Abstract
Conventional comparative genomic hybridization (CGH) profiling of neuroblastomas has identified many genomic aberrations, although the limited resolution has precluded a precise localization of sequences of interest within amplicons. To map high copy number genomic gains in clinically matched stage IV neuroblastomas, CGH analysis using a 19,200-feature cDNA microarray was used. A dedicated (freely available) algorithm was developed for rapid in silico determination of chromosomal localizations of microarray cDNA targets, and for generation of an ideogram-type profile of copy number changes. Using these methodologies, novel gene amplifications undetectable by chromosome CGH were identified, and larger MYCN amplicon sizes (in one tumor up to 6 Mb) than those previously reported in neuroblastoma were identified. The genes HPCAL1, LPIN1/KIAA0188, NAG, and NSE1/LOC151354 were found to be coamplified with MYCN. To determine whether stage IV primary tumors could be further subclassified based on their genomic copy number profiles, hierarchical clustering was performed. Cluster analysis of microarray CGH data identified three groups: 1) no amplifications evident, 2) a small MYCN amplicon as the only detectable imbalance, and 3) a large MYCN amplicon with additional gene amplifications. Application of CGH to cDNA microarray targets will help to determine both the variation of amplicon size and help better define amplification-dependent and independent pathways of progression in neuroblastoma.
Keywords: amplicons, genomic instability, genomics, N-myc, high-resolution mapping, genomic profiling
Introduction
Neuroblastoma is a pediatric malignancy with an incidence rate in the U.S. of 1 in 100,000 children per year (reviewed in Ref. [1]). About one third of neuroblastomas have gene amplification, often observed cytogenetically as double minute chromosomes (dmins), and occasionally as homogeneously staining regions (HSRs). Neuroblastoma is the best example of a human tumor with intrinsic ability to undergo high copy number gene amplification, which typically involves a large and variable amount of DNA flanking the MYCN (N-myc) region of the genome at chromosome band 2p24 (reviewed in Ref. [2]). Both the extent and complexity of the amplified sequences associated with the MYCN amplicon have been difficult to characterize using traditional cytogenetic and molecular genetic mapping methods.
Recently, comparative genomic hybridization (CGH) to metaphase chromosome targets [3] has increased our understanding of the complexities of the amplicon structures associated with MYCN amplification in neuroblastoma [2]. CGH analyses of neuroblastomas have identified genomic imbalances commonly consisting of MYCN amplification, 17q gain, and deletions at 1p36, 3p, 4p, 9p, 11q, and 14q regions [4–9]. However, the minimum resolution for determining copy number gains is probably no less than 5 to 10 Mb, which is a function of both copy number and amplicon size [3,10]. Consequently, this technique can only provide a starting point for positional cloning studies, as the low resolution of conventional cytogenetics prevents precise localization of sequences of interest. Ideally, these limits are overcome using high-resolution whole genome arrays spotted with genomic clones (BACs, PACs, cosmids). However, this method of determining the location and incidence of copy number imbalances is not yet as widely available as chromosome CGH and expression microarray technologies.
An emerging platform that addresses some of the short-comings of chromosome CGH uses microarray expression slides as target substrates for determining copy number [11]. Instead of using metaphase chromosomes, CGH is applied to arrayed short sequences of DNA bound to glass slides and probed with genomes of interest. With sufficient genetic representation on the microarray, cDNA array CGH significantly increases resolution for identification of key genes in regions of high copy number gain. However, because of the small size and reduced complexity of the cDNA target sequences, this approach may not have sufficient sensitivity to reproducibly detect low copy number imbalances. In this pilot study, we demonstrate the utility of cDNA array CGH for characterizing high copy number gene amplification in stage IV neuroblastoma tumors, to determine the diversity sequences coamplified with MYCN and the relative size of the amplicon at 2p24 using a uniformly stratified patient cohort with stage IV disease. Furthermore, we have developed dedicated software for microarray analysis and rapid in silico determination of chromosomal localizations of microarray cDNA targets that is essential for providing a comprehensive ideogram-type schematic of copy number changes. The use of these technologies in the analysis of amplification in neuroblastoma highlights the advantages and broad applicability of this system over conventional chromosome CGH.
Materials and Methods
Patient Tissue Accrual and Polymerase Chain Reaction (PCR) of MYCN Copy Number
Primary tumors were all clinically matched stage IV neuroblastomas, collected from patients at the Hospital for Sick Children. All samples were anonymous and were collected under the guidelines of the Hospital for Sick Children Research Ethics Board. The tumors were flash frozen directly after surgical removal and stored in liquid nitrogen. Genomic DNA from the tumor was extracted following standard protocols [12]. The amplification status of the MYCN gene was determined by standard quantitative PCR analyses as previously described [13]. Briefly, samples P1126, P1071, P1079, and P1008 were found to have approximately 100 or more copies of MYCN. Sample P1061 showed intermediate 25 to 50 copies of MYCN, and samples D3025, D3060, D3077-95, and D3077-97 were observed with less than 25 copies. Samples P1126, P1079, and P1071 were derived at different time points (pretreatment, posttreatment, and posttreatment inguinal lymph node metastasis, respectively) from the same patient. Table 1 lists the clinical features of the patient cohort.
Table 1.
Clinical Information for the Patient Samples Used in This Study.
| Patient sample | Gender | Age (years) | Tumor stage | Survival (months) | Primary site | Shimada histology [51] | Ploidy | 1p Deletion |
| D3025 | M | 3.97 | 4 | 9.5 | Adrenal | Unfavorable | Diploid | Deleted |
| D3060 | M | 4.42 | 4 | 9.5 | Adrenal | Unfavorable | Diploid | Deleted |
| D3077-95 | M | 1 | 4 | 9.57 | Right adrenal | N/A | Diploid | N/A |
| D3077-97 | M | 3.98 | 4 | 15 | Abdomen | Unfavorable | Aneuploid | Deleted |
| P1061 | M | 1.79 | 4 | 14.83 | Left adrenal | Unfavorable | Aneuploid | Deleted |
| P1008 | F | 2.37 | 4 | 15.37 | Right adrenal | Unfavorable | Near tetraploid | Not deleted |
| P1126 | M | 1.96 | 4 | 14 | Left adrenal | Unfavorable | Diploid | Deleted |
| P1071 | M | 1.96 | 4 | 14 | Left adrenal | Unfavorable | Diploid | Deleted |
| P1079 | M | 1.96 | 4 | 14 | Left adrenal | Unfavorable | Diploid | Deleted |
Note all samples were derived from stage IV tumors.
N/A, not available.
Chromosome CGH and Fluorescence In Situ Hybridization (FISH)
Established protocols for CGH on human metaphase chromosome spreads were followed [3]. Image capture and analysis were performed using the Vysis PathVysion and Karyotype software, respectively (Vysis, Downers Grove, IL). Telomeric and centromeric regions were excluded in the analyses due to the presence of highly repetitive genomic sequences at these sites. Ten metaphases were analyzed to create the final CGH profile with 99% confidence intervals. Where possible, FISH assessment of MYCN copy number in cytogenetic preparations from neuroblastoma primary tumors was performed as previously described [13]. A minimum of 100 interphase nuclei was evaluated in each cytogenetic sample for assessing the MYCN copy number.
cDNA Array CGH
Microarrays used in this study [Human 19k2 microarray, Clinical Genomics Centre (CGC), Toronto, Ontario, Canada] contain a total of 19,200 features (Research Genetics, Invitrogen, Huntsville, AL) including 18,980 human cDNAs and 220 positive and negative control features. The features of the 19k2 microarray are arranged on two glass slides (parts A and B), each with 32 subgrids spotted over an approximate area of 18x36 mm2. All features are placed in duplicate, for a total of 38,400 spots. A complete list of the cDNA collection used for these arrays and protocols used for array construction can be found at the University Health Network CGC web site (http://www.microarrays.ca). It should be noted that some genes that have been reported to be part of the MYCN amplicon, such as N-cym [14] and DDX1 [15], are not presently on the current configuration of the 19k2 microarray. Sequence verification studies being performed by the CGC allowed us to identify by BLAST analysis a small percentage of cDNAs contaminated with mitochondrial DNA (mtDNA). Identification of such sequences has allowed us to make appropriate changes and filters for our software suite (see below).
The procedure for cDNA array CGH is a modified version of the protocol published by Pollack et al. [11], and reviewed in Ref. [16]. Each microarray slide (part A and B) was blocked under a glass coverslip for 1 hour at 37°C with blocking solution (3% BSA, 4x SSC, 0.1% Tween-20) before hybridization with the denatured probe. Four micrograms each of high molecular weight tumor and normal genomic DNA was separately digested with EcoRI for 2 hours, purified (Qiaquick PCR kit), vacuum dried, and resuspended in 25 µl water. Random primer labeling was performed using the Bioprime Labeling kit (Invitrogen, Carlsbad, CA) according to manufacturer's instructions, with modifications to directly incorporate Cy3- and Cy5-dCTP (Amersham, Piscataway, NJ). The Cy3- and Cy5-labeled products were suppressed with 30 µg Cot-1 DNA (Invitrogen), 100 µg yeast tRNA (Invitrogen), and 20 µg poly(dAdT) (Sigma-Aldrich, St. Louis, MO), loaded onto a microcon 30 filter (Amicon, Bedford, MA), and centrifuged for 20 minutes at 5000xg. The sample was recovered with 60 µl hybridization buffer (3.4x SSC and 0.3% SDS), and denatured at 100°C for 90 seconds. The probe was immediately chilled on ice, and allowed to preanneal at 37°C for 0.5 to 1 hour. The probe was divided and added to each microarray slide (part A and B) under a glass coverslip and sealed with rubber cement. Hybridization was at 37°C for 16 to 20 hours in a moist chamber humidified with hybridization buffer. The microarrays were washed at 65°C for 5 minutes in 2x SSC, 0.03% SDS, followed by successive washes in 1x and 0.2x SSC at room temperature (5 minutes each). After air drying, the slides were scanned using an Axon GenePix 4000A confocal scanner and fluorescence intensities were quantified with GenePix Pro 3.0 software (Axon Instruments, Union City, CA). Replicate and reverse-labeling experiments were performed for selected samples. Control genomic DNAs used in these studies included three cell lines with established levels of high copy number MYCN amplifications, including the retinoblastoma cell line Y79, and the neuroblastoma cell lines IMR32 and NUB-7 (American Tissue Culture Collection, Rockville, MD).
Bioinformatics and Analyses
Custom software scripts were developed to update the chromosome localizations for the cDNAs of the 19k2 microarray. However, any gene list having GenBank accession and I.M.A.G.E. clone [17] identifiers can be updated with this approach. The algorithm involved parsing NCBI UniGene and MapViewer databases to identify up-to-date cytoband localizations. Briefly, the UniGene database (build 147) was retrieved (ftp://ftp.ncbi.nih.gov/) and used to update UniGene identifiers in the gene list based on accession number. Subsequently, the accession and updated UniGene identifiers for each cDNA were cross-referenced to sequence mapping data and additional cytoband information in the MapViewer and UniGene databases, respectively. The information updates for each cDNA in the gene list include the chromosomal distance in megabases, the corresponding cytoband (based on ISCN 800 level band resolution size measurements; [18]), and a qualitative assessment score of mapping precision based on the concordance of the results gathered from the databases. Updated gene lists for these microarrays, as well as a more detailed description of the update algorithm, can be found at the laboratory's web site (http://www.utoronto.ca/cancyto/). Table 2 depicts the genetic representation of the 19k2 microarrays.
Table 2.
Genetic Representation of the 19k2 Microarray after Updating of Localization Information.
| Number of clones with position known | Number of clones with position unknown | Total microarray clones | Microarray percentage representation | Actual genome percentage representation | |
| Chromosome 1 | 638 | 1018 | 1656 | 8.7% | 7.9% |
| Chromosome 2 | 532 | 593 | 1125 | 5.9% | 7.8% |
| Chromosome 3 | 411 | 558 | 969 | 5.1% | 6.8% |
| Chromosome 4 | 198 | 507 | 705 | 3.7% | 6.3% |
| Chromosome 5 | 380 | 432 | 812 | 4.3% | 6.0% |
| Chromosome 6 | 400 | 487 | 887 | 4.7% | 5.7% |
| Chromosome 7 | 384 | 488 | 872 | 4.6% | 5.2% |
| Chromosome 8 | 291 | 336 | 627 | 3.3% | 4.7% |
| Chromosome 9 | 319 | 402 | 721 | 3.8% | 4.4% |
| Chromosome 10 | 334 | 369 | 703 | 3.7% | 4.4% |
| Chromosome 11 | 434 | 489 | 923 | 4.8% | 4.4% |
| Chromosome 12 | 459 | 457 | 916 | 4.8% | 4.5% |
| Chromosome 13 | 166 | 178 | 344 | 1.8% | 3.6% |
| Chromosome 14 | 284 | 312 | 596 | 3.1% | 3.3% |
| Chromosome 15 | 298 | 309 | 607 | 3.2% | 3.2% |
| Chromosome 16 | 266 | 342 | 608 | 3.2% | 2.8% |
| Chromosome 17 | 405 | 415 | 820 | 4.3% | 2.8% |
| Chromosome 18 | 159 | 151 | 310 | 1.6% | 2.7% |
| Chromosome 19 | 317 | 490 | 807 | 4.3% | 2.2% |
| Chromosome 20 | 232 | 204 | 436 | 2.3% | 2.2% |
| Chromosome 21 | 92 | 114 | 206 | 1.1% | 1.4% |
| Chromosome 22 | 187 | 181 | 368 | 1.9% | 1.5% |
| Chromosome X | 211 | 327 | 538 | 2.8% | 5.1% |
| Chromosome Y | 8 | 4 | 12 | 0.1% | 1.2% |
| Unknown | - | 2412 | 2412 | 12.7% | - |
| Total | 7405 | 11575 | 18980 | 100% | 100% |
For each chromosome, the number of clones with known and unknown positions is listed, as well as its total representation on the microarray. For comparison, the last column identifies the total percentage size of each chromosome within the genome, based on ISCN chromosome size measurements [18]. Discrepancies between the microarray and actual representations are due to genetic redundancy on the microarray, the large number of clones with completely unknown positions, and that the percentage for actual chromosome genomic contribution includes non-coding sequences.
To facilitate analysis of results from multiple 19k2 microarray experiments, custom software was also developed. The software suite is currently comprised of four PC DOS-based components: 1) normalize, 2) project, 3) clusterp, and 4) profiler. The normalize software normalizes quantified Cy5 and Cy3 fluorescence intensities, with or without background subtraction, across the entire array as well as across individual subgrids. Additionally, preceding normalization, filters are applied to remove spots below user-specified thresholds for fluorescence intensity, foreground-to-background ratio, and/or spot diameter size. After normalization and filtering, duplicate spots from the microarray are averaged together. Using project, two or more normalized replicates of an experiment, including reverse-labelings, can be grouped together as a project. Additionally, project also groups together results from parts A and B of a 19k2 microarray experiment. The clusterp software uses selected project files to generate output that can be read by the Eisen Cluster software package (http://rana.lbl.gov) for two-dimensional hierarchical clustering [19,20]. Clustering was performed on genes showing hybridization in ≥80% of samples using an uncentered correlation similarity metric. Finally, profiler uses the updated chromosome localization information to generate composite chromosome plots of normalized fluorescence intensity ratios from selected projects, generating images analogous to karyotype profiles from chromosome CGH. Note that only results for cDNA sequences with known cytoband and megabase position are plotted. To make microarray CGH methods more generally accessible to the research community, this software suite and the algorithms for automated cytoband and megabase localization of microarray cDNA features are freely available online (http://www.utoronto.ca/cancyto/). All experimental results are also available online as tab-delimited text files (http://www.utoronto.ca/cancyto/NB2003/).
Results
Optimization and Performance of cDNA Array CGH
The cDNA array CGH technique was optimized initially by comparing genomic DNA from IMR32, a metastatic neuroblastoma cell line with known copy number changes, and characterized by chromosome CGH (Figure 1A), against a normal reference. Replicate cDNA array CGH experiments (n=4) including a reverse-labeling hybridization demonstrated good reproducibility and the expected MYCN amplification of sequences mapping to the 2p24 chromosomal region. The distribution of normalized fluorescence intensity ratios for all hybridization features (n=72,139 in four hybridizations) was 0.94±0.25 (mean±1 SD), establishing a 99% confidence interval limit of 0.19 to 1.69. (Figure 1B) shows the composite CGH genomic profile of IMR32 by cDNA array CGH.
Figure 1.
Validation of the cDNA array CGH technique. Genomic DNA from the neuroblastoma cell line IMR32 was used in both conventional CGH on metaphase chromosomes (inset, A) and cDNA array CGH (B). In the ideogram profiles generated for the cDNA array CGH results, the normalized fluorescence intensity ratios from individual replicates (n=4) are given in yellow, and their average is plotted in red. Note the profile for chromosome Y is not shown due to the low representation of genes from this chromosome on the microarray. Chromosome CGH of IMR32 showed expected MYCN amplification at 2p24, with an additional amplification at 2p15 (A). These changes were also observed by cDNA array CGH, but at higher resolution (B). cDNA array CGH was able to determine that the gene amplification at 2p15 corresponded to MEIS1, which was recently reported using other methods.
To semiquantitate the chromosomal distributions and relative levels of copy number imbalance by CGH using both cDNA and metaphase targets, the MYCN normalized fluorescence intensity ratios was derived from a series of 12 neuroblastomas and cell lines against copy number measurements determined by standard quantitative PCR and Southern blot methods. Southern blot analyses of the neuroblastoma cell lines IMR32 and NUB-7 [21], and the retinoblastoma cell line Y79 [22], have previously shown MYCN to have 50, 100, and 50 copies, respectively. By cDNA array CGH, the normalized fluorescence intensity ratios for MYCN spots were 6.5, 11.2, and 5.2, for IMR32, NUB-7, and Y79, respectively (not shown). Similarly, MYCN normalized fluorescence intensity ratios for the patient samples were approximately 10% of the corresponding PCR measurement. Overall, there was good correlation between the cDNA array CGH normalized fluorescence intensity ratios, and the copy number determined by PCR or Southern blot (R2=0.71). This analysis showed that with this sensitivity of detection, deletions or copy number gains below the level of 10 to 20 copies cannot be determined with certainty. Most likely, this sensitivity limit arises because of the small size of (intronless) cDNA targets on the arrays leading to basal background hybridization signals that mask the small signals arising from lower copy number imbalances. In contrast, multiple analyses (including use of dye switches) demonstrated that there was an excellent dynamic range at higher MYCN copy number with good reproducibility of copy number estimates, and with ratios that were consistent with high copy number MYCN amplification.
cDNA Array CGH of Neuroblastoma Tissues
In the next series of experiments, amplification analysis using IMR32 metaphase chromosome CGH was compared to the profiles obtained using cDNA array CGH. Chromosome CGH showed two distinct regions of amplification on chromosome 2p (Figure 1A), with the most distal centering on MYCN at 2p24, and the second more proximal location of gene amplification was at 2p15. The analysis of IMR32 by cDNA array CGH also identified these two amplification loci, and more clearly resolved the putative genes subject to amplification. At the 2p24 locus, the MYCN amplification was clearly identified by microarray CGH. In addition, the gene NSE1 was found to be coamplified within 1 Mb telomeric to MYCN in IMR32. Interestingly another gene, DNMT3A, which is situated nearly 10 Mb centromeric to MYCN, was also observed to be amplified. It is noteworthy that CGH applied to metaphase preparations was unable to differentiate the size of the 2p24-p23 amplicon in IMR32 with such precision.
The ratio analysis of the MEIS1 sequences on the cDNA array clearly identified this gene in the cytoband 2p15 to be subject to amplification (Figure 1B). The MEIS1 amplification was recently reported in IMR32 by others using different methodologies [23,24]. The normalized fluorescence intensity ratios of 5.24 and 6.85 from two distinct cDNA sequences on the microarray representing MEIS1 would predict between 50 and 70 copies. Furthermore, the analyses also indicated that a currently unknown sequence (GenBank accession N68286) within 15 kb telomeric to MEIS1 was also coamplified. These novel findings concerning the fine structure of the amplicons mapping to 2p in IMR32 have not been previously reported.
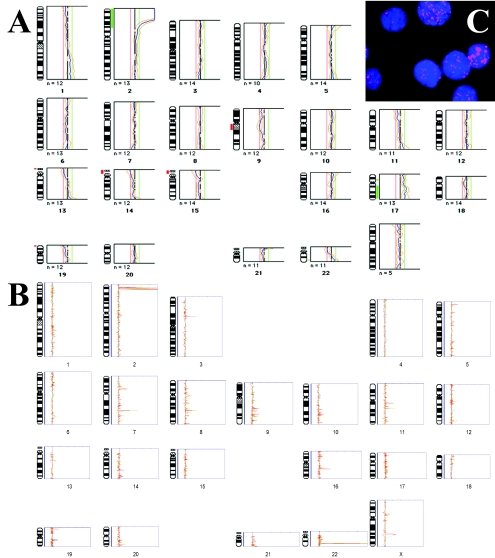
Microarray CGH was performed using genomic DNA derived from nine newly diagnosed neuroblastoma tumor samples. Of the nine studied, one (P1071) was subjected to both chromosome and cDNA array CGH, and MYCN amplification was also confirmed by interphase FISH (Figure 2, A–C). The cDNA array CGH resolved the 2p24 MYCN amplification boundaries to include the genes HPCAL1 and KIAA0188/LPIN1 that map to a 6-Mb flanking region telomeric of MYCN. The copy number gain of 22q shown by chromosome CGH was indicated to be due to amplification of the gene ST13 at 22q13 by cDNA array CGH (Figure 2, A and B, respectively). The same larger MYCN amplicon at 2p24 was also observed in samples from the same patient, P1126 and P1079, but not in any of the other patient samples. In samples P1008 and P1061, the only amplification was MYCN; however, sample P1008 showed additionally amplification of a flanking unknown sequence (accession T79037) which by BLAST sequence analysis is reported to be the gene NAG. The remaining DNA samples did not show detectable amplification of MYCN, or of any other genetic loci. A summary of the amplified genes and sequences identified by cDNA array CGH in neuroblastomas is given in Table 3.
Figure 2.
Comparison of resolution of chromosome CGH (A) and cDNA array CGH (B) techniques for determining genomic amplifications in a neuroblastoma patient P1071. Fluorescence in situ hybridization with labeled MYCN probe was used to verify copy number amplification of MYCN in interphase nuclei (C). (A) CGH on metaphase chromosomes showed amplification along the distal short arm of chromosome 2, but due to the limited resolution of the technique was not able to resolve the region to smaller than approximately 60 Mb (2pter-2p22). (B) cDNA array CGH profile of genomic imbalances in the same patient (n=3 replicates) clearly delineated the 2p24 amplification to involve MYCN as well as other 2p24 genes in the vicinity of MYCN. Although it is expected that the technical issues dealing with low detection sensitivity will be resolved in the future, this study has focused on discussion of genes showing dosage changes of greater than 25 copies.
Table 3.
Genes Showing High Copy Gain Common to all Patient Samples Identified by cDNA Array CGH and Hierarchical Clustering, Some of Which are Novel and Previously Unreported in Neuroblastomas.
| GenBank accession of clone | UniGene ID | Symbol | Cytoband | Approximate megabase from pter | Function/Evidence |
| H19826 | Hs.3618 | HPCAL1 | 2p24 | 9.7 | Calcium-binding protein with similarity to hippocalin, expressed only in the brain. |
| R77965, H23930, N45520 | Hs.81412 | LPIN1/KIAA0188 | 2p24 | 11.8 | Gene responsible for fatty liver dystrophy in mice, candidate gene for human lipodystrophy. |
| T79037 | - | unknown | 2p24 | 13.6 | Sequence has as very high similarity to genomic clone RP11-516B14 containing NAG gene. |
| T65604 | Hs.260855 | NSE1/LOC151354 | 2p24 | 14.15 | Novel member of the H-rev107 protein family. |
| R52824 | Hs.25960 | MYCN | 2p24 | 15.45 | Oncogene commonly amplified in neuroblastomas and retinoblastomas. |
| H03349 | Hs.241565 | DNMT3A | 2p24 | 24.92 | Gene transcriptional silencing by de novo DNA methylation. |
| N68286 | Hs.109391 | unknown | 2p15 | 70.81 | Unknown. |
| N95243, AA034934 | Hs.170177 | MEIS1 | 2p15 | 70.83 | Novel homeobox gene. |
Only clones that have been sequence verified are listed.
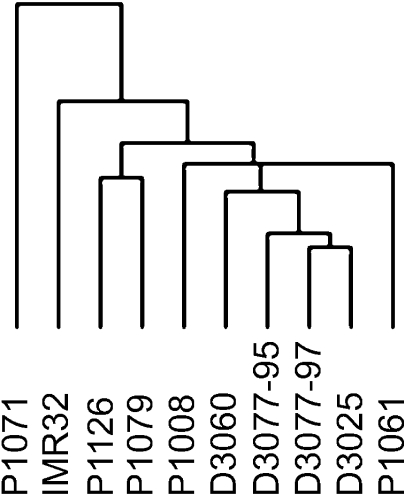
Unsupervised two-dimensional hierarchical clustering analysis was then performed on the cDNA array CGH data as a class discovery procedure for categorizing DNA copy number relationships between the neuroblastomas. The idea of clustering gene expression to detect functional relationships is conceptually similar to analyzing DNA copy number relationships in a group of tumors to empirically delineate classes of sequences subject to amplification. Hierarchical clustering of similarly amplified sequences may suggest close linkage of genes on common amplicon structures. In support of this idea, unsupervised hierarchical clustering was able to provide clear class distinction based on copy number amplification in neuroblastoma. As shown in Figure 3, the majority of primary tumor DNA samples were shown to branch together within the dendogram, away from the cell line IMR32. Within the cluster of primary tumor samples, the MYCN low- or non-amplified samples clustered closely together (D3025, D3060, D3077-95, D3077-97). Samples P1008 and P1061 coclustered separately in this group, likely due to the high copy number of MYCN. Samples from the same patient (P1126, P1079) clustered together, and apart from the other patient samples, presumably due to the higher MYCN amplification and larger MYCN amplicon. The sample P1071 was derived from an undifferentiated neuroblastoma inguinal lymph node metastasis that has additional gene amplifications not present in DNA isolated from the more differentiated samples from this same patient (P1126, P1079).
Figure 3.
Relationships between the neuroblastomas as defined by unsupervised two-dimensional hierarchical clustering of gene dosage change patterns derived from cDNA array CGH results. The full cluster results are provided as supplementary information online (http://www.utoronto.ca/cancyto/NB2003/).
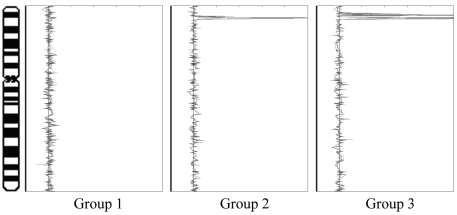
In summary, hierarchical clustering permitted classification of this sample set of stage IV neuroblastoma patient tumors into three groups based on pattern of copy number changes: 1) no amplifications evident (D3025, D3060, D3077-95, D3077-97); 2) MYCN amplification as the only evident genomic copy number change, or a small MYCN amplicon (P1061, P1008); and 3) a large MYCN amplicon with additional gene amplifications (P1126, P1071, P1079). These findings are summarized in Figure 4.
Figure 4.
Categorization of neuroblastomas based on hierarchical clustering of cDNA array CGH results, as represented by changes on chromosome 2. Groups 1, 2, and 3 are defined as having no detectable genomic amplifications; MYCN amplification as the only evident genomic copy number change, or a small MYCN amplicon; and MYCN amplification with additional amplification of a number of the genes in the vicinity of the MYCN locus; respectively.
Discussion
In this study, high-density cDNA microarray CGH profiling of 10 neuroblastomas was used to determine the precise chromosomal locations of gene amplifications in these tumors. There have been previous CGH studies of neuroblastoma using conventional metaphase chromosome preparations [4–9], but this analysis represents the first microarray genomic profiling of amplifications in neuroblastoma.
CGH analysis of metaphase chromosomes has enabled genome-wide detection of DNA copy number imbalances [3]. However, the relatively low resolution of chromosome CGH means that further experiments are necessary to map amplicon structures and to provide detailed positional mapping of amplified sequences of interest. Although CGH studies using genomic DNA microarrays (BAC, PAC, cosmid) have recently been published [25–31], these arrays are not yet generally available and/or at present do not have sufficient genomic coverage and therefore resolution for screening. Recently, several studies have shown the utility of cDNA microarray CGH for studying gene copy changes. Initially demonstrated by Pollack et al. [11], Kauraniemi et al. [32] have used a 636-feature chromosome 17-specific cDNA array for identifying gene copy amplifications by CGH in breast cancer cell lines. However, to date, there have been few reports documenting successful application of microarray CGH using cDNA targets.
To assess the sensitivity of cDNA array CGH analysis, PCR-quantified MYCN copy number was compared to amplification levels determined by cDNA array CGH. This comparison demonstrated that a minimum sensitivity of 10 to 20 copies could be obtained reproducibly, but that lower copy number alterations would escape detection. Improved sensitivity may be obtained using a recently published tyramide-based amplification protocol to augment the fluorescent signal [33]. However, one major limitation of this system is that it is not currently adaptable to two-color CGH.
After assessment of the relative sensitivity of high-density cDNA array CGH for profiling amplifications, the MYCN amplicon structure was determined using a panel of neuroblastomas. It was previously reported that the MYCN amplicon can range in size from 350 kb to 1 Mb [2]. These conclusions were based largely on enzymatic restriction mapping [34] or metaphase and chromatin fiber FISH mapping [21] techniques using different probes to define amplicon boundaries. However, with the advent of cDNA microarray technology combined with biological information from the Human Genome Project, a more precise examination of MYCN amplicon size and complexity is now possible. Our results suggest that the MYCN amplicon can be much larger than 1 Mb, and in fact we have observed up to 6 Mb in samples P1071, P1079, and P1126.
Altogether, the genes mapping to chromosome band 2p24 that were observed to be coamplified with MYCN included: hippocalcin-like 1 (HPCAL1), lipin 1 (LPIN1/KIAA0188), neuroblastoma amplified gene (NAG), and an unannotated gene (NSE1/LOC151354). Hippocalcin-like 1 has high similarity to hippocalcin on chromosome 1, which has been identified as a calcium sensor with neuronal tissue expression specificity [35,36]. Lipin 1 encodes a novel nuclear protein that although its function is currently unknown, in mouse, it is reported to be required for regulation of normal adipose tissue development [37,38]. NAG has previously been observed to be part of the MYCN amplicon, although its function is still as yet unknown [39]. The GenBank entry (accession AJ417080) for NSE1 (unpublished) indicates this gene to be a novel member of the H-rev107 protein family, whose members include the 2A protein in picornaviruses and the cellular retinoic acid receptor responder 3 (RARRES3/TIG3) protein [40,41], with putative cellular antiproliferative activity. Except for NAG, recruitment of these genes into the MYCN amplicon has not been previously reported.
In addition to those genes mapping to 2p24, amplification was also detected at other genomic locations in isolated tumors. The gene ST13 (suppressor of tumorigenicity 13), which maps to 22q13 and is reported to be an Hsp70-binding protein during steroid receptor assembly [42], was observed to be amplified in one tumor. Amplification of another gene DNA (cytosine-5) methyltransferase 3 alpha (DNMT3A), which is 10 Mb centromeric to MYCN, was observed in IMR32. This gene belongs to a family of DNA methyltransferases (DNMTs) and has a role in transcriptional regulation of gene silencing through de novo methylation of DNA CpG islands [43]. It has been reported that overexpression of DNMT3A is associated with aberrant methylation profiles in tumors as compared to normal cells [43,44]. Interestingly, amplification of mtDNA sequences was commonly observed in the tumors in the study group (data not shown). This observation suggests that increased numbers of mitochondria may be a feature of neuroblastoma oncogenesis. In keeping with this idea, it is interesting that mtDNA amplification has been reported previously in other tumors including gliomas [45–47].
To better subclassify the genomic profiles obtained in the analyses, an unsupervised hierarchical clustering approach was used to delineate three distinct groups of neuroblastomas within the study group, identified by: 1) no evident copy number change; 2) MYCN amplification as the only evident genomic copy number change, or a small MYCN amplicon; and 3) MYCN amplification concomitant with additional gene amplifications, or a large MYCN amplicon. This is interesting, given that all the samples represent clinically similar stage IV tumors. Recently, other studies have also applied clustering algorithms to subtype tumors on the basis of the patterns of imbalances present in genomic profiling data [31,48]. In one study, self-organizing maps were used to categorize stage pT2N0 prostate carcinomas analyzed by chromosome CGH [48]. Similarly, the application of a support vector machine algorithm was recently described for subclassifying dedifferentiated and pleomorphic liposarcomas using genomic microarray CGH data [31]. Interestingly, the study also demonstrated that the genomic profiling approach was more robust in delineating tumor subclasses than the corresponding expression profiling method [31].
In conclusion, cDNA array CGH can be readily used to identify and map gene amplifications at high resolution. A diversity of amplification patterns including larger more complex MYCN amplicon structures was detected in a series of neuroblastomas drawn from a group with equivalent advanced stage disease. Studies in model systems have demonstrated multiple and redundant phylogenetically conserved repair pathways involved in maintenance of genome stability (reviewed in Refs. [49,50]). Unrepaired, DNA damage results in accumulating genomic rearrangements and ensuing genomic instability, is thought to underlie the acquisition of gene amplification. In the present study, cDNA array analysis identified three diverse patterns of genomic profile associated with advanced stage neuroblastoma that (within the limitations of the study group size) did not appear to impact on disease course. This observation suggests that the inherent biological differences in genomic stability within neuroblastoma do not appear to directly impact on disease course. Presently, high-resolution, high-throughput assays such as microarray CGH can be used for detailed analysis of genomic instability in other human tumors. Furthermore, the dedicated algorithms developed for this study are freely available for improved applications of microarray analyses, based on information provided by the chromosomal band locations of the microarray cDNA sequences. In the future, increased technique sensitivity and refinements to the bioinformatics of genomic profiling will greatly facilitate an improved understanding of the biology that underlies the complex patterns of genomic imbalance identified by CGH methods.
Acknowledgement
The authors gratefully acknowledge the technical expertise of Jane Bayani.
Footnotes
Financial support for this project is generously provided by the James Birrell Fund for Neuroblastoma.
References
- 1.Olshan AF, Bunin GR, editors. Neuroblastoma. Amsterdam: Elsevier Science; 2000. Epidemiology of neuroblastoma; pp. 33–39. [Google Scholar]
- 2.George RE, Squire JA, editors. Neuroblastoma. Amsterdam: Elsevier Science; 2000. Structure of the MYCN amplicon; pp. 85–100. [Google Scholar]
- 3.Kallioniemi A, Kallioniemi OP, Sudar D, Rutovitz D, Gray JW, Waldman F, Pinkel D. Comparative genomic hybridization for molecular cytogenetic analysis of solid tumors. Science. 1992;258:818–821. doi: 10.1126/science.1359641. [DOI] [PubMed] [Google Scholar]
- 4.Van Gele M, Van Roy N, Jauch A, Laureys G, Benoit Y, Schelfhout V, De Potter CR, Brock P, Uyttebroeck A, Sciot R, Schuuring E, Versteeg R, Speleman F. Sensitive and reliable detection of genomic imbalances in human neuroblastomas using comparative genomic hybridisation analysis. Eur J Cancer. 1997;33:1979–1982. doi: 10.1016/s0959-8049(97)00289-x. [DOI] [PubMed] [Google Scholar]
- 5.Vandesompele J, Van Roy N, Van Gele M, Laureys G, Ambros P, Heimann P, Devalck C, Schuuring E, Brock P, Otten J, Gyselinck J, De Paepe A, Speleman F. Genetic heterogeneity of neuroblastoma studied by comparative genomic hybridization. Genes Chromosomes Cancer. 1998;23:141–152. doi: 10.1002/(sici)1098-2264(199810)23:2<141::aid-gcc7>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- 6.Brinkschmidt C, Poremba C, Christiansen H, Simon R, Schafer KL, Terpe HJ, Lampert F, Boecker W, Dockhorn-Dworniczak B. Comparative genomic hybridization and telomerase activity analysis identify two biologically different groups of 4s neuroblastomas. Br J Cancer. 1998;77:2223–2229. doi: 10.1038/bjc.1998.370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Breen CJ, O'Meara A, McDermott M, Mullarkey M, Stallings RL. Coordinate deletion of chromosome 3p and 11q in neuroblastoma detected by comparative genomic hybridization. Cancer Genet Cytogenet. 2000;120:44–49. doi: 10.1016/s0165-4608(99)00252-6. [DOI] [PubMed] [Google Scholar]
- 8.Vandesompele J, Speleman F, Van Roy N, Laureys G, Brinskchmidt C, Christiansen H, Lampert F, Lastowska M, Bown N, Pearson A, Nicholson JC, Ross F, Combaret V, Delattre O, Feuerstein BG, Plantaz D. Multicentre analysis of patterns of DNA gains and losses in 204 neuroblastoma tumors: how many genetic subgroups are there? Med Pediatr Oncol. 2001;36:5–10. doi: 10.1002/1096-911X(20010101)36:1<5::AID-MPO1003>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 9.Plantaz D, Vandesompele J, Van Roy N, Lastowska M, Bown N, Combaret V, Favrot MC, Delattre O, Michon J, Benard J, Hartmann O, Nicholson JC, Ross FM, Brinkschmidt C, Laureys G, Caron H, Matthay KK, Feuerstein BG, Speleman F. Comparative genomic hybridization (CGH) analysis of stage 4 neuroblastoma reveals high frequency of 11q deletion in tumors lacking MYCN amplification. Int J Cancer. 2001;91:680–686. doi: 10.1002/1097-0215(200002)9999:9999<::aid-ijc1114>3.0.co;2-r. [DOI] [PubMed] [Google Scholar]
- 10.Parente F, Gaudray P, Carle GF, Turc-Carel C. Experimental assessment of the detection limit of genomic amplification by comparative genomic hybridization CGH. Cytogenet Cell Genet. 1997;78:65–68. doi: 10.1159/000134632. [DOI] [PubMed] [Google Scholar]
- 11.Pollack JR, Perou CM, Alizadeh AA, Eisen MB, Pergamenschikov A, Williams CF, Jeffrey SS, Botstein D, Brown PO. Genome-wide analysis of DNA copy-number changes using cDNA microarrays. Nat Genet. 1999;23:41–46. doi: 10.1038/12640. [DOI] [PubMed] [Google Scholar]
- 12.Ausubel F, Brent R, Kingston R, Moore D, Seidman J, Smith J, Sturhl K. Current Protocols in Molecular Biology. New York, USA: John Wiley & Sons, Inc.; 2002. [Google Scholar]
- 13.Squire JA, Thorner P, Marrano P, Parkinson D, Ng YK, Gerrie B, Chilton-Macneill S, Zielenska M. Identification of MYCN copy number heterogeneity by direct FISH analysis of neuroblastoma preparations. Mol Diagn. 1996;1:281–289. doi: 10.1054/MODI00100281. [DOI] [PubMed] [Google Scholar]
- 14.Armstrong BC, Krystal GW. Isolation and characterization of complementary DNA for N-cym, a gene encoded by the DNA strand opposite to N-myc. Cell Growth Differ. 1992;3:385–390. [PubMed] [Google Scholar]
- 15.Squire JA, Thorner PS, Weitzman S, Maggi JD, Dirks P, Doyle J, Hale M, Godbout R. Co-amplification of MYCN and a DEAD box gene (DDX1) in primary neuroblastoma. Oncogene. 1995;10:1417–1422. [PubMed] [Google Scholar]
- 16.Beheshti B, Park PC, Braude I, Squire JA. Microarray CGH. Methods Mol Biol. 2002;204:191–207. doi: 10.1385/1-59259-300-3:191. [DOI] [PubMed] [Google Scholar]
- 17.Lennon G, Auffray C, Polymeropoulos M, Soares MB. The I.M.A.G.E. consortium: an integrated molecular analysis of genomes and their expression. Genomics. 1996;33:151–152. doi: 10.1006/geno.1996.0177. [DOI] [PubMed] [Google Scholar]
- 18.Mitelman F. ISCN (1995): International System for Human Cytogenetic Nomenclature. New York: S Karger; 1995. [Google Scholar]
- 19.Alon U, Barkai N, Notterman DA, Gish K, Ybarra S, Mack D, Levine AJ. Broad patterns of gene expression revealed by clustering analysis of tumor and normal colon tissues probed by oligonucleotide arrays. Proc Natl Acad Sci USA. 1999;96:6745–6750. doi: 10.1073/pnas.96.12.6745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pandita A, Godbout R, Zielenska M, Thorner P, Bayani J, Squire JA. Relational mapping of MYCN and DDXI in band 2p24 and analysis of amplicon arrays in double minute chromosomes and homogeneously staining regions by use of free chromatin FISH. Genes Chromosomes Cancer. 1997;20:243–252. doi: 10.1002/(sici)1098-2264(199711)20:3<243::aid-gcc4>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- 22.Godbout R, Squire J. Amplification of a DEAD box protein gene in retinoblastoma cell lines. Proc Natl Acad Sci USA. 1993;90:7578–7582. doi: 10.1073/pnas.90.16.7578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jones TA, Flomen RH, Senger G, Nizetic D, Sheer D. The homeobox gene MEIS1 is amplified in IMR-32 and highly expressed in other neuroblastoma cell lines. Eur J Cancer. 2000;36:2368–2374. doi: 10.1016/s0959-8049(00)00332-4. [DOI] [PubMed] [Google Scholar]
- 24.Spieker N, van Sluis P, Beitsma M, Boon K, van Schaik BD, van Kampen AH, Caron H, Versteeg R. The MEIS1 oncogene is highly expressed in neuroblastoma and amplified in cell line IMR32. Genomics. 2001;71:214–221. doi: 10.1006/geno.2000.6408. [DOI] [PubMed] [Google Scholar]
- 25.Snijders AM, Nowak N, Segraves R, Blackwood S, Brown N, Conroy J, Hamilton G, Hindle AK, Huey B, Kimura K, Law S, Myambo K, Palmer J, Ylstra B, Yue JP, Gray JW, Jain AN, Pinkel D, Albertson DG. Assembly of microarrays for genome-wide measurement of DNA copy number. Nat Genet. 2001;29:263–264. doi: 10.1038/ng754. [DOI] [PubMed] [Google Scholar]
- 26.Hodgson G, Hager JH, Volik S, Hariono S, Wernick M, Moore D, Nowak N, Albertson DG, Pinkel D, Collins C, Hanahan D, Gray JW. Genome scanning with array CGH delineates regional alterations in mouse islet carcinomas. Nat Genet. 2001;29:459–464. doi: 10.1038/ng771. [DOI] [PubMed] [Google Scholar]
- 27.Bruder CE, Hirvela C, Tapia-Paez I, Fransson I, Segraves R, Hamilton G, Zhang XX, Evans DG, Wallace AJ, Baser ME, Zucman-Rossi J, Hergersberg M, Boltshauser E, Papi L, Rouleau GA, Poptodorov G, Jordanova A, Rask-Andersen H, Kluwe L, Mautner V, Sainio M, Hung G, Mathiesen T, Moller C, Pulst SM, Harder H, Heiberg A, Honda M, Niimura M, Sahlen S, Blennow E, Albertson DG, Pinkel D, Dumanski JP. High resolution deletion analysis of constitutional DNA from neurofibromatosis type 2 (NF2) patients using microarray-CGH. Hum Mol Genet. 2001;10:271–282. doi: 10.1093/hmg/10.3.271. [DOI] [PubMed] [Google Scholar]
- 28.Hui AB, Lo KW, Yin XL, Poon WS, Ng HK. Detection of multiple gene amplifications in glioblastoma multiforme using array-based comparative genomic hybridization. Lab Invest. 2001;81:717–723. doi: 10.1038/labinvest.3780280. [DOI] [PubMed] [Google Scholar]
- 29.Daigo Y, Chin SF, Gorringe KL, Bobrow LG, Ponder BA, Pharoah PD, Caldas C. Degenerate oligonucleotide primed-polymerase chain reaction-based array comparative genomic hybridization for extensive amplicon profiling of breast cancers: a new approach for the molecular analysis of paraffin-embedded cancer tissue. Am J Pathol. 2001;158:1623–1631. doi: 10.1016/S0002-9440(10)64118-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wessendorf S, Fritz B, Wrobel G, Nessling M, Lampel S, Goettel D, Kuepper M, Joos S, Hopman T, Kokocinski F, Dohner H, Bentz M, Schwaenen C, Lichter P. Automated screening for genomic imbalances using matrix-based comparative genomic hybridization. Lab Invest. 2002;82:47–60. doi: 10.1038/labinvest.3780394. [DOI] [PubMed] [Google Scholar]
- 31.Fritz B, Schubert F, Wrobel G, Schwaenen C, Wessendorf S, Nessling M, Korz C, Rieker RJ, Montgomery K, Kucherlapati R, Mechtersheimer G, Eils R, Joos S, Lichter P. Microarray-based copy number and expression profiling in dedifferentiated and pleomorphic liposarcoma. Cancer Res. 2002;62:2993–2998. [PubMed] [Google Scholar]
- 32.Kauraniemi P, Barlund M, Monni O, Kallioniemi A. New amplified and highly expressed genes discovered in the ERBB2 amplicon in breast cancer by cDNA microarrays. Cancer Res. 2001;61:8235–8240. [PubMed] [Google Scholar]
- 33.Heiskanen MA, Bittner ML, Chen Y, Khan J, Adler KE, Trent JM, Meltzer PS. Detection of gene amplification by genomic hybridization to cDNA microarrays. Cancer Res. 2000;60:799–802. [PubMed] [Google Scholar]
- 34.Reiter JL, Brodeur GM. High-resolution mapping of a 130-kb core region of the MYCN amplicon in neuroblastomas. Genomics. 1996;32:97–103. doi: 10.1006/geno.1996.0081. [DOI] [PubMed] [Google Scholar]
- 35.Kobayashi M, Takamatsu K, Fujishiro M, Saitoh S, Noguchi T. Molecular cloning of a novel calcium-binding protein structurally related to hippocalcin from human brain and chromosomal mapping of its gene. Biochim Biophys Acta. 1994;1222:515–518. doi: 10.1016/0167-4889(94)90062-0. [DOI] [PubMed] [Google Scholar]
- 36.Paterlini M, Revilla V, Grant AL, Wisden W. Expression of the neuronal calcium sensor protein family in the rat brain. Neuroscience. 2000;99:205–216. doi: 10.1016/s0306-4522(00)00201-3. [DOI] [PubMed] [Google Scholar]
- 37.Nagase T, Seki N, Ishikawa K, Tanaka A, Nomura N. Prediction of the coding sequences of unidentified human genes. V. The coding sequences of 40 new genes (KIAA0161-KIAA0200) deduced by analysis of cDNA clones from human cell line KG-1. DNA Res. 1996;3:17–24. doi: 10.1093/dnares/3.1.17. [DOI] [PubMed] [Google Scholar]
- 38.Peterfy M, Phan J, Xu P, Reue K. Lipodystrophy in the fld mouse results from mutation of a new gene encoding a nuclear protein, lipin. Nat Genet. 2001;27:121–124. doi: 10.1038/83685. [DOI] [PubMed] [Google Scholar]
- 39.Wimmer K, Zhu XX, Lamb BJ, Kuick R, Ambros PF, Kovar H, Thoraval D, Motyka S, Alberts JR, Hanash SM. Co-amplification of a novel gene, NAG, with the N-myc gene in neuroblastoma. Oncogene. 1999;18:233–238. doi: 10.1038/sj.onc.1202287. [DOI] [PubMed] [Google Scholar]
- 40.DiSepio D, Ghosn C, Eckert RL, Deucher A, Robinson N, Duvic M, Chandraratna RA, Nagpal S. Identification and characterization of a retinoid-induced class II tumor suppressor/growth regulatory gene. Proc Natl Acad Sci USA. 1998;95:14811–14815. doi: 10.1073/pnas.95.25.14811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hughes PJ, Stanway G. The 2A proteins of three diverse picornaviruses are related to each other and to the H-rev107 family of proteins involved in the control of cell proliferation. J Gen Virol. 2000;81:201–207. doi: 10.1099/0022-1317-81-1-201. [DOI] [PubMed] [Google Scholar]
- 42.Prapapanich V, Chen S, Nair SC, Rimerman RA, Smith DF. Molecular cloning of human p48, a transient component of progesterone receptor complexes and an Hsp70-binding protein. Mol Endocrinol. 1996;10:420–431. doi: 10.1210/mend.10.4.8721986. [DOI] [PubMed] [Google Scholar]
- 43.Xie S, Wang Z, Okano M, Nogami M, Li Y, He WW, Okumura K, Li E. Cloning, expression and chromosome locations of the human DNMT3 gene family. Gene. 1999;236:87–95. doi: 10.1016/s0378-1119(99)00252-8. [DOI] [PubMed] [Google Scholar]
- 44.Robertson KD, Keyomarsi K, Gonzales FA, Velicescu M, Jones PA. Differential mRNA expression of the human DNA methyltransferases (DNMTs) 1, 3a and 3b during the G(0)/G(1) to S phase transition in normal and tumor cells. Nucleic Acids Res. 2000;28:2108–2113. doi: 10.1093/nar/28.10.2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liang BC. Evidence for association of mitochondrial DNA sequence amplification and nuclear localization in human low-grade gliomas. Mutat Res. 1996;354:27–33. doi: 10.1016/0027-5107(96)00004-8. [DOI] [PubMed] [Google Scholar]
- 46.Liang BC, Hays L. Mitochondrial DNA copy number changes in human gliomas. Cancer Lett. 1996;105:167–173. doi: 10.1016/0304-3835(96)04276-0. [DOI] [PubMed] [Google Scholar]
- 47.Boultwood J, Fidler C, Mills KI, Frodsham PM, Kusec R, Gaiger A, Gale RE, Linch DC, Littlewood TJ, Moss PA, Wainscoat JS. Amplification of mitochondrial DNA in acute myeloid leukaemia. Br J Haematol. 1996;95:426–431. doi: 10.1046/j.1365-2141.1996.d01-1922.x. [DOI] [PubMed] [Google Scholar]
- 48.Mattfeldt T, Wolter H, Kemmerling R, Gottfried HW, Kestler HA. Cluster analysis of comparative genomic hybridization (CGH) data using self-organizing maps: application to prostate carcinomas. Anal Cell Pathol. 2001;23:29–37. doi: 10.1155/2001/852674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kolodner RD, Putnam CD, Myung K. Maintenance of genome stability in Saccharomyces cerevisiae. Science. 2002;297:552–557. doi: 10.1126/science.1075277. [DOI] [PubMed] [Google Scholar]
- 50.Rouse J, Jackson SP. Interfaces between the detection, signaling, and repair of DNA damage. Science. 2002;297:547–551. doi: 10.1126/science.1074740. [DOI] [PubMed] [Google Scholar]
- 51.Shimada H, Chatten J, Newton WA, Jr, Sachs N, Hamoudi AB, Chiba T, Marsden HB, Misugi K. Histopathologic prognostic factors in neuroblastic tumors: definition of subtypes of ganglioneuroblastoma and an age-linked classification of neuroblastomas. J Natl Cancer Inst. 1984;73:405–416. doi: 10.1093/jnci/73.2.405. [DOI] [PubMed] [Google Scholar]