Abstract
Several pilot experiments have indicated that improvements in older NMR structures can be expected by applying modern software and new protocols (Nabuurs et al. in Proteins 55:483–186, 2004; Nederveen et al. in Proteins 59:662–672, 2005; Saccenti and Rosato in J Biomol NMR 40:251–261, 2008). A recent large scale X-ray study also has shown that modern software can significantly improve the quality of X-ray structures that were deposited more than a few years ago (Joosten et al. in J. Appl Crystallogr 42:376–384, 2009; Sanderson in Nature 459:1038–1039, 2009). Recalculation of three-dimensional coordinates requires that the original experimental data are available and complete, and are semantically and syntactically correct, or are at least correct enough to be reconstructed. For multiple reasons, including a lack of standards, the heterogeneity of the experimental data and the many NMR experiment types, it has not been practical to parse a large proportion of the originally deposited NMR experimental data files related to protein NMR structures. This has made impractical the automatic recalculation, and thus improvement, of the three dimensional coordinates of these structures. We here describe a large-scale international collaborative effort to make all deposited experimental NMR data semantically and syntactically homogeneous, and thus useful for further research. A total of 4,014 out of 5,266 entries were ‘cleaned’ in this process. For 1,387 entries, human intervention was needed. Continuous efforts in automating the parsing of both old, and newly deposited files is steadily decreasing this fraction. The cleaned data files are available from the NMR restraints grid at http://restraintsgrid.bmrb.wisc.edu.
Electronic supplementary material
The online version of this article (doi:10.1007/s10858-009-9378-z) contains supplementary material, which is available to authorized users.
Keywords: Biomolecular structure, BMRB, Restraints, Database, Nuclear magnetic resonance, PDB
Introduction
The first macromolecular X-ray structure (myoglobin) was solved in 1958 (Kendrew 1958). Thirteen years later, in 1971, the PDB was launched as a central repository for these data (Protein Data Bank 1971; Berman 2007).The idea of the PDB was to have a central data-warehouse where all structures should be deposited and from where researchers from all over the world could get free access to those valuable data. The first NMR-derived protein structures, BUSI IIa (Williamson et al. 1985) and the lac-headpiece (Kaptein et al. 1985) were published in 1985, and in 1988 the PDB accepted the first NMR structure ensemble (Driscoll et al. 1989). In the early nineties, most journals agreed that macromolecular structure data had to be deposited before the corresponding article could be published. The first X-ray reflection files were deposited in 1976 (PDB entry 155C), and X-ray reflection deposition became an obligatory aspect of the data deposition process in 2000 (Commission on Biological Macromolecules 2000). The first experimental NMR data deposited in 1991 consisted almost exclusively of NOE distance and dihedral angle restraints.
Experimental NMR data files are considerably more complex than X-ray reflection files in terms of semantics and associated syntax. In addition, NMR data assigned to specific atoms can be highly valuable even in the absence of a three-dimensional structure. It was proposed that a data bank organized by NMR data experts be instituted to collect and archive such information (Ulrich et al. 1989). The BMRB was launched in 1991 and has evolved into the recognized worldwide database for experimental NMR data (Seavey et al. 1991; Ulrich et al. 2008). In 2006, BMRB became a member of the Worldwide Protein Data Bank (wwPDB). The Advisory Committee of the Worldwide Protein Data Bank (wwPDB) recommended in 2007 that depositions of NMR structures should be accompanied by structural restraints, which was followed by the recommendation in 2008 to additionally deposit the assigned chemical shifts. The deposition of structural restraints became mandatory on February 1, 2008 (Markley et al. 2008), and the mandatory deposition of chemical shifts will be announced in 2009. By tradition, the coordinates of NMR structures, along with the raw restraints underlying the structures, have been deposited in the PDB, and the assigned chemical shifts and other experimental data have been deposited in the BMRB. Upon becoming a member of the wwPDB, the BMRB along with the European branch of the PDB (PDBe), assumed the task of curating the structural restraint data and recruited collaborators for this effort.
Experimental NMR data are highly heterogeneous, and both how certain data types are valued and which data types are actually valued are changing from year to year as the NMR research field develops. Although NOE distance restraints were the basis for the first NMR structures, currently a wide range of experimental data are used: coupling constants, chemical shifts, residual dipolar couplings, cross hydrogen bond couplings, and paramagnetic relaxation effects. As a consequence of this evolution, deposited experimental NMR data are highly heterogeneous, and owing to the lack of ontologies or common practices, these data are now hard to parse by one single computer program. Additionally, the lack of data validation possibilities in the early years of NMR allowed a massive number of errors in the deposited restraints to slip into the database. The concept of how best to represent NMR-derived structures has also evolved over the years. An initial idea, starting around 1986, held that averaging an NMR ensemble into a single structure would lead to a useful single molecular representation. However, following the introduction of validation software, such as PROCHECK and WHAT_CHECK, it was found that averaged structures often have extensive problems (Clore et al. 1986; Hooft et al. 1996; Laskowski et al. 1993; Nilges et al. 1988). Now, most structures are characterized by a family of conformers that represent both the inherent dynamics of the structure and the lack of structural restraints.
In light of these facts, we decided to take a three step approach toward remediating all experimental NMR data files. In the first step (parsing), we ensure that the data are syntactically correct. In the second step (conversion), we ensure that restraints belong to atoms that exist. In the final step (filtering), we enforce semantic correctness, which includes at least some possibility of proximity for atoms that syntactically have been connected by a NOE. The results of the second step have been stored in the Database Of Converted Restraints (DOCR), while the results of the third step have been stored in the Filtered REstraints Database (FRED). DOCR and FRED are freely available from the NMR restraints grid (NRG) at http://restraintsgrid.bmrb.wisc.edu. The initial version of the NRG included data from only 97 PDB entries (a database named “DB97”) (Doreleijers et al. 1998); in 2003 we had 545 entries (Doreleijers et al. 2003) and the previous version of the NRG included data from 1,400 entries (Doreleijers et al. 2005). Here we present the completion of the effort to include all 5,266 entries.
Methods
Data preparation
Our previous procedure for preparing NMR coordinates and restraints (Doreleijers et al. 2003, 2005) has been improved as summarized in Fig. 1. The following issues have been addressed: (a) We now retain the original positions of the hydrogen atoms. Because almost all modern files have no missing atoms, it no longer is necessary to recalculate the coordinates for the hydrogen atoms. (b) We now take advantage of the richer data in the mmCIF formatted coordinate files to carry out a more direct translation to the NMR-STAR data model. (The less informative PDB-formatted coordinate files are derived from these mmCIF master files.) In order to benefit from the latest wwPDB remediation efforts (Henrick et al. 2008), we obtain the mmCIF coordinate files from the remediated archives available from ftp://ftp.wwpdb.org. (c) Our strategy for re/deassignment of stereospecifically assigned atom(-groups) now uses overall violation analysis of the distance restraints (Doreleijers et al. 2005, 1998), instead of the per-restraint assessment used in the RECOORD project (Nederveen et al. 2005). (d) A parser was added to the Wattos software for data from the EMBOSS structure calculation program, which are present in 14 older entries. In addition, a small scale effort was made to include data in the AMBER format in collaboration with Dr. David Case (Personal Communications). AMBER-formatted NMR restraints reference atom numbers instead of the more usual atom name in combination with residue (and chain) references. Dr. Case regenerated these numbering schemes for 7 out of the 153 entries with AMBER-formatted restraints now included in the NRG. Authors of new entries subsequent to this effort are always requested to provide the numbering schemes, and many have complied. In total, 56 out of the 153 entries were converted by means of user-supplied numbering schemes. (e) Dihedral angle violations are now included, whereas before only distance violations were included. No correction for the equivalence of the Phe/Tyr sidechain atoms was attempted, which for some entries results in the reporting of very high violations that are not real. We adopted the Google Code software issue reporting mechanism to improve the quality of the NRG databases. The Google Code issue 212 available from http://code.google.com/p/nmrrestrntsgrid/issues/list details this particular issue. Therefore, these dihedral angle violations are ignored in the analyses described below.
Fig. 1.
Data flow chart showing the software tools involved in the project, Wattos and FormatConverter, and the semi-automated steps carried out by BMRB annotators. The coordinate data comes from the Worldwide Protein Data Bank (wwPDB) in an mmCIF formatted file that adheres to the PDB eXchange dictionary (pdbx). These coordinates and the restraint data file are converted to NMR-STAR and combined into a single file by Wattos. The FormatConverter then matches the two pieces of information and converts the data from NMR-STAR to CCPN, XPLOR, and CYANA formats for the Database of Converted Restraints (DOCR). Only for the Filtered REstraints Database (FRED) are the data interpreted to be consistent with the ensemble, without surplus (see text), and have the best matching stereospecificity. In order to assess whether the data are complete and well converted, the distance restraints are checked for violations and NOE completeness. Authors are only contacted for about 10% of the entries to resolve outstanding issues. BMRB never updates the original wwPDB Magnetic Resonance (MR) input restraints, but requests that the other wwPDB deposition sites do so after which BMRB processing is iterated on the updated data sets
Results from these remediation efforts, in NMR-STAR, CCPN, CYANA, and CNS data formats, are available from the DOCR and FRED databases in the NRG (Fig. 1). The vast majority of restraints (those from distance, dihedral angle and RDC measurements) are processed; those based on other types of information are not processed, because they have proved much more difficult to parse. Entries that could not be processed (fully) because of a variety of issues are tracked on the Google Code web site in the spreadsheet: http://code.google.com/p/nmrrestrntsgrid/source/browse/trunk/nmrrestrntsgrid/data/problemEntryList.csv?r=161, which is constantly updated. At the time of writing (revision 161), 221 entries were linked to 14 issues. The most common issue by far (issue 25), which is active for 154 entries, arises from incomplete parsing of AMBER data by the Wattos software. This issue leads to incomplete conversion of parsed restraints to the NMR-STAR format with the consequence that restraints could not be linked to the coordinate data. The authors of this paper are continuing to resolve these issues, and, as a result, the list of problematic entries is highly dynamic.
Conversion and data linking
The FormatConverter software (Vranken 2007; Vranken et al. 2005) imports an NMR-STAR file into the CCPN framework (Fogh et al. 2005) and subsequently links the restraint information to the coordinate data. Although the number of entries increased by nearly a factor of ten, from the 545 monomeric proteins entered in DOCR and FRED (Doreleijers et al. 2005) to the current 5,266, the number of entries (1,387) that needed a manual setting for the linking only increased by a factor of about two. Two corrections commonly were required: (a) sequence matching for proteins that contained one or more coordinated metals such as zinc or cadmium, (b) atom name matching such as H2′/H2′′/HO2′ and thymine methyl H7 s for nucleic acids. Improvements to the automated part of the workflow included: (i) better automatic matching between the atom information from the experimental data file and the molecular system description from the mmCIF file, both by code improvements and by better reference data, and (ii) more informative output about the conversion process for quicker manual curation (if required). In addition many smaller fixes were made in the code, leading to a more dependable and consistent outcome of the conversion step. The code to export NMR-STAR files was completely rewritten to produce valid and complete version 3.1 files.
Filtering
Distance restraints (DRs) with violations over 2 Å (up to a maximum of three per entry) were categorized as ‘Typos’ and left out of the FRED database as outliers. Although DRs identified as typos are sometimes real, the impact of leaving them out is expected to have a minimal impact on the overall structure. Often these restraints are errant violations that were not observed at the time of structure calculation but arose as a consequence of correcting other problems, such as typographical errors that led to a restraint being accidentally uncommented or to the incorrect mapping of one or two atom names.
In April 2006, we began to contact authors when our processing identified deposited data that led to high violations or were suspected of being incomplete. We received many positive responses, and this type of direct communication has led to improvements in processing by annotators at BMRB and to improved data sets available at the wwPDB. This procedure also caught an estimated 100 cases in which incomplete or incorrect data were sent since 2006.
Project management
A large collaborative project such as this inevitably requires the identification and remediation of issues with software developed and procedures used. Initially, the problems were identified and shared by a spreadsheet. In March, 2008, the issues were converted to a Google Code repository at: http://code.google.com/p/nmrrestrntsgrid which is used to track these issues and to link them to codes in the NRG project. Currently, almost all of the ~200 issues listed have been addressed. The documentation is conveniently described in Wiki pages at the same site. In addition, weekly video conferences and several in-person visits from JFD, WFV, and CJP to the BMRB in Madison have helped to keep this project organized which is deemed essential to maintain the databases up to date as well as reliable.
Results
NRG database overall composition
On August 3, 2009, the wwPDB contained a total of >59,000 PDB entries with ~8,000 (14%) of NMR origin (Table 1). For a growing majority of NMR entries, authors have also included restraints (5,266; 66%). Almost all of those entries, 4,800 or 90% had some restraints that could be parsed with the NRG setup, i.e. DR, dihedral angle, and RDC restraints. Most entries with restraints that could not be parsed still have these types of restraints (data not shown). All but 56 of those 4,800 entries (4,744) contain DRs, although many other NMR data types occur in addition. The homepage of the NRG website shows the overview of data types (rows) and programs (columns) in a grid, hence the name NRG.
Table 1.
Sets of PDB entries in relation to set selection criteria
| Set of entries | Counts |
|---|---|
| PDB overall | 59,330 |
| NMR with or without restraints | 7,980 |
| NMR with restraints | 5,266 |
| With parsed restraints | 4,800 |
| With parsed DRs | 4,744 |
| Set 1 < 80% restraints linked | 415 |
| Set 2 < 33% restraints after filtering left | 316 |
| Set 3 maximum DR violation > 2 Å | 353 |
| Set 4 Rms DR violation > 0.25 Å | 277 |
| Set union of 1–4: (union 1–2: 475, 3–4: 417) | 786 |
| ‘Good’ set (with parsable restraints minus set union of 1–4) | 4,014 |
Reformatted files
The BMRB, in collaboration with the NMR community and the Collaborative Computing Project for NMR (CCPN) (Vranken et al. 2005) is developing the next version of the NMR-STAR data dictionary (http://www.bmrb.wisc.edu/dictionary/htmldocs/nmr_star/dictionary.html). Many programs use the NMR-STAR format for exchanging experimental NMR data. All three databases available from the NRG user interface: (parsed data sets, DOCR, and FRED) adhere to the “developmental predecessor of NMR-STAR version 3” and will be updated to the final version 3 data dictionary when released.
Stereospecificity and surplus
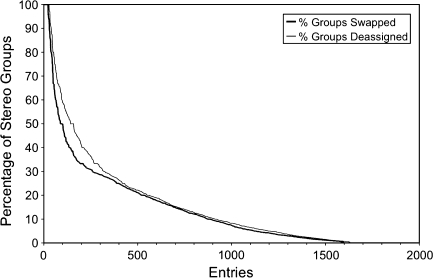
In converting data from DOCR to FRED, the atomic coordinate models in the PDB entries are synchronized so that inconsistencies between models are removed (Fig. 1). Most importantly, atoms and residues not present in all models are removed from the models in which they are present. This is a necessary but uncommon operation. More importantly, the DOCR data are interpreted for stereospecificity of the distance restraints. As a consequence of recent remediation efforts (Henrick et al. 2008), the stereospecific nomenclature of atoms in the coordinates of all but a few isolated cases (NRG Google Code issue 164), was found to be consistent with the IUPAC recommendations (Markley et al. 1998). The correctness of our interpretation of the stereospecificity of the atoms in the DRs was checked as described before (Doreleijers et al. 2005). Swapping and deassigning stereospecific assigned DRs was not needed in the vast majority of entries (Fig. 2). Nevertheless, this remains an important step needed to eliminate high DR violations in the affected files, and its application allowed many more entries to be included in the set of ‘Good’ entries described below.
Fig. 2.
The results of swapping and deassigning stereospecific assigned DRs. Most entries have none or only small percentages of these modifications indicating that the stereospecific information was treated correctly. A few entries required swapping of all stereo groups as indicative of a nomenclature problem. Floating chirality was used in another set of entries. Both issues are resolved by this procedure. The x-axis has been truncated to showcase the 1,630 entries with percentages above zero, out of a total set of 4,588 entries with converted DR
The data were subsequently filtered for what we call surplus DRs (Fig. 3). These surplus DRs do not add information to a structure calculation and can even contradict generally accepted molecular topology parameters. Twenty times more double restraints were found in the current ‘Good’ set when compared to DB545 (Doreleijers et al. 2005). This implies that less attention has been given to the application of good practices in reporting restraints for the added entries.
Fig. 3.
The results of filtering surplus restraints are shown for the four categories of surplus restraints and their sum are listed under ‘surplus’ in the legend. The average values and s.d.s are listed in the legend between brackets for each category. The percentage of double restraints in DOCR for the set of ‘finished’ entries, now 3.4%, is much higher than the 0.2% obtained previously for the set of 545 monomeric protein entries
Selection criteria for the ‘Good’ set
Four criteria were used to disqualify entries that might not have been interpreted correctly by our setup or for which the data seems to mismatch between coordinates and restraints for another reason. The first two criteria check what percentage of restraints remained after processing to DOCR and filtering to FRED. Table 1 shows that the union of those first two criteria in Sets 1 and 2 consists of a total of 475 entries. Figure 4 shows the entries sorted by these two criteria.
Fig. 4.
PDB entries with parsable NMR restraints (distance, dihedral or dipolar) from the programs X-PLOR, CYANA, Discover, or AMBER have been entered into DOCR (after conversion) and FRED (after filtering). The semi-automatic processing at BMRB fails in some entries for one or more restraints, and the percentage of restraints that successfully completed (shown here) is one way of identifying remaining problems. Note that of the entries that fail the 80% cutoff in DOCR most also fail the 33% cutoff in FRED
The second two criteria are on the maximum (not averaged over the ensemble) and rms averaged distance violations of the FRED data (Fig. 5). All together the four criteria disqualified 754 entries. The remaining 3,208 entries are identified as the ‘Good’ set. This set of ‘Good’ entries can be retrieved from the Supplemental Materials or from: http://restraintsgrid.bmrb.wisc.edu/servlet_data/viavia/mr_mysql_backupAn_2009-08-03 in the file ‘dump_file.sql’. This small file suffices to regenerate the full description and population of the metadata and analyses in the MySQL relational database (Dyer 2008) that underlies the NRG. It includes some of the metadata derived from the NMR-STAR files in NRG on stereospecificity, violation, and NOE completeness. For example, the set of good entries is retrieved by the SQL command: “SELECT * FROM DOCRFREDGoodies into OUTFILE ‘DOCRFREDGoodies.csv’;”. This set of entries fulfils the criteria set above which makes them better suited for e.g. large scale analyses and structure recalculation efforts, because they do not suffer from the serious defects observed in the lack of data consistency or high distance restraint violations. The defects that exclude entries from the ‘Good’ set result, in most cases, from our processing setup and do not mean that the entries by themselves are bad; they could just not be handled properly in the NRG setup.
Fig. 5.
PDB entries with parsable DR (NOE, hydrogen bonds, etc.) are displayed along with their violations. Distance violation cutoffs provide a way of identifying problem entries that warrant further investigation. Note that many entries that fail the 2 Å maximum cutoff also fail the 0.25 Å RMS cutoff
NOE completeness
The NOE completeness (Doreleijers et al. 1999) has been analyzed for all entries in FRED meeting the four criteria (‘Good’ set). From Fig. 6, it can be concluded that the nucleic acid entries and specifically the DNA entries in FRED have a lower overall NOE completeness than the protein only entries in FRED. The NOE completeness, as calculated here, is normalized for the expected contacts from the deposited coordinates and as such does not depend on molecule type, size, shape, or proton density. A simple explanation might be that the ribose hydrogen resonances are more overlapping and thus difficult to resolve.
Fig. 6.
The NOE completeness for proteins (45.6%) is significantly higher on average than that for RNA (34.3%) and DNA (26.8%). 313 Entries that were solved using RDCs were omitted in this plot because RDCs can be the major component of the restraints used for structure calculation. However, after this selection there are still 26 entries (24 proteins/2 DNAs) that have no NOE distances submitted as evidenced by a NOE completeness of 0% here. Entries consisting of a protein/nucleic acid complex and/or a ligand were also left out in order to prevent mixing molecule types. The NOE completeness as calculated here, is normalized for the expected contacts from the deposited coordinates and as such does not depend on molecule type, size, shape, or proton density. However, the selection of atoms that are expected to give contacts does differ somewhat between these molecule types
Conclusions
We have presented the completion of the NRG effort to include all 5,266 PDB entries with NMR restraints. The vast majority of entries (4,014) was found to fulfill reasonable criteria on consistency and agreement between restraint and coordinate data. For a significant number of ‘suspect’ validated entries we have contacted authors. This has led to improvements in our processing and more importantly in more complete and correct data sets conveniently available to all NMR spectroscopists.
This effort also provides an important stepping stone for new longitudinal analyses (studies over many entries) (Vranken 2007), and for validation with the CING software (Vuister et al. to be published, http://nmr.cmbi.ru.nl/cing and http://nmr.cmbi.ru.nl/NRG-CING), and it provides comparison datasets for structure recalculation efforts such as the recent competition with blind targets in an eNMR workshop http://www.enmr.eu/softwareworkshop. The effort resulted in the setup of a continuing effort for the Critical Assessment of automated Structure Determination from NMR data/CASD-NMR (Rosato et al. 2009).
Future perspectives
A number of clear improvements need to be addressed. (a) The parsers for the AMBER-formatted restraints need extensive overhaul so that they can fully process this class of restraints. (Google Code issue 25). (b) The NRG setup needs to be able to support the NMR-STAR and CCPN data formats directly as input, because these two formats are becoming more common (issue 209). (c) NRG processing should be integrated with deposition systems such as ADIT-NMR in order to have more efficient communication with the authors at the time of deposition (issue 210). (d) RDC restraint violations need to be calculated (issue 211). (e) Many of the dihedral angle restraint violations should be eliminated by correcting for Phe/Tyr sidechain rotation (issue 212). (f) Last but not least, the NRG data should be integrated with the main BMRB data on chemical shifts that will soon be mandatory for PDB submission.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgments
We acknowledge the many authors who contributed the results of their scientific investigations to the PDB and BMRB. They created the true resource that this secondary database relies upon. Financial support for this work came from the Netherlands Organisation for Scientific Research; NWO 700.55.443, Netherlands Bioinformatics Centre (NBIC) and EU FP6 EMBRACE grant LHSG-CT-2004-512092, EU FP6 STREP Extend-NMR grant LSHG-CT-2005-018988 (Nijmegen), BBSRC grant BB/E007511/1 (Hinxton), and the US National Library of Medicine (grant P41 LM05799) (Madison).
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Abbreviations and symbols
- BMRB
BioMagResBank
- CCPN
Collaborative Computing Project for NMR
- DB545
Set of 545 PDB entries
- DB97
Set of 97 PDB entries
- DR
Distance restraints
- DOCR
Database of converted restraints
- FRED
Filtered REstraints database
- MR
Magnetic resonance
- NRG
NMR restraints grid
- PDB
Protein Data Bank
- Pdbx
Protein Data Bank eXchange dictionary
- PDBe
Protein Data Bank in Europe
- RDC
Residual dipolar coupling
- RMS
Root mean square
- s.d.
Standard deviation
- wwPDB
Worldwide Protein Data Bank
Footnotes
Jurgen F. Doreleijers, Wim F. Vranken and Christopher Schulte contributed equally to this work.
References
- Berman H. The Protein Data Bank: a historical perspective. Acta Crystallogr A. 2007;64:88–95. doi: 10.1107/S0108767307035623. [DOI] [PubMed] [Google Scholar]
- Clore GM, Brunger AT, Karplus M, Gronenborn AM. Application of molecular dynamics with interproton distance restraints to three-dimensional protein structure determination. A model study of crambin. J Mol Biol. 1986;191:523–551. doi: 10.1016/0022-2836(86)90146-4. [DOI] [PubMed] [Google Scholar]
- Commission on Biological Macromolecules Guidelines for the deposition and release of macromolecular coordinate and experimental data. Acta Cryst. 2000;D56:2. [PubMed] [Google Scholar]
- Doreleijers JF, Rullmann JAC, Kaptein R. Quality assessment of NMR structures: a statistical survey. J Mol Biol. 1998;281:149–164. doi: 10.1006/jmbi.1998.1808. [DOI] [PubMed] [Google Scholar]
- Doreleijers JF, Raves ML, Rullmann T, Kaptein R. Completeness of NOEs in protein structure: a statistical analysis of NMR data. J Biomol NMR. 1999;14:123–132. doi: 10.1023/A:1008335423527. [DOI] [PubMed] [Google Scholar]
- Doreleijers JF, Mading S, Maziuk D, Sojourner K. BioMagResBank database with sets of experimental NMR constraints corresponding to the structures of over 1400 biomolecules deposited in the Protein Data Bank. J Biomol NMR. 2003;26:139–146. doi: 10.1023/A:1023514106644. [DOI] [PubMed] [Google Scholar]
- Doreleijers JF, Nederveen AJ, Vranken WF, Lin J. BioMagResBank databases DOCR and FRED containing converted and filtered sets of experimental NMR restraints and coordinates from over 500 protein PDB structures. J Biomol NMR. 2005;32:1–12. doi: 10.1007/s10858-005-2195-0. [DOI] [PubMed] [Google Scholar]
- Driscoll PC, Gronenborn AM, Beress L, Clore GM. Determination of the three-dimensional solution structure of the antihypertensive and antiviral protein BDS-I from the sea anemone Anemonia sulcata: a study using nuclear magnetic resonance and hybrid distance geometry-dynamical simulated annealing. Biochemistry. 1989;28:2188–2198. doi: 10.1021/bi00431a033. [DOI] [PubMed] [Google Scholar]
- Dyer R. MySQL in a nutshell. 2. Cambridge: O’Reilly; 2008. [Google Scholar]
- Fogh RH, Boucher W, Vranken WF, Pajon A, Stevens TJ, Bhat TN, Westbrook J, Ionides J, Laue E. A framework for scientific data modeling and automated software development. Bioinformatics. 2005;21:1678–1684. doi: 10.1093/bioinformatics/bti234. [DOI] [PubMed] [Google Scholar]
- Henrick K, Feng Z, Bluhm WF, Dimitropoulos D, Doreleijers JF, Dutta S, Flippen-Anderson JL, Ionides J, Kamada C, Krissinel E, Lawson CL, Markley JL, Nakamura H, Newman R, Shimizu Y, Swaminathan J, Velankar S, Ory J, Ulrich EL, Vranken WF, Westbrook J, Yamashita R, Yang H, Young J, Yousufuddin M, Berman HM. Remediation of the Protein Data Bank archive. Nucleic Acids Res. 2008;36:D426–D433. doi: 10.1093/nar/gkm937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooft RWW, Vriend G, Sander C, Abola EE. Errors in protein structures. Nature. 1996;381:272. doi: 10.1038/381272a0. [DOI] [PubMed] [Google Scholar]
- Joosten RP, Salzemann J, Bloch V, Stockinger H, Berglund A, Blanchet C, Bongcam-Rudloff E, Combet C, Costa ALD, Deleage G, Diarena M, Fabbretti R, Fettahi G, Flegel V, Gisel A, Kasam V, Kervinen T, Korpelainen E, Mattila K, Pagni M, Reichstadt M, Breton V, Ticklei IJ, Vriend G. PDB_REDO: automated re-refinement of X-ray structure models in the PDB. J Appl Crystallogr. 2009;42:376–384. doi: 10.1107/S0021889809008784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaptein R, Zuiderweg ERP, Scheek RM, Boelens R, van Gunsteren WF. A protein structure from nuclear magnetic resonance data. Lac repressor headpiece. J Mol Biol. 1985;182:179–182. doi: 10.1016/0022-2836(85)90036-1. [DOI] [PubMed] [Google Scholar]
- Kendrew JC. Architecture of a protein molecule. Nature. 1958;182:764–767. doi: 10.1038/182764a0. [DOI] [PubMed] [Google Scholar]
- Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Cryst. 1993;26:283–291. doi: 10.1107/S0021889892009944. [DOI] [Google Scholar]
- Markley JL, Bax A, Arata Y, Hilbers CW, Kaptein R, Sykes BD, Wright PE, Wüthrich K. Recommendations for the presentation of NMR structures of proteins and nucleic acids. IUPAC-IUBMB-IUPAB inter-union task group on the standardization of data bases of protein and nucleic acid structures determined by NMR spectroscopy. J Biomol NMR. 1998;12:1–23. doi: 10.1023/A:1008290618449. [DOI] [PubMed] [Google Scholar]
- Markley JL, Ulrich EL, Berman HM, Henrick K, Nakamura H, Akutsu H. BioMagResBank (BMRB) as a partner in the Worldwide Protein Data Bank (wwPDB): new policies affecting biomolecular NMR depositions. J Biomol NMR. 2008;40:153–155. doi: 10.1007/s10858-008-9221-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabuurs SB, Nederveen AJ, Vranken WF, Doreleijers JF, Bonvin AM, Vuister GW, Vriend G, Spronk CA. DRESS: a database of REfined solution NMR structures. Proteins. 2004;55:483–486. doi: 10.1002/prot.20118. [DOI] [PubMed] [Google Scholar]
- Nederveen AJ, Doreleijers JF, Vranken WF, Miller Z, Spronk CA, Nabuurs SB, Güntert P, Livny M, Markley JL, Nilges M, Ulrich EL, Kaptein R, Bonvin AM. RECOORD: a recalculated coordinate database of 500+ proteins from the PDB using restraints from the BioMagResBank. Proteins. 2005;59:662–672. doi: 10.1002/prot.20408. [DOI] [PubMed] [Google Scholar]
- Nilges M, Clore GM, Gronenborn AM. Determination of three-dimensional structures of proteins from interproton distance data by hybrid distance geometry-dynamical simulated annealing calculations. FEBS Lett. 1988;229:317–324. doi: 10.1016/0014-5793(88)81148-7. [DOI] [PubMed] [Google Scholar]
- Protein Data Bank Protein Data Bank. Nature New Biol. 1971;233:223. [Google Scholar]
- Rosato A, Bagaria A, Baker D, Bardiaux B, Cavalli A, Doreleijers JF, Giachetti A, Guerry P, Güntert P, Herrmann T, Huang YJ, Jonker H, Mao B, Malliavin TE, Montelione GT, Nilges M, Raman S, van der Schot G, Vranken WF, Vuister GW, Bonvin AM (2009) CASD-NMR: a rolling experiment for the critical assessment of automated structure determination from NMR data. Nat Meth 6(9):625–626 [DOI] [PMC free article] [PubMed]
- Saccenti E, Rosato A. The war of tools: how can NMR spectroscopists detect errors in their structures? J Biomol NMR. 2008;40:251–261. doi: 10.1007/s10858-008-9228-4. [DOI] [PubMed] [Google Scholar]
- Sanderson K. New protein structures replace the old. Nature. 2009;459:1038–1039. doi: 10.1038/4591038b. [DOI] [PubMed] [Google Scholar]
- Seavey BR, Farr EA, Westler WM, Markley JL. A relational database for sequence-specific protein NMR data. J Biomol NMR. 1991;1:217–236. doi: 10.1007/BF01875516. [DOI] [PubMed] [Google Scholar]
- Ulrich EL, Markley JL, Kyogoku Y. Creation of a nuclear magnetic resonance data repository and literature database. Protein Seq Data Anal. 1989;2:23–37. [PubMed] [Google Scholar]
- Ulrich EL, Akutsu H, Doreleijers JF, Harano Y, Ioannidis YE, Lin J, Livny M, Mading S, Maziuk D, Miller Z, Nakatani E, Schulte CF, Tolmie DE, Kent Wenger R, Yao H, Markley JL. BioMagResBank. Nucleic Acids Res. 2008;36:D402–D408. doi: 10.1093/nar/gkm957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vranken WF. A global analysis of NMR distance constraints from the PDB. J Biomol NMR. 2007;39:303–314. doi: 10.1007/s10858-007-9199-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vranken WF, Boucher W, Stevens TJ, Fogh RH, Pajon A, Llinas M, Ulrich EL, Markley JL, Ionides J, Laue E. The CCPN Data Model for NMR Spectroscopy: Development of a Software Pipeline. Proteins. 2005;59:687–696. doi: 10.1002/prot.20449. [DOI] [PubMed] [Google Scholar]
- Williamson MP, Havel TF, Wüthrich K. Solution conformation of proteinase inhibitor IIA from bull seminal plasma by 1 H nuclear magnetic resonance and distance geometry. J Mol Biol. 1985;182:295–315. doi: 10.1016/0022-2836(85)90347-X. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.