Abstract
Background
Keratins are heteropolymeric proteins that form the intermediate filament cytoskeleton in epithelial cells. The common basic structure of all keratins is organized in a central α-helical rod domain flanked by nonhelical, variable head and tail regions. Most mutations in keratins are found in the central α-helical rod domain. Keratin 9 (K9) is expressed only in the suprabasal layers of palmoplantar epidermis. Mutations in the keratin 9 gene (KRT9) have been shown to cause epidermolytic palmoplantar keratoderma (EPPK; OMIM 144200), an autosomal dominant genodermatosis characterized clinically by diffuse hyperkeratosis limited to palms and soles and histologically by epidermolysis in suprabasal layers of the epidermis.
Aim
To elucidate the genetic basis of EPPK in five Pakistani families.
Methods
Using microsatellite markers localized to the areas around the type I keratin gene cluster on chromosome 17q21, genotyping of these families was performed, followed by sequencing of the KRT9 gene.
Results
The analysis resulted in the identification of two novel (p.M157K and p.Y454H) and two recurrent (p.M157T and p.R163Q) mutations in the KRT9 of all five families. All mutations occurred within the highly conserved helix initiation or termination motif of K9.
Conclusion
The affected members of all five families possess mutations in the KRT9 gene that severely affect heterodimer formation with the type II keratin partner. The results of our study further underscore the crucial role of K9 protein in the palmoplantar epidermis.
Introduction
Keratins are a large family of structural proteins that constitute the intermediate filament cytoskeleton present in epithelial cells. They form heteropolymers consisting of heterodimers made up of type I (acidic) and type II (basic to neutral) keratin, which are expressed in a cell type-specific manner and are characteristic of different tissues at various stages of differentiation.1 Keratins are the major component of the intermediate-filament cytoskeleton in epithelial cells, and provide stability to these cells under conditions of mechanical stress. In humans, the type I and II keratin gene clusters are located at chromosome 17q21.2 and 12q13.13, respectively, and 54 functional keratin genes have been identified to date.2 Many congenital skin diseases have been attributed to keratin mutations, such as epidermolysis bullosa simplex (OMIM 131900; genes KRT5 and KRT14) and epidermolytic hyperkeratosis (OMIM 113800; genes KRT1 and KRT10).3
Palmoplantar keratoderma (PPK) is a heterogeneous group of skin disorders characterized by severe hyperkeratosis of the epidermis of the palms and soles. Epidermolytic PPK (EPPK; OMIM 144200), is an autosomal dominant genodermatosis within this family, characterized clinically by diffuse skin thickening limited to palms and soles with a well-circumscribed erythematous border, and histologically by epidermolytic hyperkeratosis, which consists of perinuclear vacuolization and cytolysis in suprabasal layers of the epidermis.4 EPPK has been mapped to chromosome 17q21 where the type I keratin gene cluster is located.5 Subsequently, mutations in the keratin 9 gene (KRT9) were identified in EPPK families.6 The keratin 9 (K9) protein is a type I keratin exclusively expressed in the suprabasal keratinocytes of the palmoplantar epidermis.5 The amino acid sequence of the type I KRT9 gene is composed of three typical structures: a head domain (153 residues), an α-helical coiled-coil-forming rod (306 residues) and a tail domain (163 residues), which are flanked by nonhelical, variable end domains.7 Pathogenic mutations have now been identified in 19 different keratin genes, all of which produce a cell-fragility phenotype. In general, most of the reported mutations in keratins can be detected within the highly conserved N-terminal and C-terminal motifs of the α-helical rod domains.8 To date, all reported mutations in the KRT9 gene occur within sequences corresponding to this conserved α-helix region at the beginning and the end of the rod.9 In particular, the highly conserved coil 1A of the rod domain is crucial for the formation of intermediate filaments and is thought to be important for heterodimerization.
In this study, we examined the molecular basis of EPPK in five Pakistani families, and identified four distinct mutations in the KRT9 gene of all five families within the coiled-coil motifs in the rod domain segments 1A and 2B.
Methods
The study was approved by the institutional ethics board and carried out in adherence to the principles of the Declaration of Helsinki. Informed consent was obtained from all participants.
Subjects
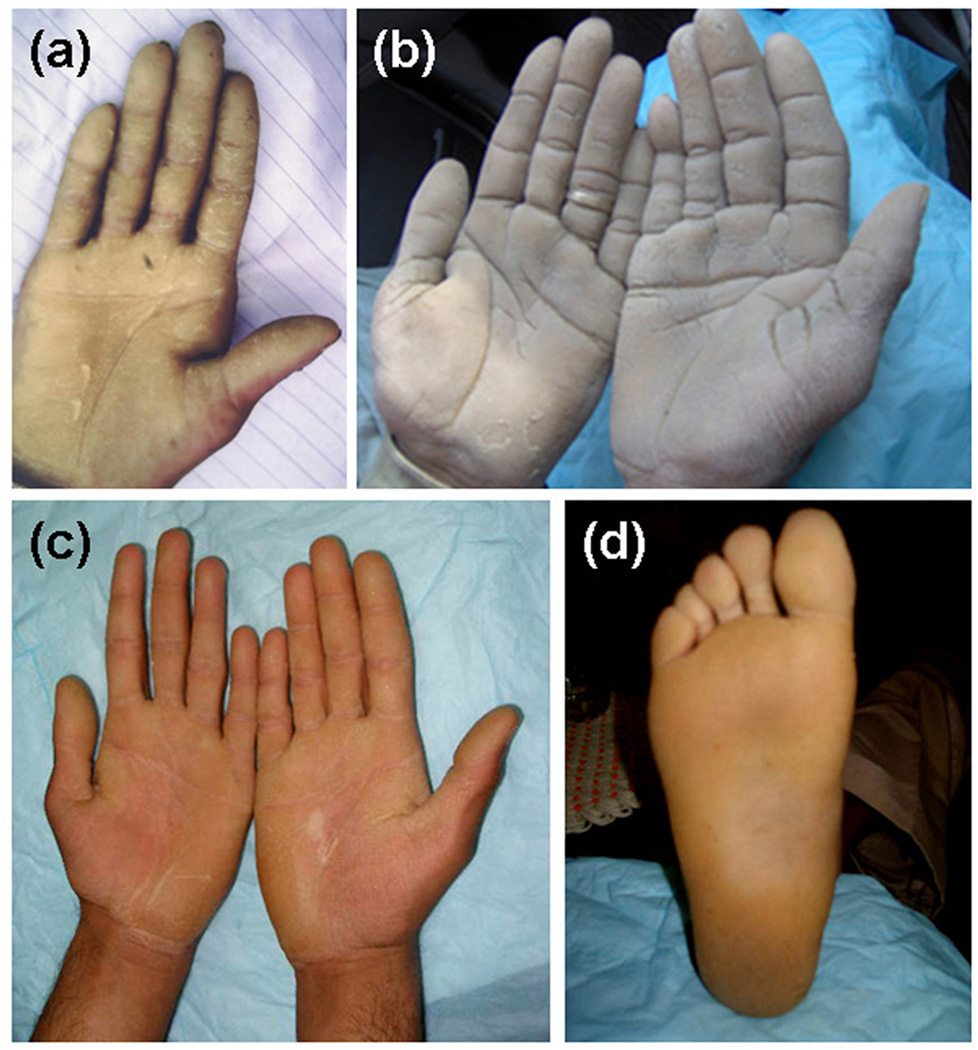
We analysed five families of Pakistani origin affected with PPK, whose pedigrees show a clear autosomal dominant inheritance pattern (Fig. 1). All affected members have diffuse hyperkeratosis with an erythematous border on the palms and soles (Fig. 2). Detailed clinical examination found no other abnormalities and there was no evidence of a high frequency of internal malignancies in these families. Skin biopsies were not available from any of the affected members.
Figure 1.
Pedigrees of five Pakistani families with epidermolytic palmoplantar keratoderma (EPPK). Haplotypes for three microsatellite markers situated around the type I keratin gene cluster are also shown.The haplotype segregating with the disease is in red.
Figure 2.
Affected members of families (a) 1 (III-5), (b) 4 (III-6) and (c,d) 5 (III-3) have diffuse palmoplantar hyperkeratosis with an erythematous border.
Genotyping
Peripheral blood samples were collected from members of these families and 100 population-matched unrelated healthy controls into tubes containing ethylene diamine tetraacetic acid (EDTA). Genomic DNA was isolated from these samples according to standard techniques. We performed genotyping only on families 1–4; we did not perform genotyping of family 5 because we could obtain DNA from only one member (III-3) of this family. Genomic DNA was amplified by PCR using primers for three polymorphic markers (D17S1818, D17S800 and D17S932) around the type I keratin gene cluster on chromosome 17q21 and three markers (D12S1629, D12S398 and D12S1651) around the type II keratin gene cluster on chromosome 12q13. The amplification conditions for each PCR assay were 94 °C for 2 min, followed by 35 cycles of 94 °C for 30 s, 55 °C for 30 s and 72 °C for 30 s, with a final extension at 72 °C for 7 min. PCR products were run on 8% polyacrylamide gels, and genotypes were assigned after visual inspection.
Mutation analysis
Using the genomic DNA from members of all five families, all exons of the KRT9 gene with the adjacent sequences of the exon–intron borders were amplified by PCR, using primers and conditions described previously.10,11 The amplified PCR products were directly sequenced in an automated sequencer (ABI Prism 310; Applied Biosystems, Foster City, CA, USA), using a commercial reaction kit (ABI Prism Big Dye Terminator Cycle Sequencing Ready Reaction Kit; PE Applied Biosystems).
Screening assays for keratin 9 gene mutations
As the mutation p.M157K in the KRT9 abolishes an NlaIII restriction enzyme site, the PCR product of exon 1 of the KRT9 was digested with this enzyme at 37 °C overnight and separated in 1.5% agarose gels. This enzyme digests the wild-type allele 9429 bp) into two bands of 266 bp and 163 bp fragments, while leaving the mutant allele undigested. To analyse the mutation p.Y454H, exon 6 of KRT9 was amplified by PCR (forward primer 5′-AGTCTTTGCAGCCCAGGAGT-3′; reverse primer 5′-GTCTTCCTGGCCTCCCTCAAGGAGGTTGTGCT-3′. The G→C substitution (shown in bold) type was introduced into the reverse primer to generate the BseYI restriction enzyme site only in the mutant allele, resulting in two bands of 265 and 299 bp. The amplification conditions were 94 °C for 2 min, followed by 35 cycles of 94 °C for 30 s, 60 °C for 30 s and 72 °C for 30 s, with a final extension at 72 °C for 7 min. The PCR products were digested with BseYI at 37 °C overnight, and separated in a 1.5% agarose gel.
Results
Four of the families analysed had linkage to the type I keratin gene cluster
As shown in Fig. 1, all affected members in families 1–4 were heterozygous for the same haplotype, suggesting that all four families analysed had linkage to the type I keratin gene cluster. Notably, the disease haplotype was different for all these families (Fig. 1). We also analysed markers around the type II keratin gene cluster on chromosome 12q13, but did not find linkage to this region in any of the families (data not shown).
Identification of four distinct mutations in the keratin 9 gene from the five families
We found pathogenic mutations in the KRT9 gene in families 1–4.
Affected members in family 1 have a novel heterozygous missense mutation consisting of a T→A transition in exon 1 (c.470T→A), resulting in a change of methionine to lysine at codon 157 (designated p.M157K; Fig. 3a). Screening with the restriction enzyme NlaIII found that all affected family members carry the mutation, but it was not found in any of the unaffected healthy family members or in 100 unrelated controls (Fig. 3b).
Figure 3.
(a) A novel mutation p.M157K in family 1. (b) Screening assay for the mutation p.M157K, resulting in digestion of the wild-type allele (429 bp) into two bands of 266 bp and 163 bp, while the mutant allele is undigested. (c) A novel mutation p.Y454H in family 4. (d) Screening assay for the mutation p.Y454H, resulting in digestion of the mutant allele only. MWM, molecular weight marker; C, control.
Sequencing of affected members in family 2 found that they were heterozygous for a T→C transition in exon 1 (c.470T→C), changing methionine to threonine at codon 157 (designated p.M157T; data not shown). This mutation was previously identified in an Irish family with EPPK10 and is a recurrent mutation.
Family 3 has a heterozygous G→A transition in exon 1 (c.488G→A), which changes arginine to glutamine at codon 163 (p.R163Q) (data not shown). This is also a recurrent mutation which was previously found in several families with EPPK.6,10,12
Affected members in family 4 possess a novel heterozygous T→C transition in exon 6 (c.1360T→C), resulting in the conversion of tyrosine to histidine at codon 454 (p.Y454H) (Fig. 3c).
Screening with the restriction enzyme BseYI was carried out to exclude a common nonconsequential polymorphism (Fig. 3d).
Finally, as the phenotype of family 5 is similar to that of the other four families with KRT9 mutations, we also sequenced this gene from the single member of family 5 for whom we had a sample, and identified a heterozygous mutation p.R163Q, which is identical to the mutation found in family 3 (data not shown).
Discussion
In this study, we identified five families of Pakistani origin with an autosomal dominant form of PPK. All the affected members have well-demarcated diffuse skin thickening over palms and soles with an erythematous border (Fig. 2), which is a characteristic feature of EPPK. Although most cases of EPPK are known to be caused by mutations in the KRT9 gene, several cases of EPPK have recently been reported in association with mutations in the KRT1 gene on chromosome 12q13.13.13–16 In these families, the results of genotyping using microsatellite markers around the type I keratin gene cluster on chromosome 17q were consistent with linkage to this region of all families analysed (Fig. 1), and subsequent mutation analysis resulted in the identification of four distinct pathogenic mutations in the KRT9 gene of these families (Fig. 3a–d and data not shown). Thus, we concluded that the affected family members have EPPK caused by a KRT9 mutation. To our knowledge, this is the first report of KRT9 mutations in Pakistani patients.
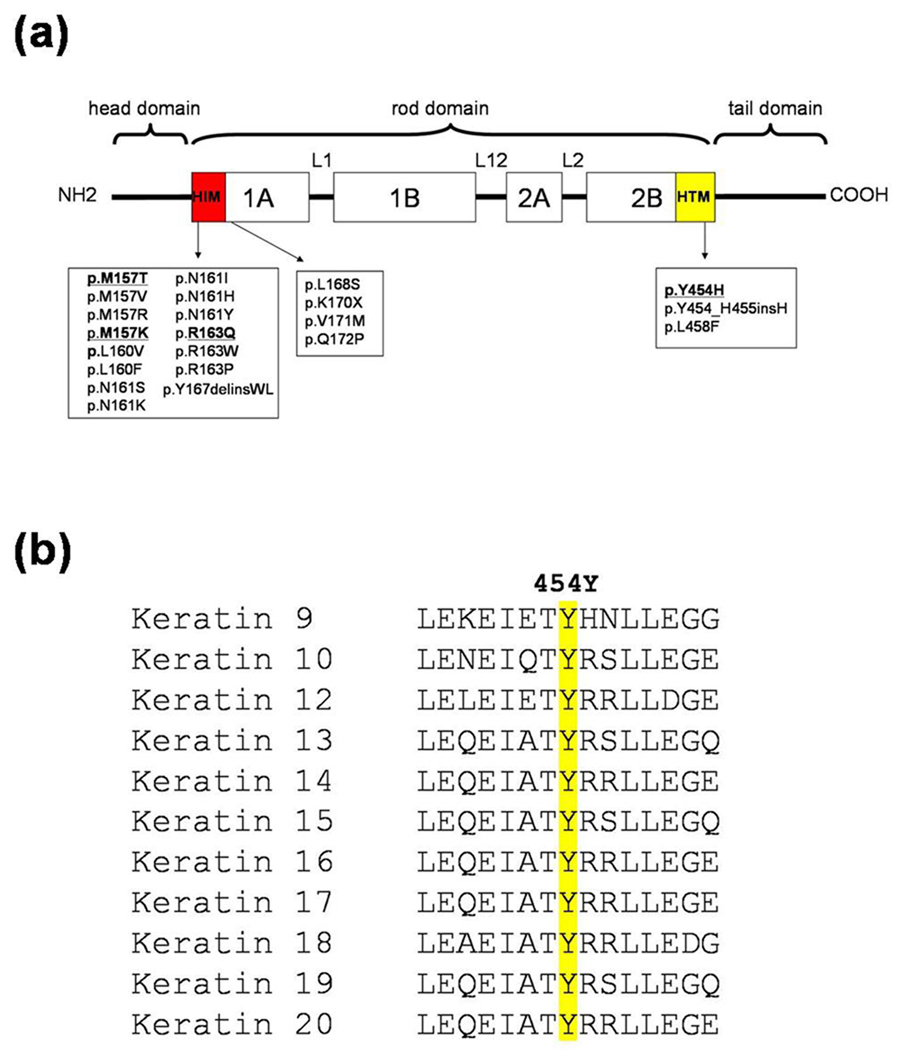
All keratins share a common structural organization consisting of a central rod domain flanked by nonhelical head and tail domains. The rod domain is composed of four α-helical segments (1A, 1B, 2A and 2B) which are connected via three nonhelical linkers (L1, L2 and L12). Two highly conserved regions at the beginning of the 1A domain and end of the 2B domain, termed the helix initiation motif (HIM) and helix termination motif (HTM), respectively, are known to be vital for heterodimer formation in keratins.17 To date, most mutations in the KRT9 have been found to cluster either within the HIM or within the amino acid residues close to the HIM (Fig. 4). In particular, several distinct missense mutations have been reported in each of the amino acid residues 157M, 160 L, 161N and 163R within the HIM, and most have been found to be recurrent in different populations (Human Intermediate Filament Database; www.interfil.org) (Fig. 4). Consistent with the previous findings, we identified one novel mutation, p.M157K, and two recurrent mutations, p.M157T and p.R163Q, in these mutational hot spots in our families 1–3 and 5 with EPPK (Fig. 3a–d). Furthermore, we also identified a novel mutation p.Y454H in family 4 (Figs 3e,f), which is only the third mutation found in the HTM of the KRT9 (Fig. 4a).9,18 The tyrosine at codon 454 of K9 is highly conserved between type I keratins (Fig. 4b). In addition, a Y→H transition at the corresponding position of keratin 14 has been shown to cause epidermolysis bullosa simplex.19 Collectively, the nonconserved mutation p.Y454H in K9 is predicted to severely disrupt the assembly of the keratin intermediate filaments.
Figure 4.
(a) Schematic representation of the K9 protein and mutations in the KRT9 gene. Mutations identified in this study are indicated in bold and underlined. HIM, helix initiation motif. HTM, helix termination motif. (b) Multiple amino acid sequence alignment of helix termination motif of type I keratins. The highly conserved tryptophan residue is in yellow.
Our findings expand the allelic series of the KRT9 mutations underlying EPPK and shed further light upon regions within the α-helical coiled-coil-forming rod that seem to be less tolerant of mutations.
Acknowledgements
We are especially grateful to the patients and their families for their participation in this study. This study was supported by USPHS NIH grant from NIH/NIAMS RO1AR44924 (to A.M.C).
Footnotes
Conflict of interest: none declared.
References
- 1.Coulombe PA, Omary MB. ‘Hard’ and ‘soft’ principles defining the structure, function and regulation of keratin intermediate filaments. Curr Opin Cell Biol. 2002;14:110–122. doi: 10.1016/s0955-0674(01)00301-5. [DOI] [PubMed] [Google Scholar]
- 2.Schweizer J, Bowden PE, Coulombe PA, et al. New consensus nomenclature for mammalian keratins. J Cell Biol. 2006;174:169–174. doi: 10.1083/jcb.200603161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lane EB, McLean WH. Keratins and skin disorders. J Pathol. 2004;204:355–366. doi: 10.1002/path.1643. [DOI] [PubMed] [Google Scholar]
- 4.Kon A, Itagaki K, Yoneda K, Takagaki K. A novel mutation of keratin 9 gene (R162P) in a Japanese family with epidermolytic palmoplantar keratoderma. Arch Dermatol Res. 2005;296:375–378. doi: 10.1007/s00403-004-0534-8. [DOI] [PubMed] [Google Scholar]
- 5.Reis A, Kuster W, Eckardt R, et al. Mapping of a gene for epidermolytic palmoplantar keratoderma to the region of the acidic keratin gene cluster at 17q12-q21. Hum Genet. 1992;90:113–116. doi: 10.1007/BF00210752. [DOI] [PubMed] [Google Scholar]
- 6.Reis A, Hennies HC, Langbein L, et al. Keratin 9 gene mutations in epidermolytic palmoplantar keratoderma (EPPK) Nat Genet. 1994;6:174–179. doi: 10.1038/ng0294-174. [DOI] [PubMed] [Google Scholar]
- 7.Langbein L, Heid HW, Moll I, et al. Molecular characterization of the body site-specific human epidermal cytokeratin 9: cDNA cloning, amino acid sequence, and tissue specificity of gene expression. Differentiation. 1993;55:57–71. doi: 10.1111/j.1432-0436.1993.tb00033.x. [DOI] [PubMed] [Google Scholar]
- 8.Smith F. The molecular genetics of keratin disorders. Am J Clin Dermatol. 2003;4:347–364. doi: 10.2165/00128071-200304050-00005. [DOI] [PubMed] [Google Scholar]
- 9.Coleman CM, Munro CS, Smith FJ, et al. Epidermolytic palmoplantar keratoderma due to a novel type of keratin mutation, a 3-bp insertion in the keratin 9 helix termination motif. Br J Dermatol. 1999;140:486–490. doi: 10.1046/j.1365-2133.1999.02715.x. [DOI] [PubMed] [Google Scholar]
- 10.Covello SP, Irvine AD, McKenna KE, et al. Mutations in keratin K9 in kindreds with epidermolytic palmoplantar keratoderma and epidemiology in Northern Ireland. J Invest Dermatol. 1998;111:1207–1209. doi: 10.1046/j.1523-1747.1998.00445.x. [DOI] [PubMed] [Google Scholar]
- 11.Zhang XN, He XH, Lai Z, et al. An insertion-deletion mutation in keratin 9 in three Chinese families with epidermolytic palmoplantar keratoderma. Br J Dermatol. 2005;152:804–806. doi: 10.1111/j.1365-2133.2005.06477.x. [DOI] [PubMed] [Google Scholar]
- 12.Wennerstrand LM, Klingberg MH, Hofer PA, et al. A de novo mutation in the keratin 9 gene in a family with epidermolytic palmoplantar keratoderma from northern Sweden. Acta Derm Venereol. 2003;83:135–137. doi: 10.1080/00015550310007517. [DOI] [PubMed] [Google Scholar]
- 13.Hatsell SJ, Eady RA, Wennerstrand L, et al. Novel splice site mutation in keratin 1 underlies mild epidermolytic palmoplantar keratoderma in three kindreds. J Invest Dermatol. 2001;116:606–609. doi: 10.1046/j.1523-1747.2001.13041234.x. [DOI] [PubMed] [Google Scholar]
- 14.Terron-Kwiatkowski A, Paller AS, Compton J, et al. Two cases of primarily palmoplantar keratoderma associated with novel mutations in keratin 1. J Invest Dermatol. 2002;119:966–971. doi: 10.1046/j.1523-1747.2002.00186.x. [DOI] [PubMed] [Google Scholar]
- 15.Terron-Kwiatkowski A, Terrinoni A, Didona B, et al. Atypical epidermolytic palmoplantar keratoderma presentation associated with a mutation in the keratin 1 gene. Br J Dermatol. 2004;150:1096–1103. doi: 10.1111/j.1365-2133.2004.05967.x. [DOI] [PubMed] [Google Scholar]
- 16.Terron-Kwiatkowski A, van Steensel MA, van Geel M, et al. Mutation S233L in the 1B domain of keratin 1 causes epidermolytic palmoplantar keratoderma with ‘tonotubular’ keratin. J Invest Dermatol. 2006;126:607–613. doi: 10.1038/sj.jid.5700152. [DOI] [PubMed] [Google Scholar]
- 17.Lu X, Lane EB. Retrovirus-mediated transgenic keratin expression in cultured fibroblasts: specific domain functions in keratin stabilization and filament formation. Cell. 1990;62:681–696. doi: 10.1016/0092-8674(90)90114-t. [DOI] [PubMed] [Google Scholar]
- 18.Kon A, Ito N, Kudo Y, et al. L457F missense mutation within the 2B rod domain of keratin 9 in a Japanese family with epidermolytic palmoplantar keratoderma. Br J Dermatol. 2006;155:624–626. doi: 10.1111/j.1365-2133.2006.07358.x. [DOI] [PubMed] [Google Scholar]
- 19.Hut PH, v d Vlies P, Jonkman MF, et al. Exempting homologous pseudogene sequences from polymerase chain reaction amplification allows genomic keratin 14 hotspot mutation analysis. J Invest Dermatol. 2000;114:616–619. doi: 10.1046/j.1523-1747.2000.00928.x. [DOI] [PubMed] [Google Scholar]