Abstract
Purpose:
Intracerebral Hemorrhage (ICH) is one of the most devastating types of strokes with mortality and morbidity rates ranging from about 51%−65% one year after diagnosis. Early hematoma expansion (HE) is a known cause of worsening neurological status of ICH patients. The goal of this study was to investigate whether non-contrast computed tomography imaging biomarkers (NCCT-IB) acquired at initial presentation can predict ICH growth in the acute stage.
Materials and Methods:
We retrospectively collected NCCT data from 200 patients with acute (<6 hours) ICH. Four NCCT-IBs (blending region, dark hole, island, and edema) were identified for each hematoma, respectively. HE status was recorded based on the clinical observation reported in the patient chart. Supervised machine learning models were developed, trained, and tested for 15 different input combinations of the NCCT-IBs to predict HE. Model performance was assessed using area under the receiver operating characteristic curve and probability for accurate diagnosis (PAD) was calculated. A 20-fold Monte-Carlo cross validation was implemented to ensure model reliability on a limited sample size of data, by running a myriad of random training/testing splits.
Results:
The developed algorithm was able to predict expansion utilizing all four inputs with an accuracy of 70.17%. Further testing of all biomarker combinations yielded PAD ranging from 0.57, to 0.70.
Conclusion:
Specific attributes of ICHs may influence the likelihood of HE and can be evaluated via a machine learning algorithm. However, certain parameters may differ in importance to reach accurate conclusions about potential expansion.
Keywords: Intracerebral hemorrhage, hematoma, machine learning, keras neural network, non-contrast computed tomography, receiver operating characteristic curve
1. INTRODUCTION
Research within this study aims to develop a method to identify patients at risk for HE, which may result in rapid neurological decline. According to the American Heart Association, ICH is a subtype of stroke which is the 3rd highest cause of death and the leading cause of disability worldwide [1]. Hemorrhagic strokes occur when one of the blood vessels within the brain ruptures causing blood to leak into the surrounding brain tissue. This bleeding induces an increase in intracranial pressure, exerting damaging force on structures within the brain. This pressure can limit blood flow, forcefully kill brain cells (axons, glial cells), and lead to herniation within the brain [17]. As the injury progresses the body will initiate responses to combat the bleeding, and further deteriorate surrounding brain cells in the process. Intracerebral hemorrhage includes five main types of hemorrhage: (1) epidural hematoma, (2) subdural hematoma, (3) subarachnoid hemorrhage (SAH), (4) intraventricular hemorrhage (IVH), (5) intracerebral/intraparenchymal hemorrhage (ICH). ICH accounts for 10–15% of all strokes and carries one of the highest morbidity and mortality rates. Half of the ICH deaths occur within the first two days following onset. At one year post onset, mortality ranges from 51% to 65% depending on the location of the hemorrhage. At six months post onset, only 20% of patients are expected to be functioning independently.[2] The incidence of hemorrhage increases exponentially with age and is higher in men than in women.[3] Despite significant advances in neuroimaging and treatment these mortality and morbidity numbers have not changed over the last 30 years.
There is currently no reliable treatment or therapy to fully combat an ICH. At best, medical professionals have utilized methods of coagulation, reducing intracranial pressure, or even a craniotomy [8]. Preventing further HE is the main objective that physicians aim to achieve in ICH cases to minimize further disease progression. Studies have shown limiting/preventing HE has an upside in decreasing hematoma volume and potentially evolving into a form of treatment for ICH [16]. In order to do so, one must understand the physiology of HE, and with that the mechanisms that are responsible. It is generally thought that a hematoma expands due to one specific blood vessel within the brain continuing to leak and adding to the volume of blood in the tissue. However, studies have produced evidence that more accurately describe what is occurring during expansion [16]. It is suggested that the initial hemorrhage actually induces secondary ruptures in vessels within close proximity of the initial tear. The event then acts as an ‘avalanche’, or chain reaction where a number of vessel walls shear and break down, adding to the hematoma volume. This can be corroborated using computed tomography angiography (CTA), which displays small white dots, or spot signs, that indicate the source of bleeding within the brain. Substantially, these images may contain multiple spot signs, indicating that the bleed is coming from multiple sources.
To be effective against such rapid and devastating effects, ICH severity diagnosis must be accurate and precise to allow for early identification of patients prone to rapid neurological decline. Computed tomography (CT) imaging is typically the standard technique used due to its widespread availability; thus, CT was utilized in this study. Other growing methods of imaging modalities include CTA, and CT perfusion. Studies have proven each to be effective in identifying biomarkers for expansion predictions [15]. Currently, these two modalities are limited to a select number of facilities and are not typically physicians ‘go-to’ imaging techniques when dealing with such a time sensitive case. Over the last decade, several non-contrast CT imaging biomarkers (NCCT-IBs) have been developed for the identification and characterization of HE (HE) after ICH, such as hypodensities, blend sign, satellite sign, island sign (IS), and shape irregularity. The aforementioned signs are indicated in Figure 1.[4,5]
Figure 1:
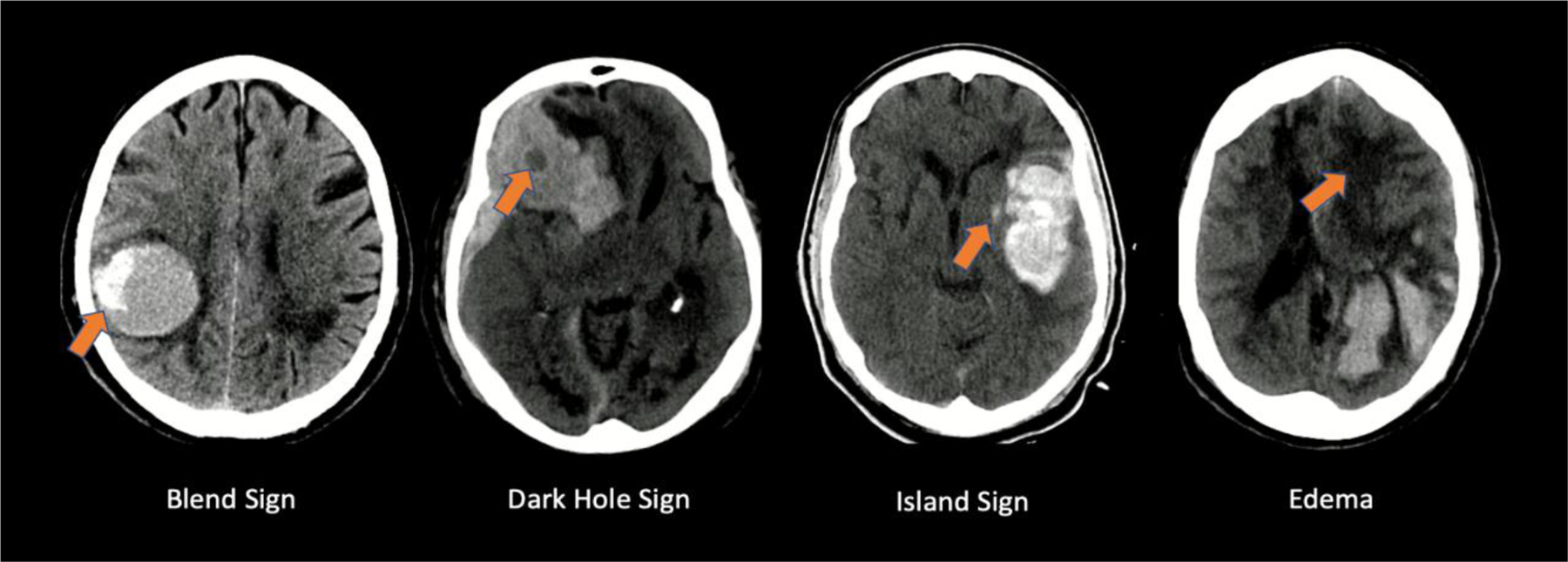
This figure demonstrates examples of specific features within non-contrast computed tomography images of hematoma cases, of which can demonstrate likelihood of hematoma expansion.
Different groups have studied the use of singular biomarkers to predict HE and have achieved moderate probability for accurate diagnosis (PAD) between 60% and 65% [5]. There is a myriad of possible permutations of imaging biomarkers that have been utilized in previous studies. One in particular describes using NCCT images within 6 hours of onset of symptoms to identify dark hole signs, blend signs, island signs, and swirl signs. Swirl signs are defined as the presence of swirling hypodensities that sweep across and through hematomas, often irregular in shape [15]. This group was able to yield a peak area under the receiver operator characteristic curve of 0.568 with 414 cases used in the analysis. Note this group statistically analyzed the data to make predictions rather than taking advantage of a deep learning network to aid in analysis and predictions. They also focused on the effectiveness of each sign independent from one another to truly analyze the predictive capabilities of these biomarkers.
Recent studies have demonstrated the ability of the AI-driven algorithms to use neuro-imaging biomarkers to improve diagnosis [6–9], assess the disease severity [10–12] and provide accurate prognosis of delayed treatment outcome [13,14]. We hypothesize that there are interdependent NCCT-IBs which could be used with machine learning algorithms to improve the diagnosis accuracy. To investigate this, we designed a study which used multiple combinations of the previously mentioned parameters as inputs for supervised machine learning models and tested the accuracy to predict HE. Specifically, this study analyzed prediction capabilities with four imaging biomarkers: blend signs, dark hole (black) signs, island signs, and edema. Each biomarker has been proven to be an indicator of HE in some capacity [18–21]. All of these signs can be identified consistently and with ease.
2. MATERIALS AND METHODS
This study was approved by the IRB at the University at Buffalo. In accordance with the approved protocol. NCCT DICOM files for 326 subjects with ICH were retrospectively collected. Each DICOM file consisted of individual CT image slices with a thickness of 5 mm. The data was originally acquired with a 512×512 pixel column detector. In-plane resolution was 0.4 mm. During acquisition, the tube voltage and current ranges were 120 to 135 kV, and 43.6 to 55.5 mGy, respectively. Overall time from scan to full reconstruction was around 1–2 minutes. Using brain tissue window level, the hematoma was located and analyzed. The presence/absence of the imaging biomarkers including blend signs, dark hole (black) signs, island signs, and edema (Figure 1) was recorded for each patient. Expansion data for each subject was provided from the acting physician on each case. This data was concatenated alongside feature presence in a binary format (1- present/expanded, 0- absent/no expansion).
Blend signs are associated with heterogenous hematomas, which is shown by differing densities of blood within the hematoma. These contrasting attenuations can be attributed to blood of different age. Hypo attenuated regions (appears bright, white) describe actively bleeding blood, where darker, hyper attenuated regions show older blood which has undergone clotting. Thus, these heterogenous hematomas can often show signs of continued bleeding, leading to HE.
Black hole signs are delineated as hypo attenuated areas fully encompassed by the remaining, more attenuated hematoma. The difference in attenuation should be clearer and more obvious than the blend signs. Similar to the concept described for blend signs, this heterogenous nature hints towards an active bleeding site with differently aged volumes of blood within the hematoma.
Island signs are defined as scattered and irregular groupings of what appear to be smaller hematoma ‘islands’ that amalgamate into a cohesive, larger hematoma. The main factor in island signs that increases the risk of expansion is the irregularity of the hematoma shape and how it originally formed in these individual ‘pockets’ of blood. Multiple studies have found that added irregularities can lead to a heightened risk of further expansion.
Edema is present in the form of significantly darker regions within the CT images of a patients’ brain, often surrounding the edges of the hematoma itself. Edema is an indicator of the many secondary injuries that follow the initial hemorrhage, further deteriorating the patient’s status and health. Thus, edema is associated with poor outcomes with respect to hematomas, giving the ability to predict further expansion. A limiting factor with strictly using edema as an indicator is its common nature in these cases. With bleeding in the brain, most cases will have edema present, complicating the machine learning algorithm’s ability to properly predict HE.
Of the cases in which HE occurred, there were 57 instances of blend signs, 40 black signs, 53 island signs, and 68 signs of edema. As for those that did not expand, 51 instances of blend signs, 44 black signs, 53 island signs, and 113 signs of edema were witnessed (Figure 2). Of the 326 cases, 116 experienced HE, and 210 did not experience HE.
Figure 2:

Project design demonstrating the step by step thought process from initial data collection to results.
Keras [22], Google LLC, Menlo Park (CA) python machine learning framework and a backend to TensorFlow, was used to develop a deep learning neural network (DNN) in Python 3.8 [23]. The network had four input nodes, which were the four signs/features mentioned above (blend, black, island, edema). This was followed by 3 dense layers with 20, 12, and 8 fully connected neurons, respectively. Each layer was activated with relu except the final layer, and the overall model was optimized with adam. This architecture is further described in Figure 3. The model was tasked with predicting whether or not a hematoma would expand based on its features. The model was compiled with the Adam optimizer and binary cross entropy as the loss function guide to the training. The data was split so that 70% was utilized for training, and 30% was utilized for testing (228 training, 98 testing). Twenty iterations of Monte Carlo cross-validation were conducted to determine the variability of the results. The model was evaluated using prediction accuracy and receiver operating characteristic (ROC) curve. Furthermore, each feature was independently tested, as well as all possible permutations of features within the same algorithm to evaluate the impact of each feature on the ability to detect HE. In order to optimize the performance of the deep learning neural network, callbacks were initiated into the model. These include a method of only saving training progresses that improve on the previous epoch, and adjusting the learning rate depending on how the model is training on the dataset. Additionally, there is an added callback to stop the training progress once the model has ended its improvements on the training set. Also, a validation split of 0.2 was used. A validation split takes a percentage of the training set (in this case 20%) and separately evaluates the loss and accuracy This adds to the efficiency and quickness of the code’s runtime.
Figure 3:

Machine Learning Algorithm Design
3. RESULTS
A total of 326 cases were analyzed by the machine learning code in order to predict HEs. The designed algorithm successfully reads and interprets the constructed datasets, predicting expansion for each given case. In the trial containing each of the four parameters, the model garnered an accuracy of 70.17%, and printed a ROC curve (Figure 4) with an area under the ROC curve (AUROC) of 0.70 as shown in Table 1. The AUROC combined with the standard deviation can be reanalyzed as the probability of accurate diagnosis (PAD). In this case, all four parameters show PAD = 70±2. As each combination of features was tested, the results varied in performance. Notably, edema alone performed the worst, showing a PAD = 57±3. For all of the independent tests (utilizing one parameter as input), the strongest feature was island sign, with a PAD of 64±3. Of the trials with three inputs, blend, black, and edema performed the best yielding a PAD of 70±4 whereas black, island, and island performed at 69±2. Furthermore, for tests with two inputs, blend and island outputted the best results alongside blend and black at 68±3 and 68±5 respectively. The lowest performance of two input parameter tests was 61±3 from using blend and edema.
Figure 4:

Receiver operator characteristic curve demonstrating algorithm performance for each permutation. Curves demonstrate better performance with multiple parameters, with peak prognosis probability predictions of 70% for all parameters, and blend, black, and edema. *A. Blend, Black, Island, Edema, B. Blend, C. Black, D. Island, E. Edema, F. Blend, Black, Island, G. Blend, Black, Edema, H. Blend, Island, Edema, I. Black, Island, Edema, J. Island, Edema, K. Black, Edema, L. Black, Island, M. Blend, Edema, N. Blend, Island, O. Blend, Black*.
Table 1:
Performance Data on each biomarker permutation. The peak accuracy and probability of accurate diagnosis occurred utilizing all four parameters, and achieved 70.17% and 70±2, respectively. Typically, as input parameters increased, as did performance.
| NCCTBI Combination | Accuracy | Mean AUROC | STD | PAD (%) |
|---|---|---|---|---|
| Blend, Black, Island, Edema | 70.17 | 0.70 | 0.02 | 70±2 |
| Blend | 64.29 | 0.63 | 0.02 | 63±2 |
| Black | 63.88 | 0.60 | 0.02 | 60±2 |
| Island | 69.18 | 0.64 | 0.03 | 64±3 |
| Edema | 64.29 | 0.57 | 0.03 | 57±3 |
| Blend, Black, Island | 69.39 | 0.69 | 0.02 | 69±2 |
| Blend, Black, Edema | 68.57 | 0.70 | 0.04 | 70±4 |
| Blend, Island, Edema | 69.80 | 0.67 | 0.04 | 67±4 |
| Black, Island, Edema | 68.37 | 0.67 | 0.02 | 67±2 |
| Island, Edema | 68.57 | 0.65 | 0.03 | 65±3 |
| Black, Edema | 68.57 | 0.64 | 0.05 | 64±5 |
| Black, Island | 66.53 | 0.67 | 0.02 | 67±2 |
| Blend, Edema | 64.29 | 0.61 | 0.03 | 61±3 |
| Blend, Island | 68.57 | 0.68 | 0.03 | 68±3 |
| Blend, Black | 65.10 | 0.68 | 0.05 | 68±5 |
4. DISCUSSION
The results of the study show improved prognosis accuracy for HE when using AI-driven methods based on multiple imaging biomarkers. Additionally, this data is able to be efficiently and effectively interpreted using the created DNN. The network was able to utilize the combinations of present biomarkers to make accurate decisions on whether or not a hematoma may expand. Typically, the more input parameters involved in consideration, the more accurate the algorithm is able to perform. In that regard, with more cases the artificial intelligence model will be able to better fit unto the dataset and make more informed decisions. Although more input parameters could complicate the model, enough cases to provide data can help increase model accuracy and overall performance.
Some adverse results, such as the performance of edema, can be contributed to the fact that a great deal of cases involved this feature (181/326). This creates challenge for the network as it is not clear how the presence of edema leads to either outcome, with 113 and 68 edemas present in no expansion and expansion cases respectively. As for a feature such as blend signs, there is a strong correlation between expansion and its presence. This is displayed when using blend as the only network input as the network was able to perform with a 64.29% accuracy. These results coincide with the percentage of blend sign in expansion (49%) and no expansion (24%). This distinct difference in percentage of presence allows the network to more easily identify patterns within the data and more closely correspond a blend sign presence with expansion. This increases when looking at hematomas with multiple markers present, as it is significantly more likely for hematomas to expand when containing more than one biomarker.
When looking at specific cases that successfully predicted expansion, specifically below in Figure 5, there are typically clear indicators of hematomas at high expansion risks. The case on the left shows a massive hematoma with a blend sign, and a number of island signs, as well as surrounding edema present. With three of the four indicators, the model was able to easily flag this as an expansion case. Looking at the case to the right, you see a much less significant hematoma with only a slight degree of edema surrounding the blood within the brain. The model predicted that this case would not expand, and had done so successfully. Here are two examples of proper prediction, with one being a true positive, and the other a true negative.
Figure 5:
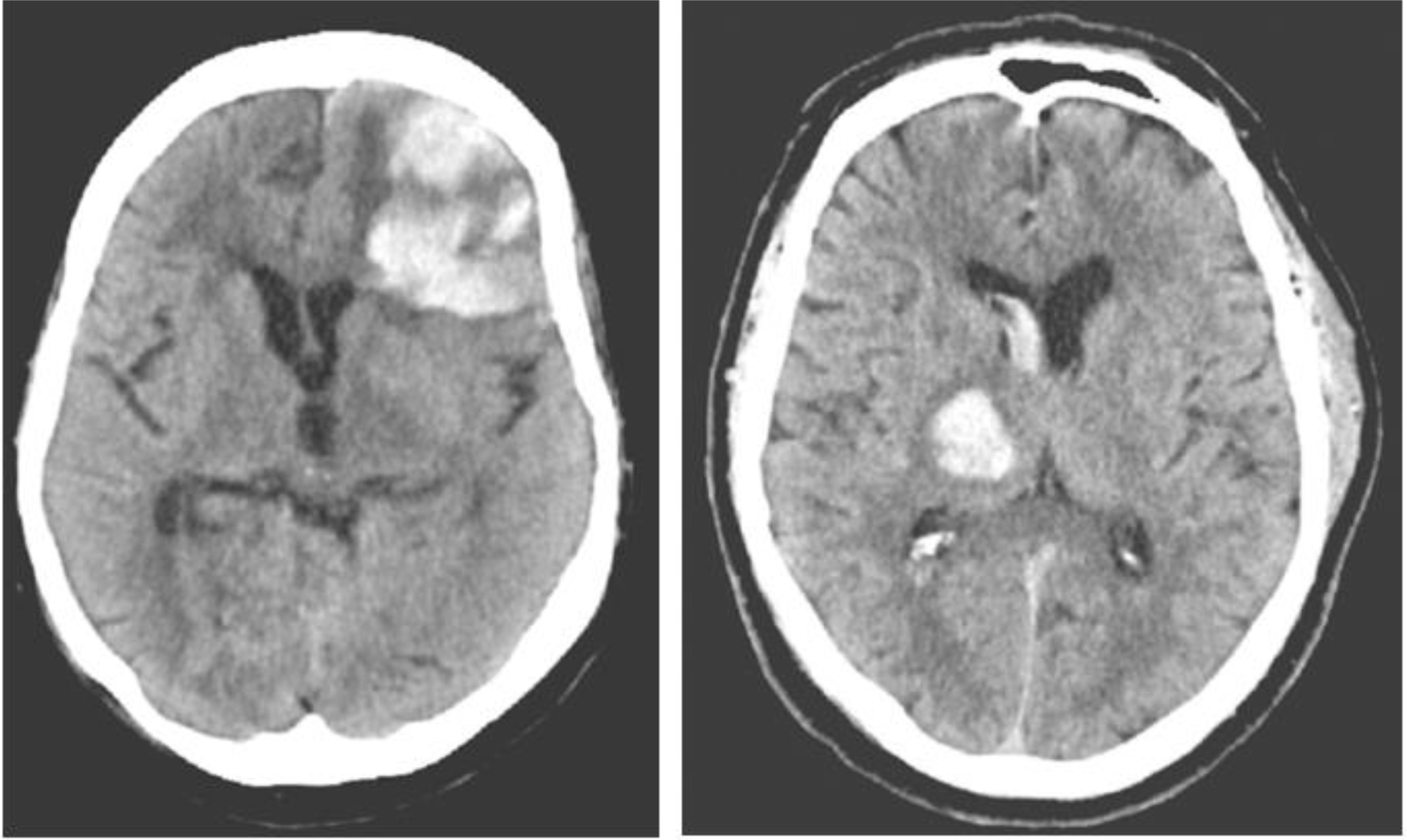
Examples of properly predicted hematoma cases.
It is important to also analyze some cases in which the algorithm predicted incorrectly in order to understand how to improve performance and mitigate the probabilities of inaccurate diagnosis. Looking at Figure 6 below, on the left is an example where the algorithm predicted expansion, when no expansion occurred (false positive). Surprisingly, this case shows a great deal of islands throughout the hematoma, as well as edema cutting in and around the masses of blood within the brain. A black spot can also be found in the upper left corner of the hematoma. Based on instincts, one may think such a heterogenous pool of blood, especially at this size would be prone to expansion. However, the data states this case did not undergo any further expansion, resulting in a false positive. According to this case, one may justify the incorrect case based on the fact that such a serious and complex hematoma may have already reached its maximum volume upon the imaging taking place. Therefore, according to the physician, the hematoma did not expand beyond this point. Perhaps if the scan were done sooner, biomarkers may have still been present, allowing for the model to identify an expansion case, which in turn would have actually been correct.
Figure 6:
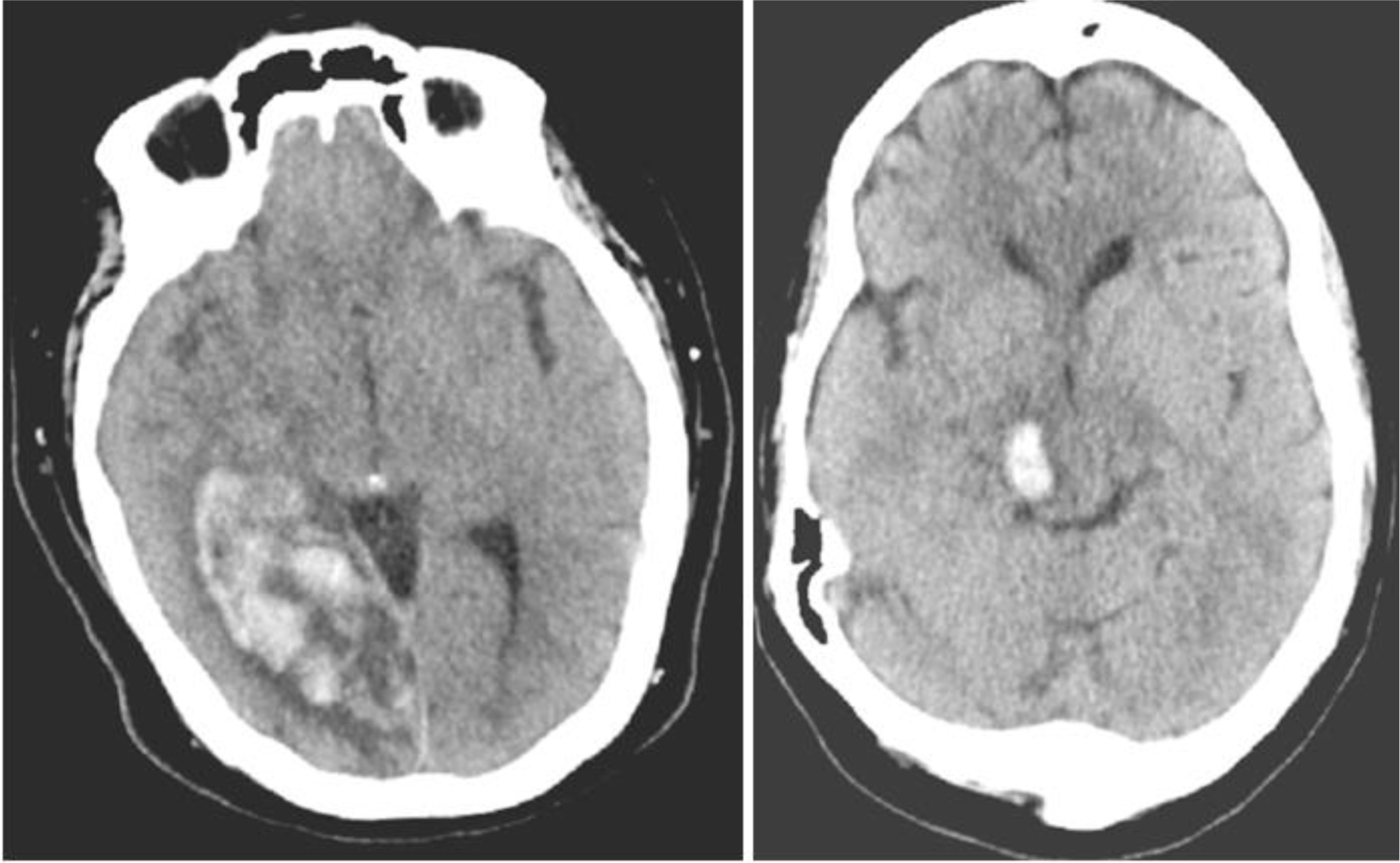
Examples of incorrect HE predictions.
The example case on the right-hand side shows a scenario where the model predicted a false negative. In looking at this case, there is a relatively smaller hematoma in comparison, with no true presence of the biomarkers been screened for. As a result, the model concluded this specific hematoma would not expand based on its training. Again, this was incorrect. One could hypothesize this specific ICH was found so early in its onset, that potentially upon the imaging the hematoma had yet to develop into a clearer state of disease with more information in and around the site of the ICH.
Within this work we demonstrated that specific attributes of intracranial hemorrhages may influence the prognosis of HE using supervised machine learning. We also demonstrate that certain parameters (i.e., blending region, dark hole, island, and edema) may be more influential to reach accurate conclusions about potential expansion. These insights into expansion predictions can lead to quick and efficiently informed decisions about hemorrhage treatments.
5. CONCLUSION
Utilizing DNN models to predict HE using biomarkers found in NCCT images shows potential for further investigation into a more automated method for HE. The network was able to predict the occurrence of an expansion with a peak accuracy of 70.17%, and probability of accurate diagnosis of 70±2%. This technology could aid physicians with probabilities of expansion, in order to assist in decision making on how to progress with treatment on a patient-to-patient basis.
6. ACKNOWLEDGEMENTS
This work was supported by the James H. Cummings Foundation and partially supported with equipment from Canon Medical Systems, Tustin, CA, NIH grants 1R01EB030092, 1R21NS123478–01 and NSF STTR # 2111865.
This work was partially supported by Canon Medical Systems Inc. This work was partially supported by Stratasys Ltd.
7. REFERENCES
- [1].Caplan LR, “Stroke Classification,” Stroke, 42(1_suppl_1), S3–S6 (2011). [DOI] [PubMed] [Google Scholar]
- [2].Counsell C, Boonyakarnkul S, Dennis M et al. , “Primary Intracerebral Hemorrhage in the Oxfordshire Community Stroke Project,” Cerebrovascular Diseases, 5(1), 26–34 (1995). [Google Scholar]
- [3].Flaherty ML, Woo D, Haverbusch M et al. , “Racial variations in location and risk of intracerebral hemorrhage,” Stroke, 36(5), 934–7 (2005). [DOI] [PubMed] [Google Scholar]
- [4].Boulouis G, Morotti A, Brouwers HB et al. , “Association Between Hypodensities Detected by Computed Tomography and HE in Patients With Intracerebral Hemorrhage,” JAMA Neurol, 73(8), 961–8 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Morotti A, Boulouis G, Romero JM et al. , “Blood pressure reduction and noncontrast CT markers of intracerebral hemorrhage expansion,” Neurology, 89(6), 548–554 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Rava RA, Mokin M, Snyder KV et al. , “Performance of angiographic parametric imaging in locating infarct core in large vessel occlusion acute ischemic stroke patients,” J Med Imaging (Bellingham), 7(1), 016001 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Rava RA, Peterson BA, Seymour SE et al. , “Validation of an artificial intelligence-driven large vessel occlusion detection algorithm for acute ischemic stroke patients,” Neuroradiol J, 34(5), 408–417 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Rava RA, Seymour SE, LaQue ME et al. , “Assessment of an Artificial Intelligence Algorithm for Detection of Intracranial Hemorrhage,” World Neurosurg, 150, e209–e217 (2021). [DOI] [PubMed] [Google Scholar]
- [9].Rava RA, Snyder KV, Mokin M et al. , “Assessment of computed tomography perfusion software in predicting spatial location and volume of infarct in acute ischemic stroke patients: a comparison of Sphere, Vitrea, and RAPID,” J Neurointerv Surg, 13(2), 130–135 (2021). [DOI] [PubMed] [Google Scholar]
- [10].Rava RA, Seymour SE, Snyder KV et al. , “Automated Collateral Flow Assessment in Patients with Acute Ischemic Stroke Using Computed Tomography with Artificial Intelligence Algorithms,” World Neurosurg, 155, e748–e760 (2021). [DOI] [PubMed] [Google Scholar]
- [11].Shiraz Bhurwani MM, Snyder KV, Waqas M et al. , “Use of quantitative angiographic methods with a data-driven model to evaluate reperfusion status (mTICI) during thrombectomy,” Neuroradiology, 63(9), 1429–1439 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Podgorsak AR, Sommer KN, Reddy A et al. , “Initial evaluation of a convolutional neural network used for noninvasive assessment of coronary artery disease severity from coronary computed tomography angiography data,” Med Phys, 47(9), 3996–4004 (2020). [DOI] [PubMed] [Google Scholar]
- [13].Podgorsak AR, Rava RA, Shiraz Bhurwani MM et al. , “Automatic radiomic feature extraction using deep learning for angiographic parametric imaging of intracranial aneurysms,” J Neurointerv Surg, 12(4), 417–421 (2020). [DOI] [PubMed] [Google Scholar]
- [14].Shiraz Bhurwani MM, Waqas M, Podgorsak AR et al. , “Feasibility study for use of angiographic parametric imaging and deep neural networks for intracranial aneurysm occlusion prediction,” J Neurointerv Surg, 12(7), 714–719 (2020). [DOI] [PubMed] [Google Scholar]
- [15].Cai J, Zhu H, Yang D, Yang R, Zhao X, Zhou J, & Gao P (2020). Accuracy of imaging markers on noncontrast computed tomography in predicting intracerebral hemorrhage expansion. Neurological research, 42(11), 973–979. 10.1080/01616412.2020.1795577 [DOI] [PubMed] [Google Scholar]
- [16].Brouwers HB, & Greenberg SM (2013). HE following acute intracerebral hemorrhage. Cerebrovascular diseases (Basel, Switzerland), 35(3), 195–201. 10.1159/000346599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Senn R, Elkind M, S, V, Montaner J, Christ-Crain M, Katan M: Potential Role of Blood Biomarkers in the Management of Nontraumatic Intracerebral Hemorrhage. Cerebrovasc Dis 2014;38:395–409. doi: 10.1159/000366470 [DOI] [PubMed] [Google Scholar]
- [18].Li Q, Zhang G, Xiong X, Wang X-C, Yang W-S, Li K-W, Wei X, Xie P, 2016. Black Hole Sign. Stroke 47, 1777–1781. doi: 10.1161/strokeaha.116.013186 [DOI] [PubMed] [Google Scholar]
- [19].Li Q, Zhang G, Huang Y-J, Dong M-X, Lv F-J, Wei X, Chen J-J, Zhang L-J, Qin X-Y, Xie P, 2015. Blend Sign on Computed Tomography. Stroke 46, 2119–2123. doi: 10.1161/strokeaha.115.009185 [DOI] [PubMed] [Google Scholar]
- [20].Li Q, Liu Q-J, Yang W-S, Wang X-C, Zhao L-B, Xiong X, Li R, Cao D, Zhu D, Wei X, Xie P, 2017. Island Sign. Stroke 48, 3019–3025. doi: 10.1161/strokeaha.117.017985 [DOI] [PubMed] [Google Scholar]
- [21].Murthy SB, Moradiya Y, Dawson J, Lees KR, Hanley DF, Ziai WC, Butcher K, Davis S, Gregson B, Lyden P, Mayer S, Muir K, Steiner T, 2015. Perihematomal Edema and Functional Outcomes in Intracerebral Hemorrhage. Stroke 46, 3088–3092. doi: 10.1161/strokeaha.115.010054 [DOI] [PubMed] [Google Scholar]
- [22].Chollet F, [Keras], (2015).
- [23].Van Rossum G, & Drake FL (2009). Python 3 Reference Manual Scotts Valley, CA: CreateSpace. [Google Scholar]


