grantham scale (green line) and the BLOSUM62 substitution matrix (red line). The black line shows the curve for a random prediction, as a reference. HGMD mutations are used to approximate a set of functionally impaired proteins, and dbSNP variations are used to approximate a set of functional proteins (see text for more details).











