Abstract
Octocorals are major contributors of terpenoid chemical diversity in the ocean. Natural products from other sessile marine animals are primarily biosynthesized by symbiotic microbes rather than by the host. Here we challenge this long-standing paradigm by describing a monophyletic lineage of animal-encoded terpene cyclases (TCs), ubiquitous in octocorals. We characterized 15 TC enzymes from nine genera, several of which produce precursors of iconic, coral-specific terpenoids such as pseudopterosin, lophotoxin and eleutherobin. X-ray crystallography reveals that coral TCs share conserved active site residues and structural features with bacterial TCs. The identification of coral TCs enabled the targeted identification of the enzyme that constructs the coral-exclusive capnellane scaffold. Several TC genes are co-localized with genes that encode enzymes known to modify terpenes. This work presents the first example of biosynthetic capacity in the kingdom Animalia that rivals the chemical complexity generated by plants, unlocking the biotechnological potential of octocorals for biomedical applications.
Introduction
With over 50,000 representatives, terpenoids are the most abundant class of natural products1. This plethora of structural diversity derives predominantly from the specialized metabolism of plants2 and microorganisms3–5 and has brought forth life-saving medicines like Taxol6 and artemisinin7. In marine environments, where plants are absent, octocorals – a subclass of sessile invertebrate animals that comprises soft corals, sea pens, and blue corals – are the most prolific source of terpenoids. With over 4,000 sesqui- and diterpenoids isolated, these compounds make up more than 12% of all reported marine natural products8. While the ecological functions of coral-derived terpenoids are largely unknown beyond serving as feeding deterrents to generalist grazers9, these compounds show bioactivity that points to promising pharmacological potential10,11. Examples include the anti-inflammatory pseudopterosin A (1) from Antillogorgia elisabethae12, the anti-metastatic polyanthellin A (3) from Briareum asbestinum13, and the neuromuscular toxin lophotoxin (5) from Leptogorgia chilensis14 (Fig. 1). Despite being known for over four decades, the biological and genetic origins of coral terpenoids have remained unsolved.
Fig. 1: Bioactive terpenoids extracted from different octocorals linked to their proposed biosynthetic precursor-terpenes.

Photos were obtained from: reefcorner.com for A. elisabethae, James St. John for B. asbestinum and Phillip Colla / Oceanlight.com for L. chilensis.
Natural products derived from most sessile marine animals such as sponges, tunicates, and bryozoans are produced by bacterial symbionts15–17, whereas reports on animals being the true producers of natural products are scarce18,19. Hence, coral terpenoid biosynthesis has been presumed to be microbial20–22. The greatest effort to elucidate terpenoid biosynthesis in octocorals has been carried out for A. elisabethae, since organic extracts from this coral contain the pseudopterosins including 1 as active constituents and are commercially used as a skin soothing ingredient in skin-care products in the cosmetic industry21. Biosynthetic studies have suggested that the source of the pseudopterosins are symbiotic dinoflagellates of the genus Symbiodinium (zooxanthellae)21. Additionally, bioactivity guided fractionation enabled the isolation of a terpene cyclase (TC) that produced the proposed terpene precursor of the pseudopterosins, elisabethatriene (2)23.However, the amino acid sequence of this enzyme was not determined and the organismal origin remained unclear23. Despite these findings, a unifying animal biosynthetic origin for terpenoid-laden octocorals, which grow in shallow tropical waters in association with symbiotic zooxanthellae as well as in deep, cold waters without photosynthetic algae, could not be ruled out.
In this work we characterize a new group of monophyletic class 1 TCs that are encoded in octocoral genomes by hidden Markov model (HMM) driven genome and transcriptome mining. These TCs produce terpene precursors for several groups of octocoral terpenoids, including 1, 3 and 5, and display high structural similarity to microbial class I TCs as determined by X-ray crystallography. Genes coding for TCs seem to be ubiquitous in octocorals and are in some cases physically co-clustered with genes encoding cytochrome P450s (CYP). These findings enable genome mining in octocorals as exemplified in our targeted characterization of the octocoral-specific capnellene synthase.
Results
Discovery and characterization of octocoral terpene cyclases.
To address the open question about the origin of terpenoid production in octocorals, we queried publicly available coral sequencing data using HMMs. Since red algal genomes were recently shown to code for microbial-type TC genes24, we hypothesized that octocorals might encode similar enzymes. Previously published HMMs were either solely based on bacterial or plant type sequences25,26, so we generated an HMM from a larger diversity of characterized TC sequences from bacteria and fungi (Supplementary Table 1) to get more generalized microbial-type class-I TC hits. Querying the genome and transcriptome of the octocoral Dendronephtha gigantea27 yielded a set of low-scoring hits, showing typical conserved type I TC motifs; refining the HMM with these sequences yielded further sequences from all other five public octocoral genomes (Supplementary Table 2) and 19 transcriptomes (Supplementary Table 3). Phylogenetic analysis showed the newly identified genes to be monophyletic and placed as a branch distinct from all previously characterized TCs (Fig. 2A, Extended Data Fig. 1). All the octocoral genes harbor typical motifs conserved in microbial and plant TCs (Supplementary Fig. 1), suggesting that the identified sequences code for a group of unexplored biosynthetic enzymes.
Fig. 2: Phylogeny of octocoral TCs and their terpene products.

A: Phylogenetic tree of type 1 TCs from different organisms. Octocoral TCs form their own distinct clade. Isoprenyl diphosphate synthases (IDSs) are included as an outgroup. B: Magnified view of the octocoral TC clade. Biochemically characterized enzymes are labeled with their respective names and the compound numbers of their respectively formed main products. M denotes production of a mixture of unidentified cyclic sesquiterpenes in the absence of diterpene cyclization. C: Structures of terpenes formed by recombinant octocoral TCs.
A subset of 14 newly identified genes, which were hypothesized to produce different products based on their phylogenetic distances, were expressed in Escherichia coli (Fig. 2B, Supplementary Fig. 2 and 3). Recombinant proteins were tested in vitro with common terpene diphosphate precursors, and the product profiles analyzed by gas chromatography-mass spectrometry (GCMS) and nuclear magnetic resonance spectroscopy (NMR, for structure elucidation, see Supplementary Note 1). All recombinant proteins showed TC activity (Supplementary Fig. 4–18). Nine of the enzymes acted as selective diterpene cyclases, converting geranylgeranyl diphosphate (GGPP) to cyclic terpene products (Fig. 2, B and C) corresponding to well-known coral terpenoid carbon backbones (Fig. 1, Extended Data Fig. 2). Four enzymes produced cembrene A (6, Supplementary Fig. 4, 8, 10 and 16), while one (RmTC-1) produced cembrene C (8, Supplementary Fig. 15), both of which bear the principal scaffold of cembranoids like 5, the largest group of octocoral diterpenoids (>1200 known representatives, Extended Data Fig. 2). Two TCs (HcTC-3 and PbTC-1) selectively formed elisabethatriene (2, Supplementary Fig. 13 and 14), the terpene precursor to 1 and related structures (>100 known representatives, Extended Data Fig. 2). The enzyme obtained from Xenia sp. (XsTC-1) showed selective formation of xeniaphyllene (7, Supplementary Fig. 18), a yet unreported terpene hydrocarbon that has been hypothesized as the biosynthetic precursor to a large group of coral specific terpenoids (>300 known representatives, Extended Data Fig. 2)28. BaTC-2 from B. asbestinum transcriptomic data produced klysimplexin R (4, Supplementary Fig. 5), exhibiting the coral specific cis-cladiellane carbon scaffold (>400 known representatives, Extended Data Fig. 2) found in 3 and its enantiomer, both reported from Briareum species11. Of the identified sesquiterpene cyclases, TmTC-1 showed highly specific product formation of α-muuroladiene (10, Supplementary Fig. 17) from farnesyl diphosphate (FPP). The four other enzymes produced sesquiterpene mixtures, two of them containing nephthene (9) as a major constituent (Supplementary Fig. 7 and 12). Nephthene was also produced as the major product by the 2-synthases HcTC-3 and PbTC-1 when incubated with FPP (Supplementary Fig. 13 and 14).
Fig. 5: Directed approach to the discovery of capnellene synthase CiTC 1.
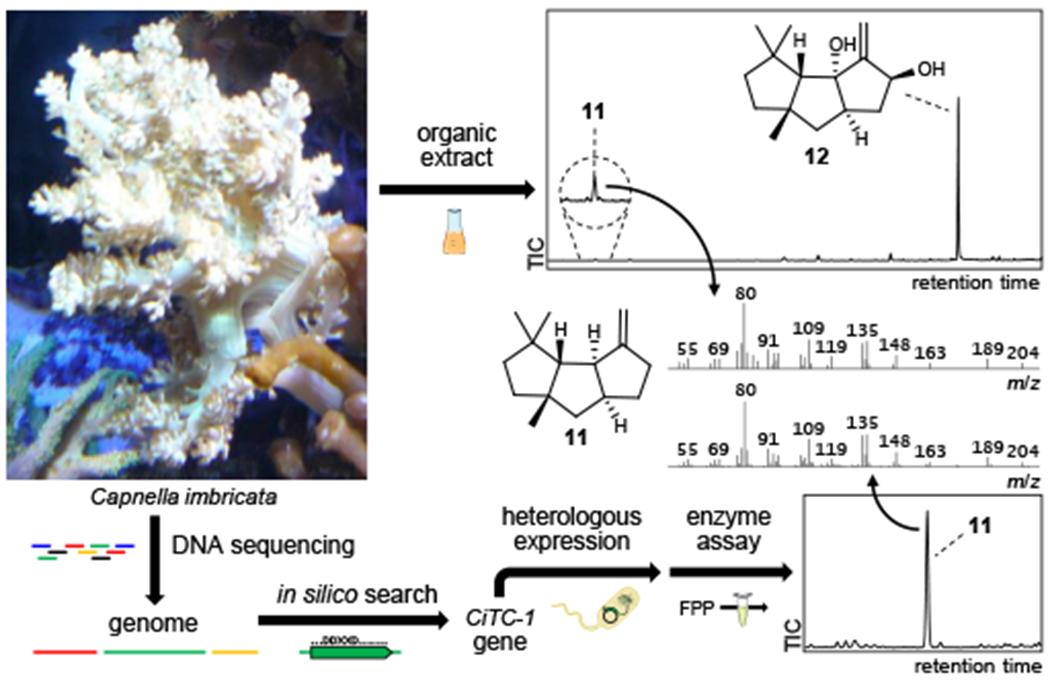
Capnella imbricata was verified to produce capnellene-8β,10α-diol (12) and its biosynthetic precursor 11. Genome mining was applied to identify the CiTC-1 gene, and the enzyme’s activity validated in vitro to produce 11. Flask and tube art by Servier (CC-BY).
Several pieces of evidence suggest that the newly described TC genes are encoded by the coral host rather than an algal or bacterial symbiont. Firstly, the genes were discovered in sequencing data from both zooxanthellate (harboring algal symbionts, e. g. Xenia sp.) and azooxanthellate corals (e. g. D. gigantea). Secondly, three of the TCs were identified in large high-quality scaffolds from Xenia sp.29 and D. gigantea27 (Supplementary Tables 2 and 3) showing classic eukaryotic features like long intergenic regions, repetitive elements, and genes containing introns (Extended Data Fig. 3, Supplementary Table 4). The genes neighboring the TC are most similar to animal sequences as their closest homologs when interrogated by BLAST, e.g. a peroxisomal biogenesis factor from the stony coral Acropora millepora or a transmembrane protein from the sea anemone Actinia tenebrosa (Extended Data Fig. 3, Supplementary Tables 5–7). Moreover, the monophyly of the coral TC clade suggests that it is inherited, which is in line with the first two points.
Enzyme structure and evolutionary origin.
While the octocoral TC clade is placed distinctly from other branches in the phylogenetic analysis (Fig. 2), all the characterized sequences exhibit the typical conserved motifs of microbial class-I TCs (Supplementary Fig. 1 and Supplementary Fig. 19). To facilitate a structural comparison, we solved the X-ray crystal structure of the Eleutherobia rubra cembrene A synthase ErTC-2 to 1.54-Å resolution with phasing of a cadmium-derivatized protein crystal. ErTC-2 crystallized as a monomer, which was supported by a Proteins, Interfaces, Structures and Assembly (PISA) server30 analysis (Supplementary Table 8). ErTC-2 assumes the typical alpha helical fold exhibited by plant and microbial type I TCs1, but shows three additional helices (D2, L, M) that are coded by an insertion (D2) and an extended C-terminus (L and M) (Fig. 3A) conserved in all characterized octocoral TCs (Supplementary Figure 19). Distinctive substrate interacting residues conserved in the coral TCs are located on helix D (aspartate rich motif DDXXE), helix H (NSD/DTE), helix K (RY), and helix G (pyrophosphate sensor R) (Fig. 3B). This three-dimensional arrangement is analogous to the microbial TCs, exemplified in the bacterial epi-isozizaene synthase31 structure (Fig. 3C). The alpha domain of taxadiene synthase32 assumes a similar overall structure to ErTC-2, but like all plant TCs, it is missing the conserved RY motif and the pyrophosphate sensor R is not located on helix G but on helix H. The overall fold of a typical animal isoprenyl diphosphate synthase (IDS) exemplified by the human FPP synthase33 is the most distinct from ErTC-2 and displays the typical two aspartate rich motifs but is lacking all the other motifs from ErTC-2. The root-mean-square deviation from alignments of ErTC-2 to microbial and plant TCs as well as IDSs agrees with these observations (Supplementary Table 9), showing that ErTC-2 is structurally most like microbial TCs. The only other known enzymes with TC activity from animals are involved in pheromone biosynthesis in different insect species and are products of insect-lineage specific recent neofunctionalizations of IDSs. Thus, they are found to clade within the IDS clade in the phylogenetic analysis (Fig. 2A) and are unrelated to the newly identified enzymes34.
Fig. 3: Structure of the coral cembrene A synthase ErTC-2 and other TCs.

A: Overall structure of ErTC-2 (PDB ID: 7s5l) showing the typical alpha-helical fold of type 1 TCs. Helices C (blue), D (teal), G (yellow), H (orange), and J (light pink) shape the active site of the enzyme. Helix D2 (green) is placed on an insertion sequence, while L and M (also green) are structural elements placed on a C-terminal extension. B: Expansion of the ErTC-2 active site showing the conserved motifs DDXXE on helix D, the “pyrophosphate sensor” arginine on Helix G, NSD/DTE on helix H, and RY on helix K. C: Structural comparison of ErTC-2 with a microbial-type TC (epi-isozizaene synthase from Streptomyces coelicolor; PDB ID 4lz0), the α-domain of a plant-type TC (taxadiene synthase from Taxus brevifolia; PDB ID 3p5r), and an IDS (FPP synthase from Homo sapiens; PDB ID 5ja0).
Octocorals are suggested to have diverged about 580 million years ago, and the distribution of genera investigated in this work (Fig. 4) spans both major octocoral clades35. The radial expansion of the monophyletic TC clade (Fig. 2A) indicates that the observed TC diversity in octocorals evolved from one ancestor gene. This observation suggests that the last common octocoral ancestor acquired an ancestral TC gene and that octocorals gained the ability to produce terpenoids before land plants emerged36. Although the phylogenetic analysis does not immediately suggest a horizontal gene transfer event, the striking structural similarities between microbial and octocoral TCs render an ancient transfer from a microbe to the last common octocoral ancestor a likely scenario. Plants are similarly thought to have acquired an ancestral gene for their typical plant type TC genes from microbes , probably an early ent-kaurene synthase2, where they too form a distinct evolutionary clade (Fig. 2A).
Fig. 4: Possible timeline of TC evolution in octocorals.
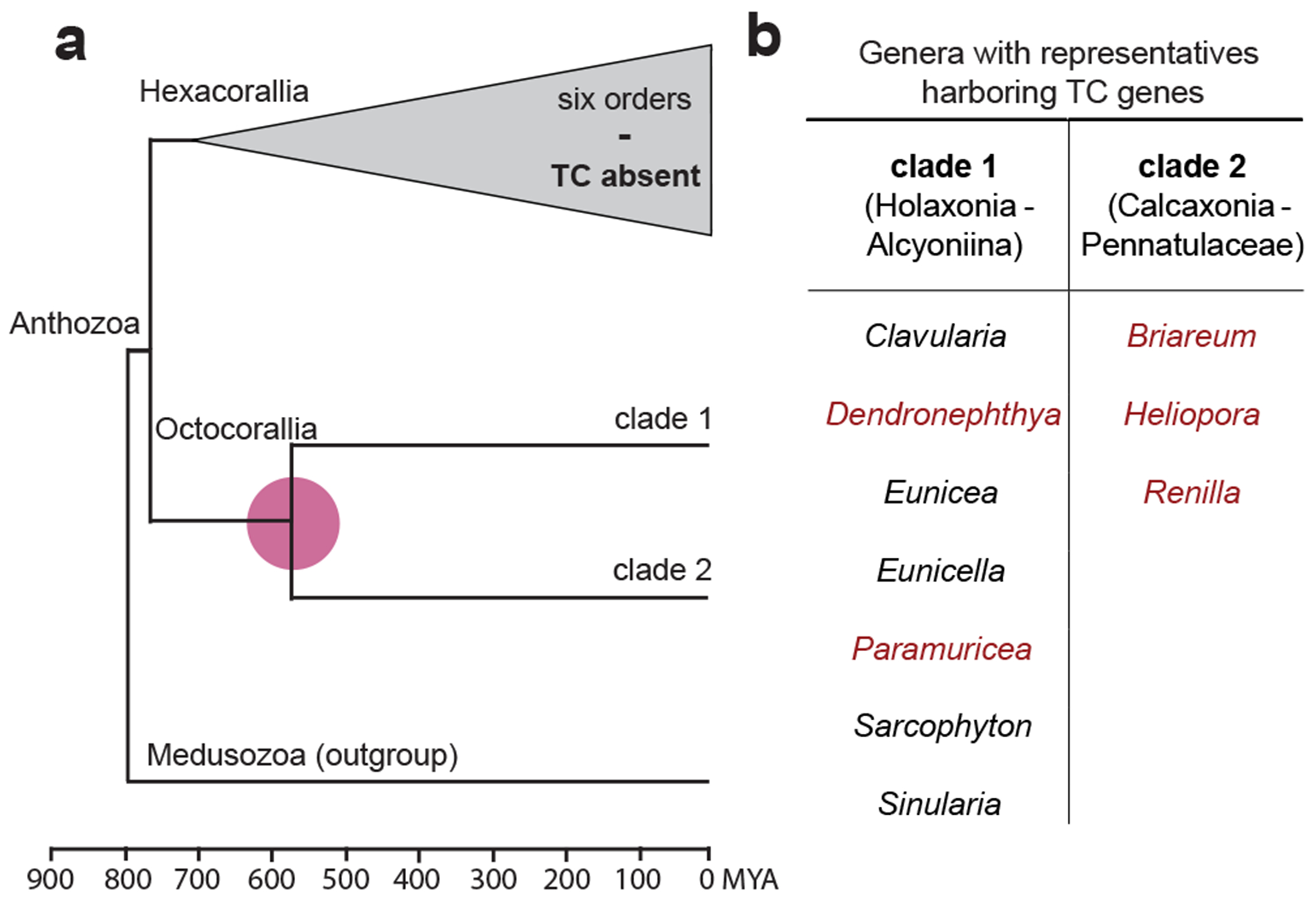
A: Schematic time-scaled phylogenomic tree for anthozoans recreated from ref. 35. Octocorals split into two major clades approximately 580 million years ago (color mark). B: List of genera from both clades with representative species shown to harbor TC genes in this work. Red genera denote those for which TCs have been biochemically characterized.
Biosynthetic gene clusters and genome mining.
Given the intriguing similarities between plant and octocoral terpenoid metabolism, we inspected the genetic neighborhoods of coral TCs for the occurrence of potential biosynthetic gene clusters37. Recent advances in plant genomics have made it apparent that plants, like bacteria and fungi, frequently cluster genes coding for enzymes that act in the same metabolic pathway38. Inspecting the genomic regions upstream and downstream of TCs in the genomes in a wider frame in Xenia sp., as well as in Trachythela sp. and Renilla muelleri revealed the presence of multiple genes coding for cytochrome P450 monooxygenases (CYPs) and short chain dehydrogenases (SDHs), classes of enzymes commonly employed in the oxidative functionalization of terpenoids39 (Extended Data Fig. 4). Notably, the contig of Xenia sp. containing a TC encodes an IDS gene that could provide the precursor GGPP, in addition to CYPs and SDHs that could convert the characterized XsTC-1 product 7 to oxidized terpenoids that are frequently isolated from Xenia spp. (Extended Data Fig 5). While these genes are not as tightly clustered as is seen in bacteria and fungi, the extent of clustering is comparable to that seen in plant genomes. Taken together, these findings suggest the existence of biosynthetic gene clusters for the production of complex secondary metabolites in metazoa.
To showcase an application of our new understanding of octocoral TCs, we sought to identify the TC involved in capnellene-8β,10α-diol (12) production in the soft coral Capnella imbricata. Natural products containing the capnellane skeleton exhibit promising anti-leukemia40 and analgesic41 properties and have only been isolated from soft corals. The biosynthetic precursor of this class of molecules is suggested to be the sesquiterpene capnellene (11)42. We acquired C. imbricata, verified that it produced 12 (Extended Data Fig. 6), and sequenced its metagenome using Illumina MiSeq technology (Fig. 5). Despite low coverage (Supplementary Figures 20 and 21), we queried the draft sequence to identify a single full-length TC homolog. The gene was expressed heterologously in E. coli, and the purified enzyme was functionally validated as the first capnellene-producing terpene synthase (Supplementary Fig. 6).
Discussion
This work describes the discovery of a lineage of TC enzymes specific to octocorals that are present in all publicly available octocoral genomes (Supplementary Table 2). Our untargeted genome mining approach enabled the identification and biochemical characterization of fifteen functional terpene cyclases, and uncovered cryptic coral secondary metabolism as illustrated by the discovery of the elisabethatriene (2) synthase PbTC-1 from the deep sea coral Paramuricea biscaya43. Compounds biosynthetically derived from 2 have not been described from this genus of octocorals before, which suggests that the newly discovered biosynthetic capacity of octocorals may be larger than represented by the thousands of known compounds. The identification of coral terpene cyclases that produce capnellene (11), elisabethatriene (2), and other high-value, coral-specific terpenes allows for biotechnological applications and future mechanistic and ecological studies.
More broadly, the discovery of widespread octocoral TCs as a new monophyletic group of class-I TCs is unprecedented in the kingdom Animalia and shows the general ability of animals to individually maintain and evolve capacities to produce complex specialized metabolites. In terms of their evolutionary history, these genes were probably acquired by the last common ancestor of octocorals, either by de novo evolution – for example through a gene duplication event – or by horizontal gene transfer. Although the phylogenetic analysis shows no clading of the octocoral TCs with bacterial sequences, several pieces of evidence suggest an ancient horizontal gene transfer. The overall protein structure and the microbial class-I TC motifs are exactly conserved in octocoral TCs, and all TCs for which genomic data is available, except for RmTC-1, are encoded as single exon genes. Their seemingly widespread maintenance and evolution by extant octocoral species hints towards high importance for octocoral evolution. Furthermore, the occurrence of TC genes and physical co-localization with genes coding for putative tailoring enzymes resembles specialized metabolic pathways in plants. This apparent convergent evolution could have arisen through similar evolutionary pressures due to an analogous sessile lifestyle, possibly to deter grazers in the absence of a calcified shell as exhibited by stony corals or strong nematocysts as utilized by the larger sea anemones44, both classes of hexacorals that seem to lack the ability of terpenoid production. This observation raises the question of whether other sessile animals that lack physical defenses also harbor the ability to produce comparably diverse bioactive compounds.
Methods
General Methods.
All chemicals and solvents were used as received from the commercial supplier (Sigma-Aldrich, Fisher or TCI). The substrates GPP, FPP, and GGPP were synthesized as reported previously45,46. Thin Layer chromatography was performed with TLC plates purchased from Merck (silica gel 60 F254, glass plates). Column chromatography was carried out using a Flash EZ prep combiflash system (Teledyne ISCO). GCMS analysis was carried out on an Agilent 7890A gas chromatograph with Agilent 5975C mass spectrometer, using a Rtx-5Sil 30 m × 0.25 mm, 0.25 μ column. Oven program was as follows: hold 70 °C for 3 min, 10 °C/min to 325 °C, hold at 325°C for 3 min. Flow rate: 1 mL/min, Injection volume: 1 μL, splitless injection, Inlet temperature: 270 °C, MS transfer line temperature: 280 °C, MS source temperature 230 °C, MS quad temperature: 150 °C. Data were analyzed using Masshunter B06.00. NMR spectra were recorded on a Bruker Avance III spectrometer (600 MHz) using a 1.7 mm inverse detection triple resonance (H-C/N/D) cryoprobe or a JEOL ECZ spectrometer (500 MHz) and were referenced against C6D6 (δ = 7.16 ppm) for 1H-NMR and C6D6 (δ = 128.06 ppm) for 13C-NMR. Data for NMR spectra are reported as follows: shift (d) in ppm, s = singlet, d = doublet, t = triplet, q = quartet, m = multiplet or unresolved, br = broad signal, J = coupling constant(s) in Hz and analyzed using MestreNova 12.0.0. Optical rotation measurements were carried out using a Jasco P-2000 polarimeter.
Identification of terpene cyclase genes.
The terpene cyclase (TC) profile HMM was made using available biochemically characterized TC genes (see Supplementary Table 1) using HMMR 3.1b247. Available genome and transcriptome assemblies were downloaded from different databases (see Supplementary Table 2 and 3). Available raw transcriptomic data with paired-end reads were downloaded from the NCBI SRA database using Fastq-dump 2.8.0 which is included in SRA-tools48 and assembled using Trinity 2.8.449. TransDecoder 5.5.050 was used to identify and translate open reading frames in transcriptomic assemblies. The translated peptide sequences were queried using the previously built profile HMM using HMMR 3.1b2. Alternatively, obtained assemblies were queried by tBLASTn searches with already identified octocoral TC amino acid sequences using Sequenceserver 1.0.1351.
Mapping of raw long reads to literature genome assembly data.
The integrity of TC containing genomic contigs from D. gigantea and X. sp was shown by mapping long sequencing raw reads back to the assemblies of the respective organism using Minimap 2.17. Expression was shown by mapping raw transcript reads back to the same assemblies using Hisat 2.2.1. The results for the genetic neighborhood of TC genes in both assemblies were visualized using the Integrated Genomics Viewer 2.8.2 (Extended Data Figure 3). Utilized datasets for mapping are from the same specimens the assemblies were reported for and are listed in Supplementary Table 4.
Phylogenetic analysis.
All sequence alignments were generated with Kalign v.2.0452 and visualized using MegaX 10.0.553. Sequence alignment figures were prepared using ESPript 3.054. The phylogenetic analysis was performed using IQ-TREE multicore version 2.0.355, substitution model LG+R6, 1000 bootstrap replicates from a sequence alignment of coral TC amino acid sequences in addition to sequences summarized in Supplementary Table 10. Plant TC sequences were manually truncated to their alpha domains.
Heterologous expression for analytical scale.
Genes for heterologous expression were purchased from TWIST Biosciences as codon optimized (Twist algorithm) sequences cloned into pET 28a (+) vectors (cut sites XhoI and NdeI). The plasmids were chemically transformed into E. coli BL21 (DE3). 15 mL LB medium (Fisher) with 50 mg/L kanamycin was inoculated with transformed bacteria and grown for 18 h at 37 °C and 180 rpm. 1 mL of these cultures was transferred to 100 mL terrific broth (Fisher, supplemented with 50 g/L kanamycin) and grown to OD600 = 0.6-0.8 at 37 °C and 180 rpm. Cultures were subsequently cooled to 18°C, induced with IPTG (400 μM) and shaken at 18 °C and 180 rpm for 18 h. The cells were pelleted by centrifugation (8000 xg, 15 min, 4 °C) and frozen at −80 °C until use or processed immediately. Pellets were resuspended in TC binding buffer (20 mM Na2HPO4, 0.5 M NaCl, 20 mM imidazole, 1 mM MgCl2, pH 7.4, 5 mL buffer per 100 mL culture). The cells were lysed by sonication (1 sec on, 1 sec off pulses for 1 min, 30 sec off, 5x) and the debris was removed by centrifugation (11000 xg, 20 min, 4 °C). The supernatant was filtered through a 0.22 μM membrane sterile filter (Merck Millipore) and loaded onto a pre equilibrated (TC binding buffer) gravity flow column containing Chelating Sepharose Fast Flow resin (GE Healthcare, 0.5 mL) loaded with Ni cations, washed with TC binding buffer (2x 2 mL) and eluted with 2.5 mL TC elution buffer (20 mM Na2HPO4, 0.5 M NaCl, 0.5 M imidazole, 1 mM MgCl2, pH 7.4). The elution fractions were loaded onto PD-10 columns (GE Healthcare) that were pre equilibrated with 25 mL TC reaction buffer (50 mM HEPES, 5 mM MgCl2, 250 mM NaCl, 10% glycerol, pH 7.8) and eluted with 3.5 mL TC reaction buffer. The protein was either used in assays immediately or flash frozen in liquid nitrogen and stored at −80°C until use.
Heterologous expression for preparative scale.
3x 1 L TB media were inoculated with 1 vol% of an E. coli overnight culture carrying the expression plasmid for the respective TC. Cultures were grown to OD600 = 0.6-0.8 at 37 °C and 180 rpm, cooled to 18 °C, induced with IPTG (400 μM) and shaken at 18 °C and 180 rpm for 18 h. The cells were pelleted by centrifugation (8000g, 15 min) and frozen at −80 °C until use or processed immediately. Pellets were resuspended in TC-binding buffer (20 mM Na2HPO4, 0.5 M NaCl, 20 mM imidazole, 1 mM MgCl2, pH 7.4, 20 mL buffer per 1 L culture). The cells were lysed by sonication (15 sec on, 45 sec off pulses for a total on-time of 7 min) and the debris was removed by centrifugation (11000 xg, 20 min, 4 °C), transferred to a new tube and centrifuged again (11000 xg, 20 min, 4 °C) after DNaseI (Roche Diagnostics, 1 mg per 20 mL supernatant) was added. The supernatant was filtered through a 0.22 μM membrane sterile filter (Merck Millipore) and loaded onto a 5 mL HisTrap-FF Ni-column (GE). The column was washed with TC binding buffer (40 mL), eluted with TC elution buffer (20 mM Na2HPO4, 0.5 M NaCl, 0.5 M imidazole, 1 mM MgCl2, pH 7.4) and the elution was collected in 2 mL fractions. Enzyme containing fractions were pooled and the buffer was exchanged in 2.5 mL batches using PD-10 columns equilibrated with TC reaction buffer (50 mM HEPES, 5 mM MgCl2, 250 mM NaCl, 10% glycerol, pH 7.8) and eluted with 3.5 mL TC-reaction buffer per column used. The protein was either used in assays immediately or flash frozen in liquid nitrogen and stored at −80 °C until use.
Analytical scale assays.
Analytical scale assays were carried out in 1.5 mL TC reaction buffer (50 mM HEPES, 5 mM MgCl2, 250 mM NaCl, 10% glycerol, pH 7.8) with an enzyme concentration of 3 μM and 0.25 mg/mL GPP, FPP or GGPP to test mono-, sesqui-, and diterpene synthase activity. The assays were run for 18 h at room temperature and extracted with hexanes (200 μL). The extracts were dried over MgSO4 and analyzed by GCMS.
Preparative scale assays.
For product isolation, GGPP (100 mg, for diterpene production) or FPP (80 mg, for sesquiterpene production) were dissolved in ammonium bicarbonate solution (15 mL, 50 mM) and added dropwise over 5h to 600 mL TC reaction buffer (50 mM HEPES, 5 mM MgCl2, 250 mM NaCl, 10% glycerol, pH 7.8) containing 1-6 μM enzyme at room temperature. The assay was run for 18 h at room temperature and extracted with pentane (3× 200 mL). The organic layer was dried over MgSO4, filtered, and concentrated in vacuo to 0.5 mL. The crude product was purified by silica gel column chromatography. Fractions were checked by TLC (stained with phosphomolybdic acid stain) and product containing fractions were pooled, concentrated to dryness, and analyzed by NMR spectroscopy.
Protein purification for crystallography experiments.
The plasmid containing ErTC-2 were chemically transformed into E. coli BL21 (DE3). The transformed cells were selected on LB-agar plates with 50 μg/mL kanamycin. A single colony was inoculated into a 10 mL LB starter culture and grown at 37 °C and 220 rpm shaking overnight. The starter culture was then inoculated into 2 L TB media, grown at 37 °C and 220 rpm shaking until OD600 reaches 1.0. The growth temperature was then reduced to 18 °C, and the culture was induced by IPTG at 0.4 mM final concentration. Cells were harvested 16 h after induction by centrifugation (4,500 xg, 10 mins, 4 °C). The cell pellets were flash-frozen in liquid nitrogen and saved in −80 °C freezer. Cell pellets were resuspended in lysis buffer (50 mM TRIS pH 7.8, 150 mM NaCl, 2 mM MgCl2, and 0.5 mM TCEP). The suspension was incubated with 0.2 mg/mL lysozyme and 0.5 mM PMSF on ice for 10 min and then sonicated for 4 min (65% amplitude, three-second interval between one-second sonication). The sonicated cell suspension was clarified by centrifugation (28,000 xg, 40 mins, 4 °C). The supernatant from the centrifugation was loaded on a 5 mL HisTrap-FF Ni-column (GE) pre-equilibrated with lysis buffer. The column was further washed with six column volumes (CV) of wash buffer (lysis buffer with 50 mM imidazole). The column was then eluted with step-gradients (lysis buffer with 100 mM, 150 mM, 200 mM, and 250 mM imidazole; five CV for each imidazole concentration). The eluate fractions with over 95% TC, assessed by SDS-PAGE gels, were concentrated by Amicon ultra centrifugal filters (10 kD molecular weight cut-off (MWCO), EMD-Millipore) to 5 mL. The protein was further purified on a Superdex 200 16/60 size exclusion column (GE Healthcare Life Sciences) in the lysis buffer. The monomeric peak from gel filtration was collected and concentrated by Amicon ultra centrifugal filters (10 kD MWCO) to a final concentration of 20-25 mg/mL. The protein was flash-frozen in 100 μL aliquots and stored in −80 °C freezer.
Crystallization of ErTC-2 and obtaining Cd(II)-ErTC-2 used for phasing.
ErTC-2 was crystallized by sitting drop crystallization at room temperature. 1.0 μL 20 mg/mL ErTC-2 in the lysis buffer was mixed with 1.0 μL well solution (0.10 M Tris pH 8.5, 28 % (w/v) PEG 3350, and 0.28 M MgCl2) to make a 2 μL sitting drop in a sealed well with 500 μL well solution. Transparent plate crystals grew overnight. The crystals were then transferred to a cryogenic solution (0.10 M Tris pH 8.5, 28 % (w/v) PEG 3350, 0.28 M MgCl2, and 20 % (v/v) ethylene glycol) and flash-cooled in liquid nitrogen. To generate crystals of ErTC-2 with Cd(II) bound, ErTC-2 crystals were transferred to a 2-uL drop containing 0.10 M Tris pH 8.5, 30 % (w/v) PEG 3350, 0.28 M MgCl2, and 10 mM CdCl2, and soaked for 2 hrs. Cd(II)-derivatized crystals were then transferred to a cryogenic solution (0.10 M Tris pH 8.5, 28 % (w/v) PEG 3350, 0.28 M MgCl2, and 20 % (v/v) ethylene glycol) and flash-cooled in liquid nitrogen.
Data collection and processing of ErTC-2.
A data set of Cd(II)-ErTC-2 was collected at UCSD X-ray Crystallography Facility on a Bruker Microstar APEX II CCD diffractometer equipped with Cu Ka radiation (1.54178 Å) at a temperature of 100 K. The data set was collected on one Cd(II)-ErTC-2 crystal in a single wedge of 4320º to capture Cd anomalous signal. The data were integrated using the Bruker SAINT 8.40A software and scaled using SADABS 2016/2. Data statistics are listed in Supplementary Table 11. Data set of ErTC-2 was collected at Stanford Synchrotron Radiation Lightsource (Menlo Park, California, USA) on beamline 12-2 using a Pilatus 6M PAD detector at a temperature of 100 K. Resolution cutoffs were chosen as completeness ~ 90%. Data were indexed, integrated, and scaled in XDS 56. Data statistics are listed in Supplementary Table 11.
Structure determination and refinement of ErTC-2.
The structure of ErTC-2 was phased by single-wavelength anomalous diffraction (SAD) with the dataset of Cd(II)-ErTC-2 using Crank257 pipeline implemented in CCP458. Seven heavy atom sites for cadmium were found using SHELXD59 and further refined in PEAKMAX. Initial phases were solved to 2.57-Å resolution with a FOM of 0.243. The initial phase was subjected to density modification by Parrot60 and Multicomb61. The density-modified experimental map, which has a FOM of 0.470, was sufficient for Buccaneer62 to build an initial model. Contiguous electron density for one terpene synthase monomer in the asymmetric unit allowed for further manual model building in Coot63. The completed model was then used for solving the high resolution ErTC-2 structure. The structure of ErTC-2 was determined to 1.58-Å resolution by rigid-body refinement from the protein part of the Cd(II)-ErTC-2 structure. The atomic coordinates and B-factors was iteratively refined in Phenix Refine64 with model building and manual adjustment of model in Coot. Anisotropic B factors for protein atoms were refinement in the last few rounds of refinement. Water molecules were added manually throughout model building using Fo-Fc electron density contoured to 3.0σ as criteria. The refinement statistics are in Supplementary Table 11, and the final model of the ErTC-2 structure contains residues listed in Supplementary Table 11.
Capnella DNA barcoding.
The mitochrondrial mutS (msh1) locus was used for DNA barcoding because it is a commonly used locus unique to corals65, allowing for clean amplification even in the presence of zooxanthellae. A single coral polyp was macerated in 50 μL dimethyl sulfoxide, which was used at a concentration of 1% in a PCR using primers 5’-GCCATTATGGTTAACTATTAC-’366 and 5’-TSGAGCAAAAGCCACTCC-3’67, and 5Prime HotMasterMix polymerase (Quanta Bio) according to the manufacturer’s instructions. The amplicon was Sanger sequenced using the former primer and verified to correspond to C. imbricata using NCBI MOLE-BLAST. For the sequence see Supplementary Note 2.
C. imbricata genome sequencing and assembly.
DNA was isolated from flash-frozen ground coral using the MoBio (now Qiagen) Powersoil kit according to the manufacturer’s instructions. The DNA was sheared in a cup-horn sonicator (Qsonica Q700 with 431MPX horn), libraries were generated using the NEBNext DNA Library Prep Master Mix Set according to the manufacturer’s instructions, the library was multiplexed with non-coral samples and sequenced on an Illumina MiSeq using v3 chemistry in paired-end 300 mode.
3379935 paired-end reads were recovered and trimmed using Trimmomatic 0.3968 using the parameters ILLUMINACLIP:TruSeq2-PE.fa:2:30:10:3:true CROP:299 HEADCROP:10 SLIDINGWINDOW:3:20 TRAILING:10 MINLEN:40. The 3097810 remaining paired reads and 246738 and 39341 unpaired forward and reverse reads respectively, were assembled using Spades 3.15.369, using kmer sizes 21,33,55. Higher kmer sizes did not appear to influence the quality of the assembly. Assigning taxonomy to contigs larger than 1 kbp using Autometa 1.0.270 classified 42%. Of those 42% classified, ~94% were of coral origin, ~2% was assigned non-coral animal, ~1% was assigned dinoflagellate and 0.8% was assigned bacteria. 80 Mbp of the assembly was contained in contigs larger 1 kbp, which is likely only about a quarter of the full genome (compare to 276 Mbp for D. gigantea27). Nonetheless, the full gene coding for CiTC-1 was contained on a 2.4 kbp contig. A coverage plot is included as Supplementary Figure 24 and contig statistics are shown in Supplementary Figure 25, both images were generated using Blobtools 2.6.1.
Analysis of C. imbricata terpene content by GC-MS.
~50 mg of C. imbricata was chopped fine with a razor blade and left to soak in 1 mL dichloromethane for 1 h. The suspension was filtered through a cotton pipette filter, washed with 1 mL distilled water, and the organic layer was used directly for GC-MS analysis on an Agilent 7890A gas chromatograph with Agilent 5975C mass spectrometer, using a HP-5MS 30 m × 0.25 mm, 0.25 μ column. Oven program was as follows: hold 60 °C for 3 min, 5 °C/min to 120 °C, 1 °C/min to 125 °C, hold 125 °C for 10 min, 10 °C/min to 300 °C, hold at 300 °C for 2.5 min. Flow rate: 0.44 mL/min, Injection volume: 1 μL, Splitless injection, Inlet temperature: 250 °C, MS transfer line temperature: 280 °C, MS source temperature 230 °C, MS quad temperature: 150 °C.
Data availability:
Shotgun sequencing reads for C. imbricata are available at NCBI-SRA under project accession PRJNA761189. The crystal structure of ErTC-2 has been deposited at PDB under the accession number 7S5L. NMR and EI-MS spectra of characterized compounds are available in the SI. Structure elucidation data are shown in Supplementary Note 1. Sequences used in this study are listed in Supplementary Note 2. Accession numbers of proteins used for HMM building are listed in Supplementary Table 1. Accession data for the literature genomic and transcriptomic data used in this study is summarized in Supplementary Table 2 and 3 respectively. Accession data for raw genomic data used for mapping is listed in Supplementary Table 4. Accession numbers for proteins used for the phylogenetic analysis are listed in Supplementary Table 10. The PDB accession numbers for reference protein structures in Fig. 3 are: 4lz0, 3p5r and 5ja0.
Extended Data
Extended Data Fig. 1. Radial depiction of the phylogenetic tree from Fig. 2.

The IDS clade is used as outgroup and bootstrap values for the branches are shown. The representative enzymes used for structure comparison in Fig. 3 are indicated by arrows.
Extended Data Fig. 2. Examples of octocoral derived terpenoid structures sharing a common terpene carbon backbone8.
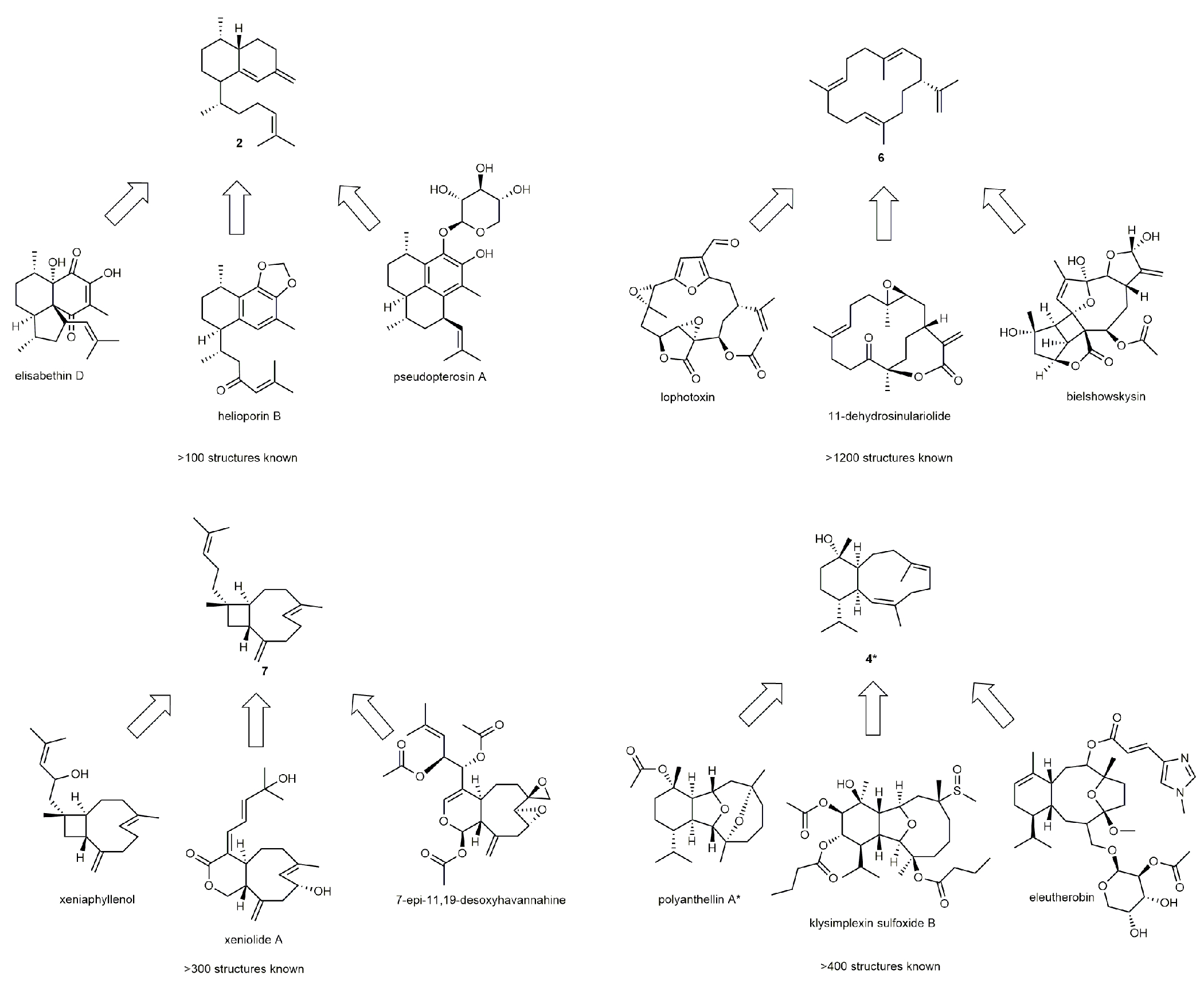
Structures marked with an asterisk have been found as both enantiomers.
Extended Data Fig. 3. Genetic context of octocoral TCs.

Mapped transcripts (top), gene annotations (middle) and mapped long-reads (bottom) for the indicated regions are shown. a) Context of XsTC-1 from Xenia sp. b) Context of DgTC-1 from D. gigantea. c) Context of DgTC-2 from D. gigantea. All genes are coded on long eukaryotic contigs (see Supplementary Table 4) with continuous long read coverage between 60 and 130 (Oxford Nanopore for a, PacBio for b and c). While the expression of DgTC-1 and DgTC-2 is low, XsTC-1 is highly expressed. For gene IDs see Supplementary Tables 5–7. The co-occurrence of two different bases in the same position (marked by colors) results from the heterozygosity of D. gigantea and Xenia sp.
Extended Data Fig. 4. Octocoral gene clusters.
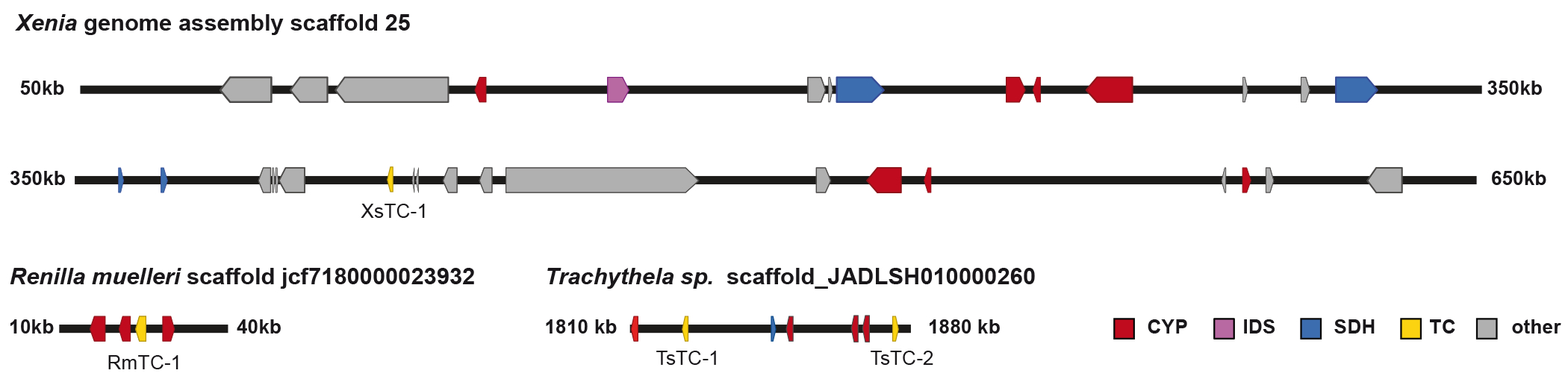
Genomic contigs from Xenia sp., Renilla muelleri and Trachythela sp. show the physical co-localization of TCs with genes coding for CYP and SHD enzymes. Genes on the Xenia scaffold that are marked in grey are either annotated as unknown proteins or show similarity to enzymes with non-biosynthetic functions.
CYP = cytochrome P450 monooxygenase, IDS = isoprenyl diphosphate synthase, SDH = short-chain dehydrogenase, TC = terpene cyclase.
Extended Data Fig. 5. Putative biochemical pathway from 7 to oxidized Xenia diterpenoids.
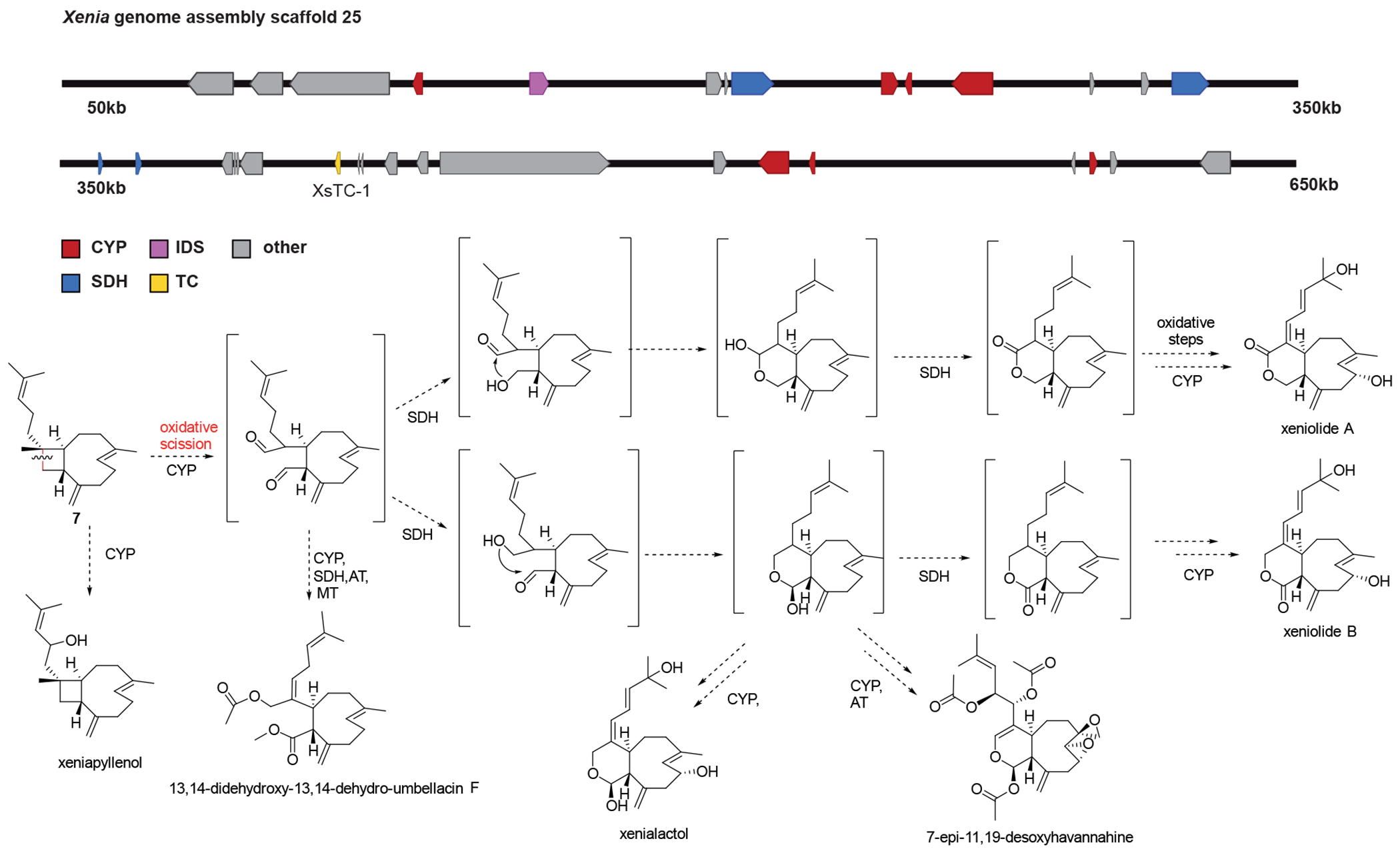
A: Putative terpenoid cluster including Cytochrome P450 genes and short chain dehydrogenase genes additionally to the characterized terpene cyclase XsTC-1, which produces 7. B: Putative overview how different compounds isolated from Xenia spp. and closely related corals could be explained from 7 using CYP and SHD chemistry. The oxidative cleavage of the cyclobutene ring in 7 has been proposed in the literature28. The end points of these putative biochemical pathways all represent isolated molecules from different Xenia spp.8 and could all result from CYP and SDH chemistry following the outlined scheme. Acylations and methylations are also observed in some compounds but could be the result of non-clustered specific or promiscuous enzymatic activity.
CYP = cytochrome P450 monooxygenase, IDS = isoprenyl diphosphate synthase, SDH = short-chain dehydrogenase, TC = terpene cyclase, MT = methyl transferase, AT = acyl transferase.
Extended Data Fig. 6. Analysis of a dichloromethane extract of Capnella imbricata by GC-MS.
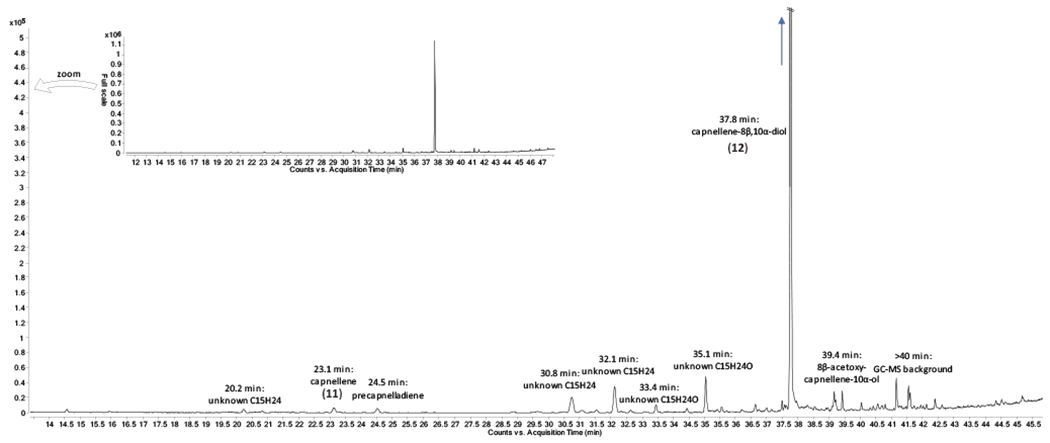
Signals for capnellene (11), capnellene-8β,10α-diol (12) and putative terpenoid compounds are labeled.
Supplementary Material
Supplementary Figures 1-21, Supplementary Notes 1 and 2, Supplementary Tables 1-11, Supplementary References 1-51
Acknowledgments:
We thank our UC San Diego colleagues W. Fenical, L. Aluwihare, and B. Duggan for critical feedback, access to GC-MS equipment, and assistance with NMR measurements, respectively, C. Delbeek (California Academy of Sciences) for access to C. imbricata, J. Keasling and J. Blake-Hedges (UC Berkeley) for assisting in genome sequencing, J. Bailey and the UCSD Crystallography Facility for collecting the X-ray diffraction data used for phasing, and J.P. Noel and G. Louie (Salk Institute for Biological Studies) for beamtime coordination. Use of the Stanford Synchrotron Radiation Lightsource is supported by the US Department of Energy, Office of Science, Office of Basic Energy Sciences under contract number DE-AC02-76SF00515, the Department of Energy Office of Biological and Environmental Research, and by the NIH and NIGMS (including P41GM103393). This work was supported by National Institutes of Health awards F32GM129960 to T.d.R. and R01GM085770 to B.S.M. and a Leopoldina postdoctoral fellowship (LPDS 2019-04) to I.B.
Footnotes
Competing interests:
The authors declare no competing interests.
References
- 1.Christianson DW Structural and Chemical Biology of Terpenoid Cyclases. Chem. Rev 117, 11570–11648 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Karunanithi PS & Zerbe P Terpene Synthases as Metabolic Gatekeepers in the Evolution of Plant Terpenoid Chemical Diversity. Front. Plant Sci 10, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dickschat JS Bacterial terpene cyclases. Nat. Prod. Rep 33, 87–110 (2016). [DOI] [PubMed] [Google Scholar]
- 4.Quin MB, Flynn CM & Schmidt-Dannert C Traversing the fungal terpenome. Nat. Prod. Rep 31, 1449–1473 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen X et al. Terpene synthase genes in eukaryotes beyond plants and fungi: Occurrence in social amoebae. Proc. Natl. Acad. Sci 113, 12132–12137 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schiff PB, Fant J & Horwitz SB Promotion of microtubule assembly in vitro by taxol. Nature 277, 665–667 (1979). [DOI] [PubMed] [Google Scholar]
- 7.Eckstein-Ludwig U et al. Artemisinins target the SERCA of Plasmodium falciparum. Nature 424, 957–961 (2003). [DOI] [PubMed] [Google Scholar]
- 8.Lyu C et al. CMNPD: a comprehensive marine natural products database towards facilitating drug discovery from the ocean. Nucleic Acids Res. 49, D509–D515 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Giordano G et al. Volatile secondary metabolites as aposematic olfactory signals and defensive weapons in aquatic environments. Proc. Natl. Acad. Sci 114, 3451–3456 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Berrue F & Kerr RG Diterpenes from gorgonian corals. Nat. Prod. Rep 26, 681–710 (2009). [DOI] [PubMed] [Google Scholar]
- 11.Li G, Dickschat JS & Guo Y-W Diving into the world of marine 2,11-cyclized cembranoids: a summary of new compounds and their biological activities. Nat. Prod. Rep 37, 1367–1383 (2020). [DOI] [PubMed] [Google Scholar]
- 12.Look SA, Fenical W, Jacobs RS & Clardy J The pseudopterosins: anti-inflammatory and analgesic natural products from the sea whip Pseudopterogorgia elisabethae. Proc. Natl. Acad. Sci 83, 6238–6240 (1986). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hassan HM et al. Pachycladins A−E, Prostate Cancer Invasion and Migration Inhibitory Eunicellin-Based Diterpenoids from the Red Sea Soft Coral Cladiella pachyclados. J. Nat. Prod 73, 848–853 (2010). [DOI] [PubMed] [Google Scholar]
- 14.Fenical W, Okuda RK, Bandurraga MM, Culver P & Jacobs RS Lophotoxin: a novel neuromuscular toxin from Pacific sea whips of the genus Lophogorgia. Science 212, 1512–1514 (1981). [DOI] [PubMed] [Google Scholar]
- 15.Wilson MC et al. An environmental bacterial taxon with a large and distinct metabolic repertoire. Nature 506, 58–62 (2014). [DOI] [PubMed] [Google Scholar]
- 16.Smith TE et al. Accessing chemical diversity from the uncultivated symbionts of small marine animals. Nat. Chem. Biol 14, 179–185 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sudek S et al. Identification of the Putative Bryostatin Polyketide Synthase Gene Cluster from “Candidatus Endobugula sertula”, the Uncultivated Microbial Symbiont of the Marine Bryozoan Bugula neritina. J. Nat. Prod 70, 67–74 (2007). [DOI] [PubMed] [Google Scholar]
- 18.Torres JP & Schmidt EW The biosynthetic diversity of the animal world. J. Biol. Chem 294, 17684–17692 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shinzato C et al. Using the Acropora digitifera genome to understand coral responses to environmental change. Nature 476, 320–323 (2011). [DOI] [PubMed] [Google Scholar]
- 20.McCauley EP et al. Highlights of marine natural products having parallel scaffolds found from marine-derived bacteria, sponges, and tunicates. J. Antibiot 73, 504–525 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mydlarz LD, Jacobs RS, Boehnlein J & Kerr RG Pseudopterosin Biosynthesis in Symbiodinium sp., the Dinoflagellate Symbiont of Pseudopterogorgia elisabethae. Chem. Biol 10, 1051–1056 (2003). [DOI] [PubMed] [Google Scholar]
- 22.Frenz-Ross JL, Enticknap JJ & Kerr RG The Effect of Bleaching on the Terpene Chemistry of Plexaurella fusifera: Evidence that Zooxanthellae Are Not Responsible for Sesquiterpene Production. Mar. Biotechnol 10, 572–578 (2008). [DOI] [PubMed] [Google Scholar]
- 23.Kohl AC & Kerr RG Identification and characterization of the pseudopterosin diterpene cyclase, elisabethatriene synthase, from the marine gorgonian, Pseudopterogorgia elisabethae. Arch. Biochem. Biophys 424, 97–104 (2004). [DOI] [PubMed] [Google Scholar]
- 24.Wei G et al. Terpene Biosynthesis in Red Algae Is Catalyzed by Microbial Type But Not Typical Plant Terpene Synthases. Plant Physiol. 179, 382–390 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yamada Y et al. Terpene synthases are widely distributed in bacteria. Proc. Natl. Acad. Sci 112, 857–862 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mistry J et al. Pfam: The protein families database in 2021. Nucleic Acids Res. 49, D412–D419 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jeon Y et al. The Draft Genome of an Octocoral, Dendronephthya gigantea. Genome Biol. Evol 11, 949–953 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huber T, Weisheit L & Magauer T Synthesis of Xenia diterpenoids and related metabolites isolated from marine organisms. Beilstein J. Org. Chem 11, 2521–2539 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hu M, Zheng X, Fan C-M & Zheng Y Lineage dynamics of the endosymbiotic cell type in the soft coral Xenia. Nature 582, 534–538 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Krissinel E & Henrick K Inference of Macromolecular Assemblies from Crystalline State. J. Mol. Biol 372, 774–797 (2007). [DOI] [PubMed] [Google Scholar]
- 31.Li R et al. Reprogramming the Chemodiversity of Terpenoid Cyclization by Remolding the Active Site Contour of epi-Isozizaene Synthase. Biochemistry 53, 1155–1168 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Köksal M, Jin Y, Coates RM, Croteau R & Christianson DW Taxadiene synthase structure and evolution of modular architecture in terpene biosynthesis. Nature 469, 116–120 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Park J, Zielinski M, Magder A, Tsantrizos YS & Berghuis AM Human farnesyl pyrophosphate synthase is allosterically inhibited by its own product. Nat. Commun 8, 14132 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lancaster J et al. De novo formation of an aggregation pheromone precursor by an isoprenyl diphosphate synthase-related terpene synthase in the harlequin bug. Proc. Natl. Acad. Sci 115, E8634–E8641 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McFadden CS et al. Phylogenomics, Origin, and Diversification of Anthozoans (Phylum Cnidaria). Syst. Biol 70, 635–647 (2021). [DOI] [PubMed] [Google Scholar]
- 36.Morris JL et al. The timescale of early land plant evolution. Proc. Natl. Acad. Sci 115, E2274–E2283 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Medema MH, de Rond T & Moore BS Mining genomes to illuminate the specialized chemistry of life. Nat. Rev. Genet 22, 553–571 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nützmann H-W, Huang A & Osbourn A Plant metabolic clusters – from genetics to genomics. New Phytologist 211, 771–789 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.De La Peña R & Sattely ES Rerouting plant terpene biosynthesis enables momilactone pathway elucidation. Nat. Chem. Biol 17, 205–212 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Morris LA, Jaspars M, Adamson K, Woods S & Wallace HM The capnellenes revisited: New structures and new biological activity. Tetrahedron 54, 12953–12958 (1998). [Google Scholar]
- 41.Jean Y-H et al. Capnellene, a natural marine compound derived from soft coral, attenuates chronic constriction injury-induced neuropathic pain in rats. Br. J. Pharmacol 158, 713–725 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ayanoglu E, Gebreyesus T, Beechan CM, Djerassi C & Kaisin M Terpenoids LXXV. Δ9(12)-capnellene, a new sesquiterpene hydrocarbon from the soft coral capnella imbricata. Tetrahedron Lett. 19, 1671–1674 (1978). [Google Scholar]
- 43.DeLeo DM et al. Gene expression profiling reveals deep-sea coral response to the Deepwater Horizon oil spill. Mol. Ecol 27, 4066–4077 (2018). [DOI] [PubMed] [Google Scholar]
- 44.Todaro D & Watson GM Force-dependent discharge of nematocysts in the sea anemone Haliplanella luciae (Verrill). Biol. Open 1, 582–587 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Methods References
- 45.Woodside AB, Huang Z, Poulter CD Trisammonium geranyl diphosphate, Org. Synth 66, 211 (1988). [Google Scholar]
- 46.Jo Davisson V, Woodside AB & Dale Poulter CD, Synthesis of allylic and homoallylic isoprenoid pyrophosphates. in Methods Enzymol. 110, 130–144 (1985). [DOI] [PubMed] [Google Scholar]
- 47.Eddy SR Accelerated Profile HMM Searches. PLOS Comput. Biol 7, e1002195 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.SRA-Tools - NCBI. http://ncbi.github.io/sra-tools/.
- 49.Grabherr MG et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol 29, 644–652 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Haas BJ et al. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat. Protoc 8, 1494–1512 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Priyam A et al. Sequenceserver: A Modern Graphical User Interface for Custom BLAST Databases. Mol. Biol. Evol 36, 2922–2924 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lassmann T, Frings O & Sonnhammer ELL Kalign2: high-performance multiple alignment of protein and nucleotide sequences allowing external features. Nucleic Acids Res. 37, 858–865 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kumar S, Stecher G, Li M, Knyaz C & Tamura K MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol 35, 1547–1549 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Robert X & Gouet P Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 42, W320–W324 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Minh BQ et al. IQ-TREE 2: New Models and Efficient Methods for Phylogenetic Inference in the Genomic Era. Mol. Biol. Evol 37, 1530–1534 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kabsch W XDS. Acta Cryst. D 66, 125–132 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Skubák P & Pannu NS Automatic protein structure solution from weak X-ray data. Nat. Commun 4, 2777 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Winn MD et al. Overview of the CCP4 suite and current developments. Acta Cryst. D 67, 235–242 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schneider TR & Sheldrick GM Substructure solution with SHELXD. Acta Cryst. D 58, 1772–1779 (2002). [DOI] [PubMed] [Google Scholar]
- 60.Cowtan K Recent developments in classical density modification. Acta Cryst. D 66, 470–478 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Skubák P, Waterreus W-J & Pannu NS Multivariate phase combination improves automated crystallographic model building. Acta Cryst. D 66, 783–788 (2010). [DOI] [PubMed] [Google Scholar]
- 62.Cowtan K The Buccaneer software for automated model building. 1. Tracing protein chains. Acta Cryst. D 62, 1002–1011 (2006). [DOI] [PubMed] [Google Scholar]
- 63.Emsley P, Lohkamp B, Scott WG & Cowtan K Features and development of Coot. Acta Cryst. D 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Liebschner D et al. Macromolecular structure determination using X-rays, neutrons and electrons: recent developments in Phenix. Acta Cryst. D 75, 861–877 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pont-Kingdon GA et al. A coral mitochondrial mutS gene. Nature 375, 109–111 (1995). [DOI] [PubMed] [Google Scholar]
- 66.France SC & Hoover LL DNA sequences of the mitochondrial COI gene have low levels of divergence among deep-sea octocorals (Cnidaria: Anthozoa). Hydrobiologia 471, 149–155 (2002). [Google Scholar]
- 67.Sánchez JA, McFadden CS, France SC & Lasker HR Molecular phylogenetic analyses of shallow-water Caribbean octocorals. Mar. Biol 142, 975–987 (2003). [Google Scholar]
- 68.Bolger AM, Lohse M & Usadel B Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bankevich A et al. SPAdes: A New Genome Assembly Algorithm and Its Applications to Single-Cell Sequencing. J. Comp. Biol 19, 455–477 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Miller IJ et al. Autometa: automated extraction of microbial genomes from individual shotgun metagenomes. Nucleic Acids Res. 47, e57 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures 1-21, Supplementary Notes 1 and 2, Supplementary Tables 1-11, Supplementary References 1-51
Data Availability Statement
Shotgun sequencing reads for C. imbricata are available at NCBI-SRA under project accession PRJNA761189. The crystal structure of ErTC-2 has been deposited at PDB under the accession number 7S5L. NMR and EI-MS spectra of characterized compounds are available in the SI. Structure elucidation data are shown in Supplementary Note 1. Sequences used in this study are listed in Supplementary Note 2. Accession numbers of proteins used for HMM building are listed in Supplementary Table 1. Accession data for the literature genomic and transcriptomic data used in this study is summarized in Supplementary Table 2 and 3 respectively. Accession data for raw genomic data used for mapping is listed in Supplementary Table 4. Accession numbers for proteins used for the phylogenetic analysis are listed in Supplementary Table 10. The PDB accession numbers for reference protein structures in Fig. 3 are: 4lz0, 3p5r and 5ja0.


