Abstract
Crohn’s disease (CD) is a debilitating inflammatory bowel disease with no known cure. Computational analysis of hematoxylin and eosin (H&E) stained colon biopsy whole slide images (WSIs) from CD patients provides the opportunity to discover unknown and complex relationships between tissue cellular features and disease severity. While there have been works using cell nuclei-derived features for predicting slide-level traits, this has not been performed on CD H&E WSIs for classifying normal tissue from CD patients vs active CD and assessing slide label-predictive performance while using both separate and combined information from pseudo-segmentation labels of nuclei from neutrophils, eosinophils, epithelial cells, lymphocytes, plasma cells, and connective cells. We used 413 WSIs of CD patient biopsies and calculated normalized histograms of nucleus density for the six cell classes for each WSI. We used a support vector machine to classify the truncated singular value decomposition representations of the normalized histograms as normal or active CD with four-fold cross-validation in rounds where nucleus types were first compared individually, the best was selected, and further types were added each round. We found that neutrophils were the most predictive individual nucleus type, with an AUC of 0.92 ± 0.0003 on the withheld test set. Adding information improved cross-validation performance for the first two rounds and on the withheld test set for the first three rounds, though performance metrics did not increase substantially beyond when neutrophils were used alone.
Keywords: Crohn’s Disease, Cell Nucleus Density, Pseudo Labels, Classification, Cell Nucleus Segmentation, Machine Learning, H&E
1. INTRODUCTION
More than 3 million people in the United States have been diagnosed with inflammatory bowel disease (IBD), which includes Crohn’s disease (CD) and ulcerative colitis1. Although CD is becoming more commonly diagnosed, its precise cause is still to be determined2. Among other tests, analysis of biopsies plays a key role in CD diagnosis, traditionally performed under a light microscope by a trained pathologist2. The advent of whole slide imaging has allowed for the digitization of microscope slides as high-resolution gigapixel images. The analysis of these gigapixel images is now an active field of study in digital pathology, with computational methods frequently involving deep learning3,4.
Pathologist-assigned disease severity scores for CD biopsies are often given at the slide level, though the disease features that resulted in the scoring might not present homogeneously across the slide. More fine-grained labels may allow computational methods to better learn biopsy subtleties, but acquiring such labels is more time-intensive and costly, sparking interest in automatic segmentation methods. For IBD, the segmentation of glands on H&E5 and multimodal imaging data6 has been investigated for predicting disease grading of the slide6. At a smaller scale, nucleus segmentation can be used to investigate the cells within biopsies. For the colon, interest in this space has been illustrated by the organization of the Colon Nuclei Identification and Counting (CoNIC) Challenge, which used a dataset of 4,981 patches with dimensions 256 × 256, from images at 20× objective magnification7. The challenge was introduced to engage the community and improve algorithmic nucleus segmentation7.
A sensible approach for analysis of imaged slides is to extract features from the nuclei and then use these features to make predictions about known traits of the biopsy. This process can be generally applied to images of tissue-containing slides and has been used on lung cancer tasks8,9. For IBD, sub-microscopic scale features from nuclei have been used to differentiate between high and low-risk samples for development of neoplasia10. Specifically for Crohn’s disease, segmented nuclei have been used as a component in determining desirable anatomical locations for biopsies with respect to a present ulcer11. In ulcerative colitis, deep learning has been used to assess eosinophil density which was found to correlate with certain disease features but not with histologic activity12 which refers to mucosal neutrophils existing in specific locations within the tissue13. A recent IBD study looked at 24 samples, used imaging mass cytometry to generate highly multiplexed images, segmented nuclei into 13 classes, and performed an analysis to find differences between cases and controls14. However, they did not have distinct myeloid cell classes clearly separating eosinophils and neutrophils14, which are both markers of interest in IBD15.
Comparisons of the predictive value of separate and mixed densities from neutrophils, eosinophils, lymphocytes, plasma cells, epithelial cells, and connective cells for slide-level CD severity binary classification have not been previously investigated using H&E WSIs. The purpose of this study was to investigate predictiveness for differentiating normal tissue from CD patients and active (mild, moderate, severe) CD using only pseudo-label-derived input for six cell types and to determine how combining information from different cell types impacted performance. In Figure 1. we visualize our approach for aggregating slide-level nucleus information before performing classification. In this pursuit, we segmented nuclei using the baseline deep learning model from the CoNIC Challenge 20227, which was trained on external data, computed slide-level normalized histograms of nucleus density, performed dimensionality reduction, and used a support vector machine (SVM) for binary classification in a series of rounds, where we first assessed cell types individually, and then added types incrementally to the best performer(s) from each round.
Figure 1.
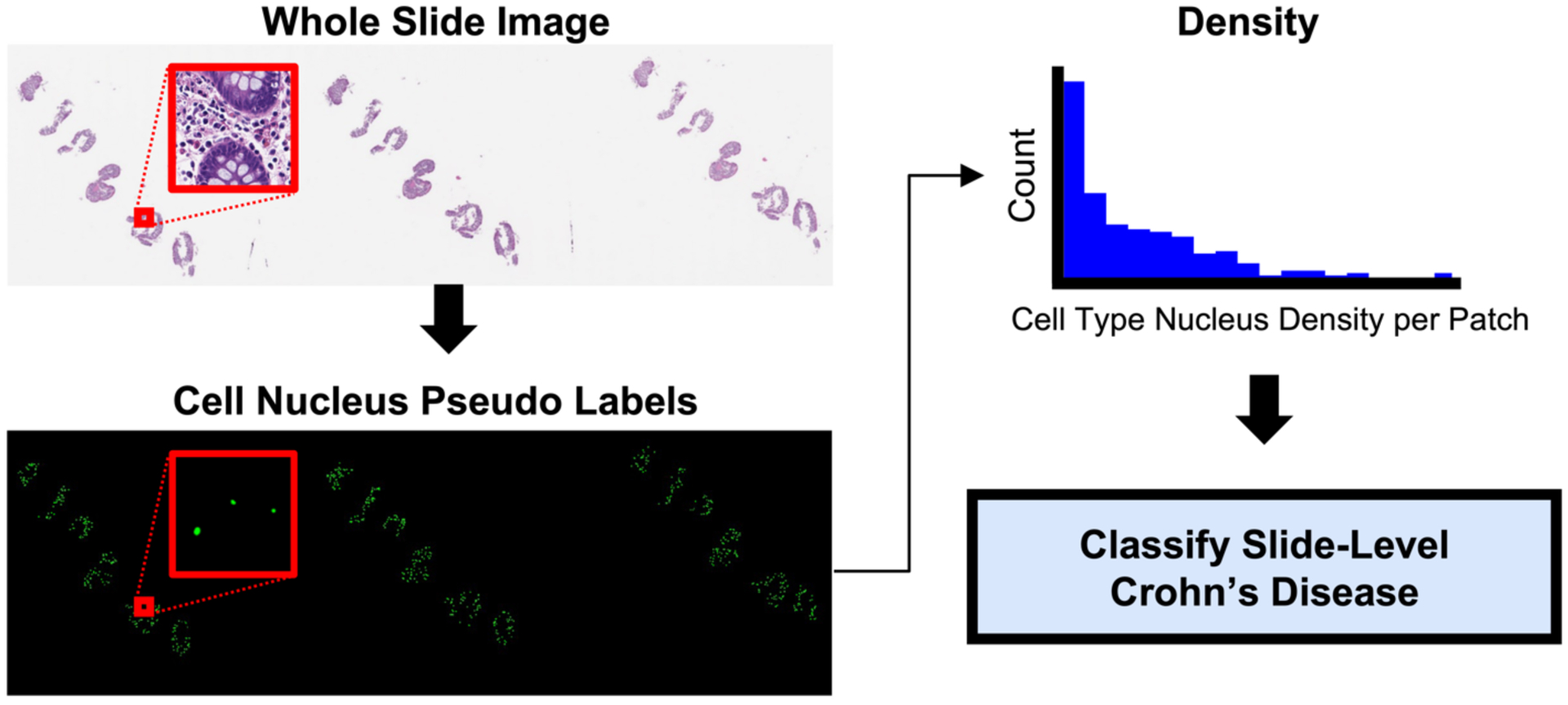
WSIs were patched and segmented for six types of cell nuclei. The density for each nucleus type was separately computed at the patch level, and then aggregated into a normalized histogram to represent the nucleus type’s density at the slide-level. The normalized histograms were then reduced in dimensionality with truncated singular value decomposition before using a SVM to classify examples as normal tissue from CD patients or active CD. By using cell nucleus pseudo labels and calculating slide-level nucleus density, we retained global information while greatly reducing the dimensionality of gigapixel WSIs.
2. METHODS
2.1. Data
Our dataset of 1,115 H&E whole slide images was retrieved in deidentified form from Vanderbilt University Medical Center under Institutional Review Board (IRB) approval corresponding to both Vanderbilt IRB #191738 and #191777. From these 1,115 images, we selected a subset of data that included 81 subjects and 413 WSIs, all from CD patients where biopsies came from four anatomical region classes: right colon, transverse colon, left colon, sigmoid/rectosigmoid colon. This dataset included 247 images of biopsies with a pathology score of normal, and 166 with a pathology score of mild, moderate, or severe disease activity. Splitting the data into these two groups allowed us to investigate the data in a binary classification manner, where 60% of the slides had normal-appearing tissue and 40% depicted active CD. The images were collected at 20 × objective magnification.
Data that were not used from the full dataset of 1,115 images were removed based on the following set of exclusions: 1) subjects not labeled as having CD, 2) anatomical regions that were either non-colon regions, or uncategorized/various surgical regions, 3) slides graded as quiescent CD, or with an unknown pathology score, 4) slides in which the nucleus segmentation failed (e.g., sharpie on the slide).
2.2. Image processing and feature extraction
Methods for image processing and classification are illustrated in Figure 2. The images had regions containing tissue divided into patches of size 256 × 256 pixels using tools from the CLAM pipeline4. These patches were then segmented for six types of cell nuclei (neutrophils, eosinophils, lymphocytes, plasma cells, epithelial cells, and connective cells) using the pretrained baseline cell nucleus segmentation model from the CoNIC Challenge 20227. Pseudo labels were generated using a workstation equipped with an NVIDIA Quadro RTX 5000 GPU with 16 GB of RAM.
Figure 2.
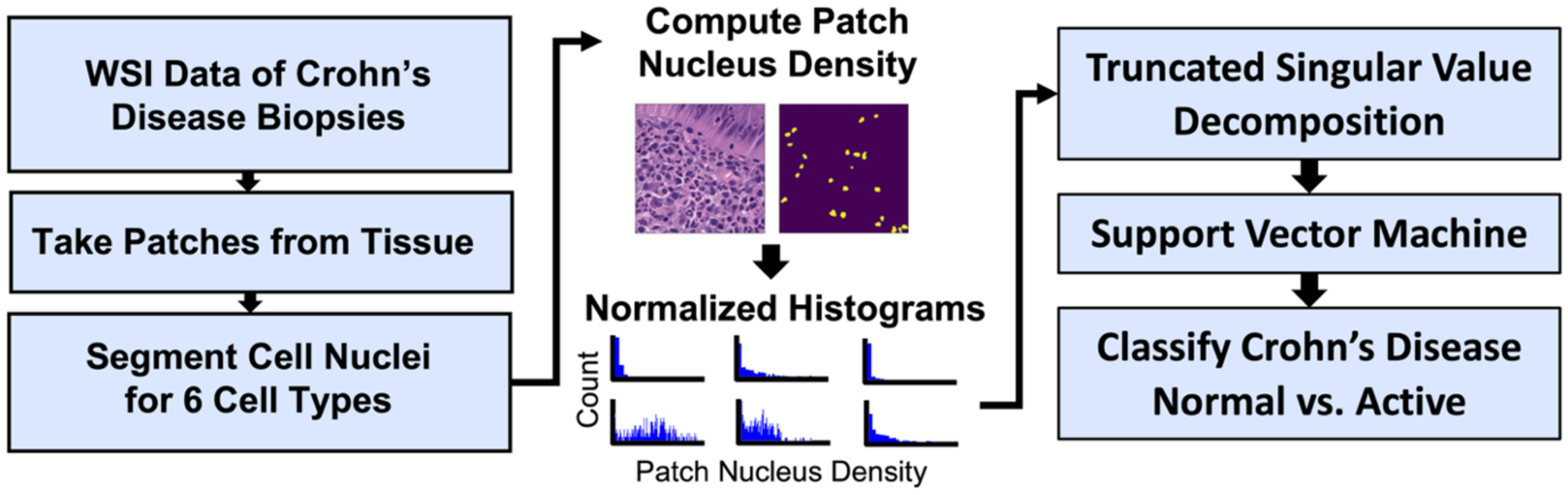
Whole slide images were patched around tissue regions and had cell nuclei segmented. The density of cell nucleus types in each patch was computed and aggregated into slide-level normalized histograms (scaled only for visualization in this figure), which were then reduced in dimensionality using TSVD. Components from TSVD were used for binary classification of normal vs active disease using a support vector machine.
For each of the six cell types, we calculated the nucleus density at each non-overlapping patch from CLAM. Density here is defined as the number of pixels of the nucleus type divided by the number of pixels in a patch, which constrained the patch density between zero and one. Patch densities were then aggregated at the slide level and represented as a histogram, where the x-axis denoted nucleus type density. The number of bins was set at 1024 for each cell type, which allowed a ratio of one bin to 64 pixels in each patch. To account for the varying amount of tissue across slides, each histogram bin was normalized based on the number of patches in the image, which is a rough estimate for the amount of segmented tissue in the WSI, as only tissue containing areas were patched. This process gave us normalized histograms where the number of bins was always 1024, the maximum possible patch nucleus density was one (x-axis), and the maximum possible count was one (y-axis), regardless of the cell type or other factors.
Because 1024 was the constant number of bins across cell nucleus types and images, many bins were often empty. We addressed this by performing dimensionality reduction using truncated singular value decomposition (TSVD)16. We used 20 TSVD components as they explained at least 80% of the variance in the training data from fold zero on all of the cell types individually. For each fold, a separate TSVD transformation was fit to the corresponding training data, and then applied to the fold’s validation data.
2.3. Feature aggregation and classification via support vector machine
In the dataset, there was at least one image per subject, however many subjects had more than one WSI. All subjects had CD, though biopsies either depicted normal tissue or active disease tissue, thus images from the subjects had binary labels of normal CD or active CD. Using a random seed, the data were split 80:20 on unique subjects into two groups. The seed was selected such that the balance of corresponding WSIs was as close to 60% normal CD and 40% active CD in both groups as possible. The larger group was used for cross-validation and the smaller as the withheld test set. Again, using a random seed, the cross-validation data were split into four folds on the subject level. Each fold in the cross-validation data was stratified over the two image classes. This resulted in the balance of each cross-validation fold, and the withheld test set, being approximately 60% normal images and 40% active disease images.
To assess for separability between normal and active disease biopsies, we used an SVM with a radial basis function kernel for binary classification. We performed four-fold cross-validation, where threshold moving was computed for each fold using Youden’s index, based on its training set predictions. We did this in rounds; the best cell type for the classification task was selected and used as the starting point for the next round, where other cell types were added in one at a time and compared. Adding in cell types was done by concatenating the TSVD components. The best was selected and this process was repeated, resulting in 6 rounds. Round 1 experiments had 20 features, and each subsequent round had 20 more features. Final testing was performed on the withheld test set, evaluating all models that were generated during cross-validation.
3. RESULTS
3.1. Quantitative results on Round 1
Table 1. gives numerical details for all experiments that were performed. When only comparing individual cell nucleus types against each other in Round 1, we were able to see that neutrophils provided the most predictive value in terms of area under the receiver operating characteristic curve (AUC) and average precision (AP), with both metrics sitting at or above 0.91 on cross-validation and the withheld test set. Additionally, neutrophils showed the most stability, with almost identical scores on both cross-validation and the withheld test set, differing only by the standard deviation—this phenomenon happened separately for AUC and AP.
Table 1.
Results from each round are shown for cross-validation and the withheld test data.
| Cross-Validation | Withheld Test | ||||
|---|---|---|---|---|---|
| AUC | AP | AUC | AP | ||
|
Round 1 20 fts |
N | 0.92 ± 0.0540 | 0.91 ± 0.0486 | 0.92 ± 0.0003 | 0.91 ± 0.0004 |
| C | 0.83 ± 0.0520 | 0.78 ± 0.0750 | 0.83 ± 0.0055 | 0.81 ± 0.0074 | |
| L | 0.81 ± 0.0515 | 0.77 ± 0.0530 | 0.81 ± 0.0059 | 0.70 ± 0.0030 | |
| EPI | 0.78 ± 0.0599 | 0.76 ± 0.0609 | 0.79 ± 0.0036 | 0.72 ± 0.0132 | |
| P | 0.79 ± 0.0732 | 0.71 ± 0.0945 | 0.72 ± 0.0059 | 0.66 ± 0.0115 | |
| EOS | 0.75 ± 0.0386 | 0.70 ± 0.0913 | 0.81 ± 0.0059 | 0.81 ± 0.0065 | |
|
Round 2 40 fts |
N, L | 0.93 ± 0.0432 | 0.92 ± 0.0403 | 0.93 ± 0.0030 | 0.92 ± 0.0022 |
| N, C | 0.92 ± 0.0565 | 0.92 ± 0.0460 | 0.91 ± 0.0028 | 0.90 ± 0.0021 | |
| N, P | 0.92 ± 0.0471 | 0.91 ± 0.0413 | 0.91 ± 0.0023 | 0.91 ± 0.0021 | |
| N, EPI | 0.91 ± 0.0424 | 0.91 ± 0.0429 | 0.93 ± 0.0008 | 0.91 ± 0.0008 | |
| N, EOS | 0.90 ± 0.0321 | 0.90 ± 0.0351 | 0.91 ± 0.0082 | 0.91 ± 0.0055 | |
|
Round 3 60 fts |
N, L, P | 0.92 ± 0.0425 | 0.92 ± 0.0373 | 0.92 ± 0.0023 | 0.92 ± 0.0010 |
| N, L, C | 0.92 ± 0.0507 | 0.92 ± 0.0421 | 0.91 ± 0.0016 | 0.91 ± 0.0029 | |
| N, L, EPI | 0.92 ± 0.0361 | 0.91 ± 0.0414 | 0.94 ± 0.0040 | 0.92 ± 0.0066 | |
| N, L, EOS | 0.91 ± 0.0314 | 0.91 ± 0.0304 | 0.91 ± 0.0076 | 0.92 ± 0.0049 | |
|
Round 4 80 fts |
N, L, P, C | 0.92 ± 0.0504 | 0.92 ± 0.0386 | 0.90 ± 0.0027 | 0.90 ± 0.0022 |
| N, L, P, EPI | 0.91 ± 0.0351 | 0.91 ± 0.0314 | 0.93 ± 0.0031 | 0.92 ± 0.0028 | |
| N, L, P, EOS | 0.91 ± 0.0271 | 0.90 ± 0.0272 | 0.91 ± 0.0082 | 0.92 ± 0.0063 | |
|
Round 5 100 fts |
N, L, P, C, EPI | 0.92 ± 0.0489 | 0.92 ± 0.0363 | 0.91 ± 0.0041 | 0.90 ± 0.0027 |
| N, L, P, C, EOS | 0.90 ± 0.0397 | 0.91 ± 0.0232 | 0.90 ± 0.0068 | 0.90 ± 0.0063 | |
|
Round 6 120 fts |
All | 0.90 ± 0.0408 | 0.91 ± 0.0294 | 0.91 ± 0.0053 | 0.91 ± 0.0047 |
AUC stands for ROC AUC, AP stands for average precision, N stands for neutrophil, C stands for connective, L stands for lymphocyte, EPI stands for epithelial, P stands for plasma, and EOS stands for eosinophil. The best performer(s) at each round were picked by equally weighting AUC and AP. Only cross-validation best performers were used for selecting the combinations of cell types in the subsequent rounds. The AUC and AP for the best performer(s) are bolded; the bolding does not denote the best individual AUC or AP from a round. Bolding is always on the same cell type for AUC and AP within a set, indicating the round’s best performer.
While neutrophils were the most predictive, other cell types were also predictive, though not to the same degree. Non-neutrophil AUC scores ranged from 0.75 ± 0.0386 (eosinophil) to 0.83 ± 0.0520 (connective) on cross-validation, and from 0.72 ± 0.0059 (plasma) to 0.83 ± 0.0055 (connective) on the withheld test set. Looking at AP scores, we see that non-neutrophil performance ranged from 0.70 ± 0.0913 (eosinophil) to 0.78 ± 0.0750 (connective) on cross-validation, and from 0.66 ± 0.0115 (plasma) to 0.81 ± 0.0065 (eosinophil) on the withheld test set.
Inspecting the cross-validation and withheld test set receiver operating characteristic (ROC) and precision-recall curves (Figure 3), neutrophils consistently stood out as the most predictive cell nucleus type, even when accounting for the shaded standard deviation. For non-neutrophil cell type curves, there was a large amount of overlap on cross-validation, especially when noting the shaded standard deviations. The withheld test set curves for non-neutrophil cell types show more clear delineation of performance, however because of the curve overlap on cross-validation, where evaluation is on separate data for the four folds, the withheld test set performance for these cell nucleus types may be misleading if taken to imply overall generalizability.
Figure 3.
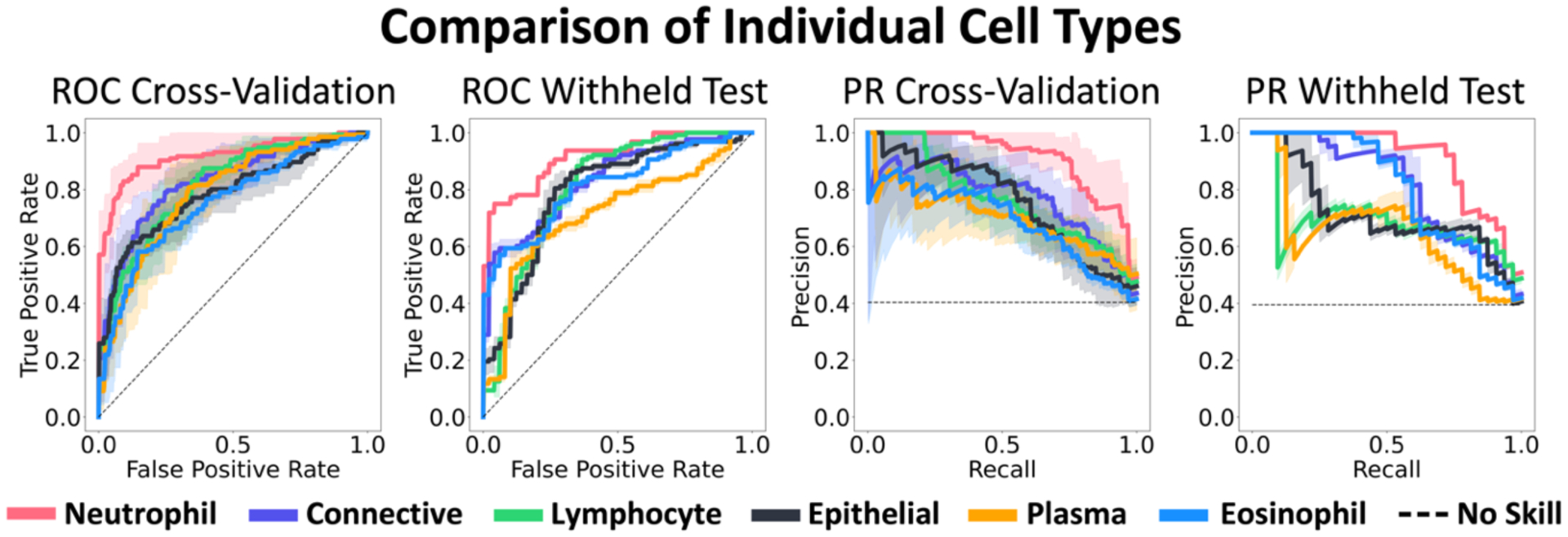
Average ROC and precision-recall (PR) curves are shown from cross-validation and the withheld test set. Shaded regions represent ± one standard deviation. When comparing the six cell types by themselves, neutrophils consistently performed best.
3.2. Quantitative results on remaining rounds
As we added information in each round to the best performer(s) (Table 1.), we saw increased performance, though subtle, on cross-validation up to the second round, before tapering off. On the withheld test set, we saw a more marked increase in performance through the third round, before a decrease. This decrease in performance may be due to the increase in dimensionality, as each subsequent round added 20 features. The best performer(s) differed between cross-validation and the withheld test data on the third and fourth rounds but were the same on all other rounds. The highest scoring combinations of cell types only marginally surpassed performance of neutrophils by themselves.
3.3. Qualitative results
We performed a uniform manifold and projection (UMAP) clustering of the neutrophil normalized histogram representation of all the data to assess separability of the two images classes, which can be seen in Figure 4. In the UMAP, we found that the normal and active disease samples generally separated into two distinct but overlapping clusters. To interpret the WSIs corresponding to the clustering, we pulled the maximum neutrophil density CLAM patch (256 × 256 pixels) from each image and incorporated them into the clustering visualization. We found that these max neutrophil patches in the normal cluster more often clearly depicted crypts and edges of tissue than the active disease cluster.
Figure 4.
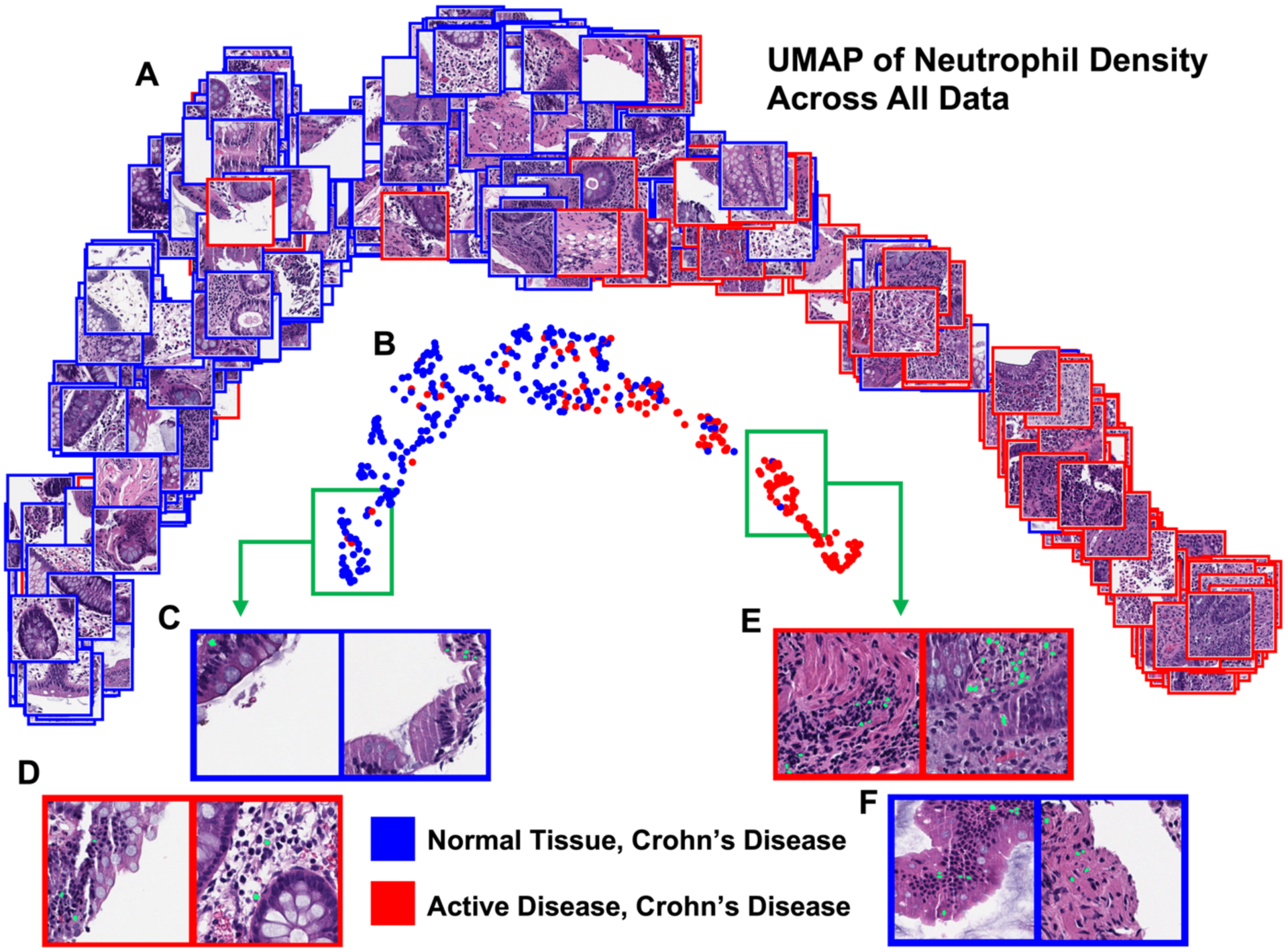
UMAP of the normalized histogram representation of WSI neutrophil density for the entire dataset. All patches in this figure depict the 256 × 256 pixel CLAM patch with the maximum neutrophil density from the corresponding WSI. The UMAP displayed with patches depicts more clearly visible crypts and tissue edges in the normal cluster max patches than the active disease cluster (A). The UMAP displayed with circle markers highlights the separability of the two classes (B). Example patches are shown with their predicted neutrophil segmentation in green (C, D, E, F). Looking deep into the normal cluster, expected patches (C) and active disease outliers (D) show tissue edges and few segmented neutrophils. In the active disease cluster, expected patches (E) and normal tissue CD outliers (F) both show a larger number of segmented neutrophils, with fewer tissue edges visible (F).
4. CONCLUSIONS
The separate use of nucleus types showed that WSI neutrophil density was very predictive for differentiating normal from active (mild, moderate, severe) disease, when looking at biopsies from CD patients. The other cell types (lymphocytes, plasma cells, epithelial cells, eosinophils, and connective cells) showed varying degrees of predictiveness, implying that WSI density of these nucleus types on their own store information about slide-level CD severity. Combining cell types was expected to improve the ability of the SVM to differentiate between classes. While we saw some marginal increase in performance, we did not see large performance boosts beyond the performance of neutrophils alone. Future work will investigate different ways to fuse information from cell nucleus types to increase predictiveness.
This work is limited in that all input to our SVM was derived from predicted cell nucleus segmentations produced by a deep learning model trained on external data, and so our conclusions are dependent upon the reliability of the initial predictions. Despite this, we saw promising predictive ability—during development a UMAP of neutrophil nucleus density led to the discovery of a mislabeled sample, which shows that an approach like the one demonstrated in Figure 4. could be used as a quality assurance method for large amounts of data.
ACKNOWLEDGMENTS
This research was supported by the Leona M. and Harry B. Helmsley Charitable Trust grant G-1903-03793 and G- 2103-05128, NSF CAREER 1452485, NSF 2040462, and in part using the resources of the Advanced Computing Center for Research and Education (ACCRE) at Vanderbilt University, Nashville, TN. This project was supported in part by the National Center for Research Resources, Grant UL1 RR024975-01, and is now at the National Center for Advancing Translational Sciences, Grant 2 UL1 TR000445-06, the National Institute of Diabetes and Digestive and Kidney Diseases, the Department of Veterans Affairs I01BX004366, and I01CX002171. The de-identified imaging dataset(s) used for the analysis described were obtained from ImageVU, a research resource supported by the VICTR CTSA award (ULTR000445 from NCATS/NIH), Vanderbilt University Medical Center institutional funding and Patient-Centered Outcomes Research Institute (PCORI; contract CDRN-1306-04869). We would like to acknowledge the VUMC Digestive Disease Research Center supported by NIH grant P30DK058404. This work is supported by NIH grant T32GM007347 and R01DK103831. We extend gratitude to NVIDIA for their support by means of the NVIDIA hardware grant.
REFERENCES
- [1].Dahlhamer JM, Zammitti EP, Ward BW, Wheaton AG and Croft JB, “Prevalence of Inflammatory Bowel Disease Among Adults Aged ≥18 Years — United States, 2015,” Mortality Weekly Report 65(42), 1166–1169 (2016). [DOI] [PubMed] [Google Scholar]
- [2].Roda G, Chien Ng S, Kotze PG, Argollo M, Panaccione R, Spinelli A, Kaser A, Peyrin-Biroulet L and Danese S, “Crohn’s disease,” Nature Reviews Disease Primers 6(1), 22 (2020). [DOI] [PubMed] [Google Scholar]
- [3].Zarella MD, Bowman D, Aeffner F, Farahani N, Xthona A, Absar SF, Parwani A, Bui M and Hartman DJ, “A Practical Guide to Whole Slide Imaging: A White Paper From the Digital Pathology Association,” Archives of Pathology & Laboratory Medicine 143(2), 222–234 (2019). [DOI] [PubMed] [Google Scholar]
- [4].Lu MY, Williamson DFK, Chen TY, Chen RJ, Barbieri M and Mahmood F, “Data-efficient and weakly supervised computational pathology on whole-slide images.,” Nat Biomed Eng 5(6), 555–570 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Ma Z, Swiderska-Chadaj Z, Ing N, Salemi H, McGovern D, Knudsen B and Gertych A, “Semantic Segmentation of Colon Glands in Inflammatory Bowel Disease Biopsies,” 379–392 (2019). [Google Scholar]
- [6].Pradhan P, Meyer T, Vieth M, Stallmach A, Waldner M, Schmitt M, Popp J and Bocklitz T, “Semantic Segmentation of Non-linear Multimodal Images for Disease Grading of Inflammatory Bowel Disease: A SegNet-based Application,” Proceedings of the 8th International Conference on Pattern Recognition Applications and Methods, 396–405, SCITEPRESS - Science and Technology Publications; (2019). [Google Scholar]
- [7].Graham S, Jahanifar M, Vu QD, Hadjigeorghiou G, Leech T, Snead D, Raza SEA, Minhas F and Rajpoot N, “CoNIC: Colon Nuclei Identification and Counting Challenge 2022” (2021).
- [8].Alsubaie N, Sirinukunwattana K, Raza SEA, Snead D and Rajpoot NM, “A bottom-up approach for tumour differentiation in whole slide images of lung adenocarcinoma,” 6 March 2018, 10, SPIE-Intl Soc Optical Eng. [Google Scholar]
- [9].Lu C, Bera K, Wang X, Prasanna P, Xu J, Janowczyk A, Beig N, Yang M, Fu P, Lewis J, Choi H, Schmid RA, Berezowska S, Schalper K, Rimm D, Velcheti V and Madabhushi A, “A prognostic model for overall survival of patients with early-stage non-small cell lung cancer: a multicentre, retrospective study,” The Lancet Digital Health 2(11), e594–e606 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Uttam S, Hashash JG, LaFace J, Binion D, Regueiro M, Hartman DJ, Brand RE and Liu Y, “Three-Dimensional Nanoscale Nuclear Architecture Mapping of Rectal Biopsies Detects Colorectal Neoplasia in Patients with Inflammatory Bowel Disease.,” Cancer Prev Res (Phila) 12(8), 527–538 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Novak G, Stevens T, van Viegen T, Bossuyt P, Štabuc B, Jeyarajah J, Zou G, Gaemers IC, McKee TD, Fu F, Shackelton LM, Khanna R, van den Brink GR, Sandborn WJ, Feagan BG, Pai RK, Jairath V, vande Casteele N and D’Haens G, “Evaluation of optimal biopsy location for assessment of histological activity, transcriptomic and immunohistochemical analyses in patients with active Crohn’s disease,” Alimentary Pharmacology & Therapeutics 49(11), 1401–1409 (2019). [DOI] [PubMed] [Google Scholar]
- [12].vande Casteele N, Leighton JA, Pasha SF, Cusimano F, Mookhoek A, Hagen CE, Rosty C, Pai RK and Pai RK, “Utilizing Deep Learning to Analyze Whole Slide Images of Colonic Biopsies for Associations Between Eosinophil Density and Clinicopathologic Features in Active Ulcerative Colitis,” Inflammatory Bowel Diseases 28(4), 539–546 (2022). [DOI] [PubMed] [Google Scholar]
- [13].Pai RK, Lauwers GY and Pai RK, “Measuring Histologic Activity in Inflammatory Bowel Disease: Why and How,” Advances in Anatomic Pathology 29(1), 37–47 (2022). [DOI] [PubMed] [Google Scholar]
- [14].Kondo A, Ma S, Lee MYY, Ortiz V, Traum D, Schug J, Wilkins B, Terry NA, Lee H and Kaestner KH, “Highly Multiplexed Image Analysis of Intestinal Tissue Sections in Patients With Inflammatory Bowel Disease,” Gastroenterology 161(6), 1940–1952 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Alhmoud T, Gremida A, Colom Steele D, Fallahi I, Tuqan W, Nandy N, Ismail M, Aburajab Altamimi B, Xiong M-J, Kerwin A and Martin D, “Outcomes of inflammatory bowel disease in patients with eosinophil-predominant colonic inflammation,” BMJ Open Gastroenterology 7(1), e000373 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Hansen PC, “Solution of Ill-Posed Problems by Means of Truncated SVD,” 179–192 (1988). [Google Scholar]


