Abstract
Multiplex immunofluorescence (MxIF) is an emerging imaging technology whose downstream molecular analytics highly rely upon the effectiveness of cell segmentation. In practice, multiple membrane markers (e.g., NaKATPase, PanCK and β-catenin) are employed to stain membranes for different cell types, so as to achieve a more comprehensive cell segmentation since no single marker fits all cell types. However, prevalent watershed-based image processing might yield inferior capability for modeling complicated relationships between markers. For example, some markers can be misleading due to questionable stain quality. In this paper, we propose a deep learning based membrane segmentation method to aggregate complementary information that is uniquely provided by large scale MxIF markers. We aim to segment tubular membrane structure in MxIF data using global (membrane markers z-stack projection image) and local (separate individual markers) information to maximize topology preservation with deep learning. Specifically, we investigate the feasibility of four SOTA 2D deep networks and four volumetric-based loss functions. We conducted a comprehensive ablation study to assess the sensitivity of the proposed method with various combinations of input channels. Beyond using adjusted rand index (ARI) as the evaluation metric, which was inspired by the clDice, we propose a novel volumetric metric that is specific for skeletal structure, denoted as . In total, 80 membrane MxIF images were manually traced for 5-fold cross-validation. Our model outperforms the baseline with a 20.2% and 41.3% increase in and ARI performance, which is significant (p<0.05) using the Wilcoxon signed rank test. Our work explores a promising direction for advancing MxIF imaging cell segmentation with deep learning membrane segmentation. Tools are available at https://github.com/MASILab/MxIF_Membrane_Segmentation.
Keywords: MxIF, 2D, Membrane Segmentation, Skeleton, Topological-Preserving
1. INTRODUCTION
Crohn’s disease (CD) is a complicated inflammatory bowel disease (IBD) of the gastrointestinal tract, characterized by chronic, relapsing, and remitting bowel inflammation 1. The prevalence of IBD is increasing, this has resulted in higher medical costs. In 2015, the medical cost of CD was estimated to be $3.48 billion per year and is expected to increase to $3.72 billion per year in 2025 in total US national costs 2. The Gut Cell Atlas (GCA), an initiative funded by The Leona M. and Harry B. Helmsley Charitable Trust, seeks to create reference maps of all cells in the terminal ileum and ascending colon to understand the human gut focused on comparing Crohn’s disease patients to healthy controls (https://www.gutcellatlas.helmsleytrust.org/). The GCA project provides a unique opportunity to define different human cell types with gene and protein expression as a function of anatomical location and physiological perturbations of IBD.
Multiplexed immunofluorescence (MxIF) is an emerging technique that allows multiple protein markers to be assessed on a single tissue section with repeated rounds of stain, imaging, stripping, and re-staining 3,4. Our site 5 obtained formalin-fixed paraffin-embedded tissues from the terminal ileum (TI) and ascending colon (AC), which were subsequently stained and imaged via MxIF (including a total of 27 markers that were distributed in 19 rounds) to understand cell composition, functional state, and cell-to-cell spatial distribution 5
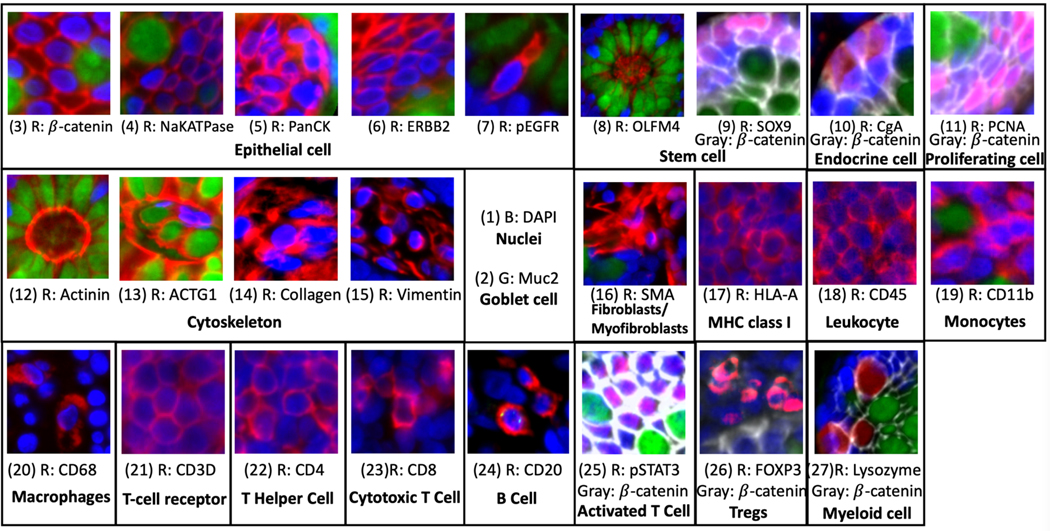
Figure 1 presents 27 MxIF markers on randomly selected regions of a sample tissue and shows how 19 different cell types and internal cellular structures are identified. The membrane outlines the cell boundary and helps the whole cell identification (i.e., Cytoplasm other than nucleus). Figure 1 implies that not all MxIF markers are suited for outlining cell boundaries. Empirically, we selected 9 markers, as shown in Figure 2 (A-B), to serve as membrane markers. However, no particular marker can stain all membranes as cell boundaries. Merging different membrane markers provides complementary global structural information but might hide the underlying local spatial information from specific markers. Moreover, the merged signal cannot overlay seamlessly due to the fluorescence staining lens distortion and variation in field illumination 6. In this work, we aim to segment the membrane of MxIF images that integrate global (projection images) and local (separate individual markers) information to maximize topology preservation. Because the membrane in the MxIF stain could cross over the nuclei, we do not intend to segment the nuclei to break any membrane connectivity.
Figure 1.
This figure presents the 27 markers that are used in our MxIF study. The red channel indicates the marker show positive. The patches with blue channel (DAPI), green channel (MUC2), and gray channel (β-catenin) are collected from epithelium, while the patches with only blue and red channels are acquired from stroma. The magenta regions represent the overlay between red and blue channels.
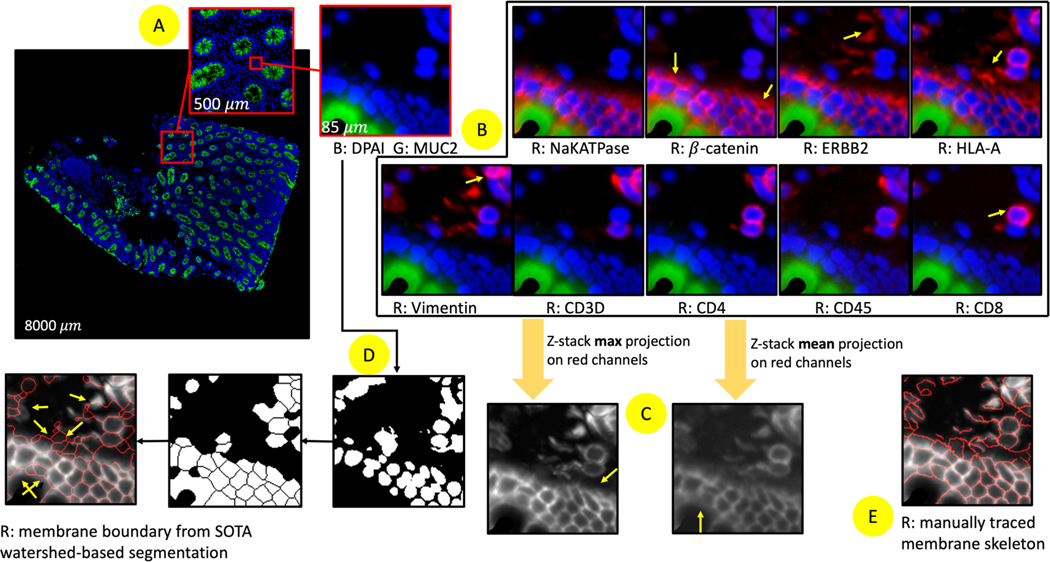
Figure 2.
(A) one random tissue section on DAPI and MUC2 stain is selected. (B) Nine markers highlight cell membrane structures in the same region of interest. There is no particular marker that can stain all cell membrane boundaries. Some nuclei might not be stained (right yellow arrow in β-catenin). (C) Merging all marker channels provides complementary structural information across different markers but might hide or over enhance the underlying information. (D) Recent MxIF cell segmentation methods focus on segmenting DAPI images first with watershed segmentation, leading to the mismatch between membrane topology. (E) The proposed method segments the membrane skeleton of MxIF images that integrates global (projection images) and local (separate individual markers) information to maximize the topology preservation.
The current de facto standard MxIF cell segmentation methods are Voronoi diagram-based 7,8, and wavelet/watershed-based 9–12, which focus on segmenting DAPI images first and then using DAPI binary masks as anchor seed points to further segment the whole cells. McKinley et al. extended the watershed method and segmented cytoplasm and membrane with a Frangi vesselness filter that detected tubular structures on combined markers (NaKATPase, pan-cytokeratin (PanCK), β-catenin, and Villin) 13. Han et al., proposed a deep learning cell segmentation model for MxIF, where the model was trained with pseudo labels on nuclei seeded watershed annotation from NaKATPase images only 14. Gerdes et al. 15 proposed to utilize connected component analysis to segment nuclei and membrane 16–18. However, similar to 13, only few membrane markers are involved and processed (i.e., a combination of NaKATPase, PanCK, and E-cadherin). The existing Vororoni and watershed morphology approaches might cause topological mismatch between membrane segmentation and raw images Figure 2 (D-E). For example, some nuclei cannot always be consistently stained due to batch effects, which would impact the robustness of the watershed segmentation. Moreover, none of the above works have used as extensive of a number of membrane markers as this study.
In this paper, we investigate the feasibility of utilizing four state-of-the-art (SOTA) 2D deep networks and four volumetric-based loss functions to segment the membrane skeleton. The contribution of this work is three-fold:
We develop and comprehensively evaluate the deep membrane segmentation frameworks for large-scale MxIF multi-channel data.
We propose a novel metric that is topology preserving and skeleton-based, denoted as to fill the gap of lacking objective metrics for membrane skeleton segmentation.
We perform the first deep learning membrane segmentation study with large-scale MxIF markers.
2. METHODS
Objective.
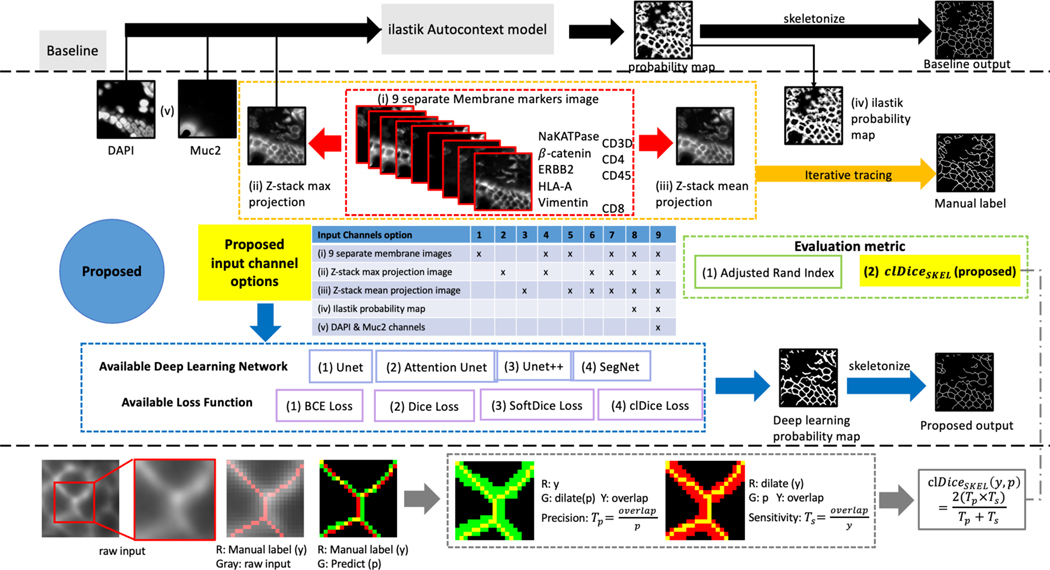
The target annotation of this work is simplified as a skeleton to maximize topology preservation of all membranes. Instead of segmenting the whole merged membrane highlighted area that is more than 1-pixel width, the proposed model learns skeletons of cell membranes for downstream analytics. Figure 3 outlines the membrane segmentation workflow, which includes gold standard annotation, baseline, and proposed approaches.
Figure 3.
The manual label is created by iteratively tracing over nine separate membrane markers on the same ROI and their z-stack max/mean projection images (instead of including the DAPI and MUC2 channels). The baseline method is an ilastik Autocontext model. We investigated the feasibility of four SOTA 2D deep networks and four volumetric-based loss functions. Moreover, we conducted a comprehensive ablation study to assess the sensitivity of the proposed method with various combinations of input channels. Beyond using the existing Adjusted Rand Index based evaluation metric, we propose a novel metric, , to assess topology preserving and skeleton-based membrane segmentation in MxIF.
Dataset and data pre-processing.
Six sample biopsies were collected from 2 CD patients and 1 healthy control from the AC and TI with IRB approval (Vanderbilt IRB #191738 and #191777). All data were de-identified under Institutional Review Board approval. The MxIF markers were acquired with 20× magnification and stained in the following order – DAPI (first round), MUC2, β-catenin 04, CD4, CD3D, HLA-A, CD8, NaKATPase, Vimentin, ERBB2, and CD45. The standard DAPI-based registration and autofluorescence correction were applied 19. A semi-automatic data quality check was to ensure marker alignment efficiency 20. We computed the tissue masks that covered the tissue pixels across all staining rounds. The masks were applied to the images and preprocessed with group-wise linear normalization. To prepare membrane annotation data, we split each marker image into 128×128 patches (86 μ𝑚). Except for DAPI and MUC2, the rest of the markers were z-stacked and generated max/mean projection images. Finally, 80 regions of interest (ROIs) were selected for further annotation and learning.
Manual annotation.
A senior digital pathology researcher manually contoured all membrane skeletons of ROIs using MITK Workbench v2022.04 21. As shown in Figure 3, the membrane annotation was first traced mainly at the z-stack max/mean projection images, then iteratively optimized via the nine individual channels, especially for the hard cases and weak signal regions. In summary, it took about 20 minutes to trace one image patch.
Membrane skeleton segmentation.
The baseline membrane segmentation model is implemented by an ilastik random forest pixel classification approach 22. Eight RGB patches (R: z-stack max projection image, G: MUC2, B: DAPI) are interactively traced over the training output from the ilastik feature selection intermediate results by a domain expert in cell and molecular biology. The baseline ilastik model would produce a membrane probability mask per input, following skeletonized morphological operation 23.
In recent years, deep learning has rapidly become the SOTA basis by showing enhanced performance on 2D and 3D semantic segmentation tasks in various medical image analysis applications 24. To train a deep learning-based membrane segmentation framework, we propose to investigate nine input options by employing five different sets of inputs: (i) nine individual and separate membrane markers, (ii) z-stack max projection image, (iii) z-stack mean projection image, (iv) ilastik probability map (intermediate output from the baseline, referenced from 14), and (v) DAPI & MUC2. We utilized four SOTA deep neural networks to generate 2D membrane skeletons: U-Net 25, U-Net++ 26, Attention U-Net 27, and SegNet 28. We also evaluated four different loss functions to perform gradient descent: binary cross-entropy (BCE) Loss 29, Dice Loss 30, SoftDice Loss (Dice Loss with Laplacian smoothing) 31, and clDice Loss (combining SoftDice with a topology-preserving Loss function specific for tubular structure segmentation) 31. Similar to the baseline, the final membrane from the proposed deep learning framework was converted to a skeleton by skeletonizing network output as shown in Figure 3.
Evaluation metric.
We use the adjusted rand index (ARI) to validate the segmentation topology performance to measure the similarity between two cell segmentation clusters. The clusters can be generated from a filled membrane skeleton followed by indexing each closed contour. In addition to ARI, to validate the skeleton’s volumetric performance, the target and predicted skeletons may have multiple paths to segment a broad membrane region without necessarily being fully overlayed but sharing a similar topology. Thus, the regular Dice or clDice metrics 31 might not suit for evaluating the volumetric overlapping for such scenarios. We extend the clDice and propose a new metric, , to estimate the volumetric similarity of a ground truth skeleton y and the predicted skeleton p while maintaining the topology (an illustrative example is shown at the bottom of Figure 3), as defined in the equation (1):
| (1) |
where D is a dilation 32 function that dilate the input skeleton. 𝑇p interprets the topology precision, and 𝑇s determines the topology sensitivity as defined in equation (2):
| (2) |
Experiment design.
Two primary validation purposes were designed: (1) to investigate the best deep neural network and loss function pair, we trained models with different combinations over three deep learning frameworks (U-Net++, Attention U-Net, and SegNet) and four loss functions. We only implemented U-Net with the SoftDice Loss case. All models used the same input channel option (11 channels that contained z-stack max, z-stack mean, and nine separate membrane images) to reduce the complexity of the experimental configuration. And (2) to validate the sensitivity of model input channels (in total, nine options as described in Figure 3), we tested U-Net++, Attention U-Net, and SegNet with SoftDice Loss equipped only.
All deep neural networks were implemented in PyTorch 1.8 and Python 3.8. The networks followed standard architectures, which can be downloaded from a public GitHub repository (https://github.com/Andy-zhujunwen/UNET-ZOO). The models were trained on a high-performance computing datacenter with GPU nodes equipped with NVIDIA Titan Xp 12GB graphic cards. The training batch size of U-Net, U-Net++, and Attention UNet is 32, while the batch Size is set as 4 for SegNet to saturate the GPU. We empirically set 25 iterations and α = 0.01 (the ratio of SoftDice loss and topology-preserving Loss) for clDice Loss. We conducted a 5-fold cross-validation for each model with static training/validation split per fold. No duplicate validation data were allowed across folds. Each network was trained by 100 epochs and the highest score on validation data decided the best model to save. A 3×3 isotropic structuring kernel dilates each pixel in function D of metric.
3. RESULTS
Exploring the best deep learning framework.
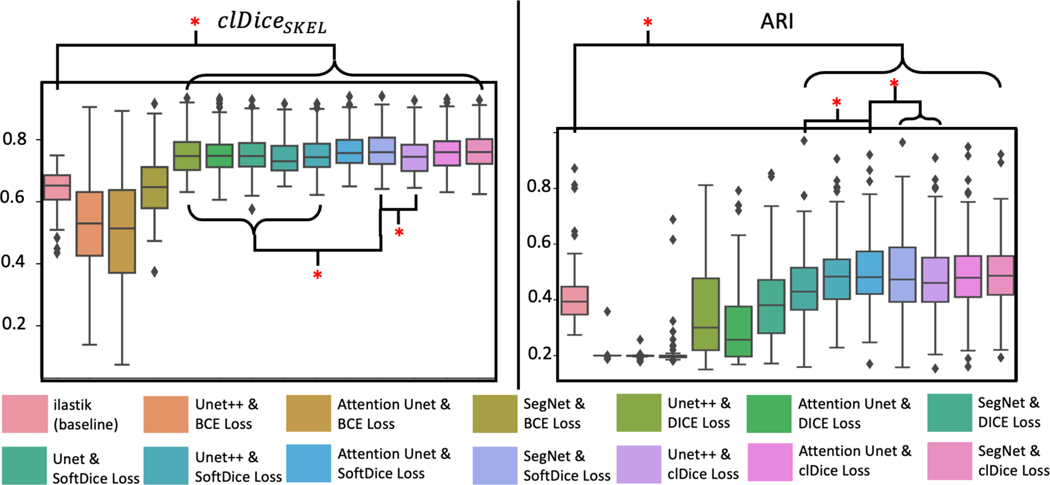
The quantitate results on 5-fold cross validation across the ilastik baseline and 13 proposed deep learning works are shown in Figure 4. In summary, for volumetric performance, the SegNet with SoftDice Loss has the best score (mean ± std: 0.757±0.068) that outperforms the baseline by 20.2%. For topology performance, Attention U-Net with SoftDice Loss (mean ± std: 0.309 ± 0.143) outperforms the baseline by 41.3%. The Wilcoxon sign rank test was employed as the statistical model, where the significant difference (p-value < 0.05) is marked. Figure 5 demonstrates the qualitative results that include good/average/bad across all models. The relevant score is marked for reference.
Figure 4.
The quantitate results on 5-fold cross-validation across the ilastik baseline method and 13 proposed deep learning-based approaches. For the results, SegNet and SoftDice Loss leads to the best average results. U-Net++ and SoftDice Loss conduct the best overall results for the Rand Index results. The Wilcoxon signed rank test is calculated with significant differences marked in red asterisks (p<0.05).
Figure 5.
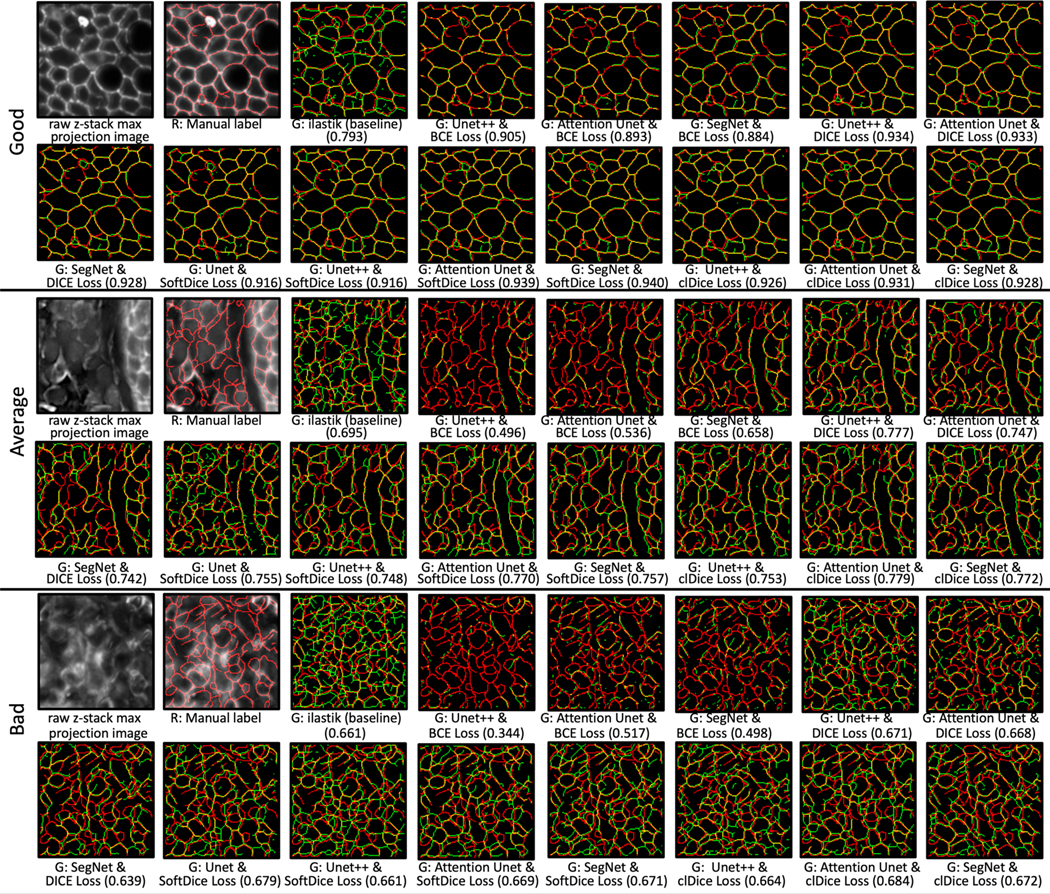
The qualitative results on three randomly selected MxIF image patches with good/average/bad SKEL-clDice performance. Segmentation results (in green) are overlayed on manual labels (in red). The yellow skeletons represent the overlay between red and green channels. The relevant values of different techniques are presented (in parentheses) for reference.
Sensitivity analysis on different input channel.
Table 1 demonstrates the different deep learning frameworks with training by different input channel options when they are all using the SoftDice Loss. The Wilcoxon sign rank test was computed for each method. Table 1 shows that two of the input options (6 & 7, where input channels integrate z-stack max and mean projection image with/without individual markers) have better performance on average with a significant difference (p-value < 0.05).
Table 1.
Sensitivity analysis results (mean ± standard deviation) from different input channel options on individual deep learning frameworks. The Wilcoxon sign rank test is calculated per column, where the reference of each test and significant difference (p-value < 0.05) with other input options per method is highlighted with ‘*’.
| ARI | ||||||
|---|---|---|---|---|---|---|
| Input options | U-Net++ | Attention U-Net | SegNet | U-Net++ | Attention U-Net | SegNet |
| 1: 9 separate makers | 0.750 ±0.064 | 0.759±0.066(*) | 0.761±0.065(*) | 0.313±0.135 | 0.313±0.139 | 0.292±0.165 |
| 2: z-stack max projection image | 0.738±0.071(*) | 0.757±0.068(*) | 0.754±0.072(*) | 0.288±0.160(*) | 0.318±0.147 | 0.280±0.167(*) |
| 3: z-stack mean projection image | 0.745±0.058(*) | 0.761±0.059(*) | 0.760±0.061(*) | 0.314±0.139(ref) | 0.322±0.132(ref) | 0.298±0.154 |
| 4: 1 & 2 | 0.754±0.060(ref) | 0.763±0.063(*) | 0.764±0.066 | 0.296±0.145(*) | 0.306±0.132(*) | 0.294±0.165 |
| 5: 1 & 3 | 0.748±0.061(*) | 0.759±0.062(*) | 0.763±0.062 | 0.275±0.126(*) | 0.303±0.140(*) | 0.295±0.138 |
| 6: 2 & 3 | 0.750±0.064 | 0.768±0.061(ref) | 0.766±0.067 | 0.303±0.139 | 0.321±0.145 | 0.295±0.167 |
| 7: 1 & 2 & 3 | 0.750±0.061(*) | 0.764±0.061 | 0.766±0.064(ref) | 0.304±0.141 | 0.309±0.143 | 0.286±0.157(*) |
| 8: 7 & ilastik probability map | 0.744±0.066(*) | 0.758±0.064(*) | 0.765± 0.063 |
0.304± 0.144(*) | 0.295±0.151(*) | 0.305±0.153 (ref) |
| 9: 8 & DAPI & Muc2 | 0.740±0.059(*) | 0.756±0.064(*) | 0.762±0.065(*) | 0.274±0.138 | 0.303±0.129(*) | 0.284± 0.135(*) |
4. DISCUSSION AND CONCLUSION
From Figure 4, the SegNet with SoftDice Loss leads to better performance over other configurations. Figure 5 shows that the baseline ilastik model is prone to over-segment the membrane. We also find that the epithelial region brings good segmentation results. The bad segmentation performance patch is mainly on the stromal section. Furthermore, the predicted skeleton contains a loose segment end that causes low ARI scores, even when the topology of the skeleton shape matches the ground truth label. The sensitivity analysis results show a moderate difference between employing or not utilizing individual markers. Moreover, we observe that adding DAPI channels degrades the overall skeleton volumetric and topology performance, indicating that the membrane across nuclei scenario is not random.
In summary, this is the first comprehensive work that studies deep learning-based membrane segmentation on large-scale MxIF marker images. We exploit the efficiency of how global and local spatial information from the markers can help membrane segmentation. Our work explores a promising direction for advancing further MxIF imaging cell segmentation to maximize topology preservation. Interestingly, there is no significant difference between SoftDice Loss and clDice Loss (aim to preserve tubular topology), which is worth further investigating on other topological loss functions (i.e., evaluating 33,34) as the next steps. Our proposed has the potential to be extended as a loss function and cooperate with SoftDice Loss. Advancing the post-processing on generated membrane skeleton mask (i.e., bridge the loose, remove island, and avoid over-segmenting) should improve the overall cell clusters topology performance. Finally, we could extend the work by working on DAPI and MUC2 channels and analyzing how to outline the cell when membrane signals across the DAPI and identify the cell when nuclei staining is absent.
5. ACKNOWLEDGEMENT
This research was supported by the Leona M. and Harry B. Helmsley Charitable Trust grant G-1903-03793 and G-2103-05128, NSF CAREER 1452485, NSF 2040462, and in part using the resources of the Advanced Computing Center for Research and Education (ACCRE) at Vanderbilt University, Nashville, TN. This project was supported in part by the National Center for Research Resources, Grant UL1 RR024975-01, and is now at the National Center for Advancing Translational Sciences, Grant 2 UL1 TR000445-06, the National Institute of Diabetes and Digestive and Kidney Diseases, the Department of Veterans Affairs I01BX004366, and I01CX002171. The de-identified imaging dataset(s) used for the analysis described were obtained from ImageVU, a research resource supported by the VICTR CTSA award (ULTR000445 from NCATS/NIH), Vanderbilt University Medical Center institutional funding and Patient-Centered Outcomes Research Institute (PCORI; contract CDRN-1306-04869). This work is supported by NIH grant T32GM007347 and grant R01DK103831. We extend gratitude to NVIDIA for their support by means of the NVIDIA hardware grant.
REFERENCES
- [1].Baumgart DC and Sandborn WJ, “Crohn’s disease,” The Lancet 380(9853), 1590–1605 (2012). [DOI] [PubMed] [Google Scholar]
- [2].Hamdeh S, Aziz M, Altayar O, Olyaee M, Murad MH and Hanauer SB, “Early vs late use of anti-TNFa therapy in adult patients with Crohn disease: a systematic review and meta-analysis,” Inflamm Bowel Dis 26(12), 1808–1818 (2020). [DOI] [PubMed] [Google Scholar]
- [3].Lin J-R, Fallahi-Sichani M. and Sorger PK, “Highly multiplexed imaging of single cells using a high-throughput cyclic immunofluorescence method,” Nat Commun 6(1), 1–7 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Stack EC, Wang C, Roman KA and Hoyt CC, “Multiplexed immunohistochemistry, imaging, and quantitation: a review, with an assessment of Tyramide signal amplification, multispectral imaging and multiplex analysis,” Methods 70(1), 46–58 (2014). [DOI] [PubMed] [Google Scholar]
- [5].Bao S, Chiron S, Tang Y, Heiser CN, Southard-Smith AN, Lee HH, Ramirez MA, Huo Y, Washington MK and Scoville EA, “A cross-platform informatics system for the Gut Cell Atlas: integrating clinical, anatomical and histological data,” Medical Imaging 2021: Imaging Informatics for Healthcare, Research, and Applications 11601, 8–15, SPIE (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Berry S, Giraldo N, Green B, Engle E, Xu H, Ogurtsova A, Wang D, Stein JE, Nguyen P. and Topalian S, “The ‘AstroPath’platform for spatially resolved, single cell analysis of the tumor microenvironment (TME) using multispectral immunofluorescence (mIF),” Cancer Res 80(16_Supplement), 6584 (2020). [Google Scholar]
- [7].Phillips D, Schürch CM, Khodadoust MS, Kim YH, Nolan GP and Jiang S, “Highly multiplexed phenotyping of immunoregulatory proteins in the tumor microenvironment by CODEX tissue imaging,” Front Immunol 12, 687673 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Berens ME, Sood A, Barnholtz-Sloan JS, Graf JF, Cho S, Kim S, Kiefer J, Byron SA, Halperin RF and Nasser S, “Multiscale, multimodal analysis of tumor heterogeneity in IDH1 mutant vs wild-type diffuse gliomas,” PLoS One 14(12), e0219724 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Wiesmann V, Bergler M, Palmisano R, Prinzen M, Franz D. and Wittenberg T, “Using simulated fluorescence cell micrographs for the evaluation of cell image segmentation algorithms,” BMC Bioinformatics 18(1), 1–12 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Rashid R, Gaglia G, Chen Y-A, Lin J-R, Du Z, Maliga Z, Schapiro D, Yapp C, Muhlich J. and Sokolov A, “Highly multiplexed immunofluorescence images and single-cell data of immune markers in tonsil and lung cancer,” Sci Data 6(1), 1–10 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Schapiro D, Sokolov A, Yapp C, Chen Y-A, Muhlich JL, Hess J, Creason AL, Nirmal AJ, Baker GJ and Nariya MK, “MCMICRO: A scalable, modular image-processing pipeline for multiplexed tissue imaging,” Nat Methods 19(3), 311–315 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].McKinley ET, Shao J, Ellis ST, Heiser CN, Roland JT, Macedonia MC, Vega PN, Shin S, Coffey RJ and Lau KS, “MIRIAM: A machine and deep learning single‐cell segmentation and quantification pipeline for multi‐dimensional tissue images,” Cytometry Part A (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].McKinley ET, Sui Y, Al-Kofahi Y, Millis BA, Tyska MJ, Roland JT, Santamaria-Pang A, Ohland CL, Jobin C. and Franklin JL, “Optimized multiplex immunofluorescence single-cell analysis reveals tuft cell heterogeneity,” JCI Insight 2(11) (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Han W, Cheung AM, Yaffe MJ and Martel AL, “Cell segmentation for immunofluorescence multiplexed images using two-stage domain adaptation and weakly labeled data for pre-training,” Sci Rep 12(1), 1–14 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Gerdes MJ, Sevinsky CJ, Sood A, Adak S, Bello MO, Bordwell A, Can A, Corwin A, Dinn S. and Filkins RJ, “Highly multiplexed single-cell analysis of formalin-fixed, paraffin-embedded cancer tissue,” Proceedings of the National Academy of Sciences 110(29), 11982–11987 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Sood A, Sui Y, McDonough E, Santamaría-Pang A, Al-Kofahi Y, Pang Z, Jahrling PB, Kuhn JH and Ginty F, “Comparison of multiplexed immunofluorescence imaging to chromogenic immunohistochemistry of skin biomarkers in response to monkeypox virus infection,” Viruses 12(8), 787 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Graf JF and Zavodszky MI, “Characterizing the heterogeneity of tumor tissues from spatially resolved molecular measures,” PLoS One 12(11), e0188878 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Cheung AM-Y, Wang D, Liu K, Hope T, Murray M, Ginty F, Nofech-Mozes S, Martel AL and Yaffe MJ, “Quantitative single-cell analysis of immunofluorescence protein multiplex images illustrates biomarker spatial heterogeneity within breast cancer subtypes,” Breast Cancer Research 23(1), 1–17 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Lin J-R, Fallahi-Sichani M. and Sorger PK, “Highly multiplexed imaging of single cells using a high-throughput cyclic immunofluorescence method,” Nat Commun 6(1), 1–7 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Bao S, Li J, Cui C, Tang Y, Deng R, Remedios LW, Lee HH, Chiron S, Patterson NH and Lau KS, “MxIF Q-score: Biology-Informed Quality Assurance for Multiplexed Immunofluorescence Imaging,” International Workshop on Medical Optical Imaging and Virtual Microscopy Image Analysis, 42–52, Springer; (2022). [Google Scholar]
- [21].Wolf I, Vetter M, Wegner I, Nolden M, Bottger T, Hastenteufel M, Schobinger M, Kunert T. and Meinzer H-P, “The medical imaging interaction toolkit (MITK): a toolkit facilitating the creation of interactive software by extending VTK and ITK,” Medical Imaging 2004: Visualization, Image-Guided Procedures, and Display 5367, 16–27, SPIE (2004). [Google Scholar]
- [22].Sommer C, Straehle C, Koethe U. and Hamprecht FA, “Ilastik: Interactive learning and segmentation toolkit,” 2011 IEEE international symposium on biomedical imaging: From nano to macro, 230–233, IEEE; (2011). [Google Scholar]
- [23].Zhang TY and Suen CY, “A fast parallel algorithm for thinning digital patterns,” Commun ACM 27(3), 236–239 (1984). [Google Scholar]
- [24].Shen D, Wu G. and Suk H-I, “Deep learning in medical image analysis,” Annu Rev Biomed Eng 19, 221 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Ronneberger O, Fischer P. and Brox T, “U-net: Convolutional networks for biomedical image segmentation,” International Conference on Medical image computing and computer-assisted intervention, 234–241, Springer; (2015). [Google Scholar]
- [26].Zhou Z, Rahman Siddiquee MM, Tajbakhsh N. and Liang J, “Unet++: A nested u-net architecture for medical image segmentation,” [Deep learning in medical image analysis and multimodal learning for clinical decision support], Springer, 3–11 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Oktay O, Schlemper J, Folgoc L. le Lee M, Heinrich M, Misawa K, Mori K, McDonagh S, Hammerla NY and Kainz B, “Attention u-net: Learning where to look for the pancreas,” arXiv preprint arXiv:1804.03999 (2018). [Google Scholar]
- [28].Badrinarayanan V, Kendall A. and Cipolla R, “Segnet: A deep convolutional encoder-decoder architecture for image segmentation,” IEEE Trans Pattern Anal Mach Intell 39(12), 2481–2495 (2017). [DOI] [PubMed] [Google Scholar]
- [29].Long J, Shelhamer E. and Darrell T, “Fully convolutional networks for semantic segmentation,” Proceedings of the IEEE conference on computer vision and pattern recognition, 3431–3440 (2015). [DOI] [PubMed] [Google Scholar]
- [30].Milletari F, Navab N. and Ahmadi S-A, “V-net: Fully convolutional neural networks for volumetric medical image segmentation,” 2016 fourth international conference on 3D vision (3DV), 565–571, IEEE; (2016). [Google Scholar]
- [31].Shit S, Paetzold JC, Sekuboyina A, Ezhov I, Unger A, Zhylka A, Pluim JPW, Bauer U. and Menze BH, “clDice-a novel topology-preserving loss function for tubular structure segmentation,” Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, 16560–16569 (2021). [Google Scholar]
- [32].Howse J, [OpenCV computer vision with python], Packt Publishing Birmingham; (2013). [Google Scholar]
- [33].Hu X, Li F, Samaras D. and Chen C, “Topology-preserving deep image segmentation,” Adv Neural Inf Process Syst 32 (2019). [Google Scholar]
- [34].Wang F, Liu H, Samaras D. and Chen C, “Topogan: A topology-aware generative adversarial network,” European Conference on Computer Vision, 118–136, Springer; (2020). [Google Scholar]