Summary
Polycystic ovarian syndrome (PCOS) is an endocrine syndrome that affects a large portion of women worldwide. This proteogenomic and functional study aimed to uncover candidate therapeutic targets for PCOS. We comprehensively investigated the causal association between circulating proteins and PCOS using two-sample Mendelian randomization analysis. Cis-protein quantitative trait loci were derived from six genome-wide association studies (GWASs) on plasma proteome. Genetic associations with PCOS were obtained from a large-scale GWAS meta-analysis, FinnGen cohort, and UK Biobank. Colocalization analyses were performed to prioritize the causal role of candidate proteins. Protein-protein interaction (PPI) and druggability evaluation assessed the druggability of candidate proteins. We evaluated the enrichment of tier 1 and 2 candidate proteins in individuals with PCOS and a mouse model and explored the potential application of the identified drug target. Genetically predicted levels of 65 proteins exhibited associations with PCOS risk, with 30 proteins showing elevated levels and 35 proteins showing decreased levels linked to higher susceptibility. PPI analyses revealed that FSHB, POSTN, CCN2, and CXCL11 interacted with targets of current PCOS medications. Eighty medications targeting 20 proteins showed their potential for repurposing as therapeutic targets for PCOS. EGLN1 levels were elevated in granulosa cells and the plasma of individuals with PCOS and in the plasma and ovaries of dehydroepiandrosterone (DHEA)-induced PCOS mouse model. As an EGLN1 inhibitor, administration of roxadustat in the PCOS mouse model elucidated the EGLN1-HIF1α-ferroptosis axis in inducing PCOS and validated its therapeutic effect in PCOS. Our study identifies candidate proteins causally associated with PCOS risk and suggests that targeting EGLN1 provides a promising treatment strategy.
Keywords: polycystic ovarian syndrome, plasma proteome, drug target, Mendelian randomization, GWAS, EGLN1, ferroptosis, roxadustat
This study utilizes genetic variation associated with circulating protein levels to identify drug targets for polycystic ovarian syndrome (PCOS) through Mendelian randomization of druggable genome. A total of 65 proteins are casually associated with PCOS risk, and targeting the EGLN1-HIF1α-ferroptosis axis with roxadustat represents a promising therapeutic approach for PCOS.
Introduction
As a prevalent heterogeneous syndrome affecting women at reproductive age, polycystic ovary syndrome (PCOS) (MIM: 184700) is characterized with hyperandrogenism, ovulatory dysfunction, and polycystic ovarian morphology.1 The global prevalence of PCOS hovers around 10%–13% among women.2 PCOS causes various health complications across female lifespan, including infertility, diabetes mellitus, and cardiovascular disease. Current treatment approaches for PCOS primarily focus on managing symptoms rather than addressing the underlying cause. Current less-than-optimal treatment regimens plague individuals with PCOS. The complex biological etiology of PCOS poses challenges for the development of effective drugs. It is imperative to identify non-invasive early-diagnostic biomarkers and develop therapeutics for PCOS.
Circulating proteins play pivotal roles in human health maintenance and serve as a predominant type of prospective drug target.3 Several circulating proteins, particularly those involved in regulating insulin resistance, have been implicated in PCOS risk, including sex-hormone-binding globulin, chemerin, irisin, lipocalin-2, and others.4,5,6,7 However, limitations such as insufficient protein amounts, inadequate population sizes, and difficulty conducting randomized control trials have hindered efforts to explore the causal associations between thousands of proteins and PCOS.
Emerging large-scale proteomic studies have unearthed a multitude of protein quantitative trait loci (pQTL) covering various circulating proteins. Using pQTL and genome-wide association study (GWAS) data, two-sample Mendelian randomization (MR) can assess whether protein levels of the proxy of genetic variants affect disease risk while minimizing the impact of confounding factors.8 Owing to advances in high-throughput proteomic techniques, we can leverage genetic data from the human blood proteome and adopted MR-based strategies.9,10 This enables us to enhance our comprehension of PCOS genetic architecture, elucidate the causal effects of plasma proteins on PCOS risk, and identify promising drug targets for PCOS.
In this study, we aimed to identify which plasma proteins could be used as potential therapeutic targets for intervening in PCOS. First, we performed a proteome-wide MR analysis by integrating human plasma proteome with genome data to identify PCOS-related proteins. The primary findings were subjected to external MR validation, subsequent Bayesian colocalization analysis, and summary-data-based MR (SMR) analysis. Then, we performed druggability evaluation and mapped the interaction network among the identified proteins and the targets of current PCOS medications. Furthermore, we validated levels of candidate proteins in individuals with PCOS and a mouse model, administrated roxadustat in the mouse model to assess its efficacy as a potential drug inhibiting the EGLN1-HIF1α-ferroptosis pathway for treating PCOS.
Material and methods
The overall study design is depicted in Figure 1. Briefly, we leveraged pQTL data from six large-scale proteomic studies and investigated their associations with PCOS using a two-stage proteome-wide MR framework. Bayesian colocalization, SMR, and heterogeneity in dependent instruments (HEIDI) tests were employed to provide additional insights into associations between plasma protein and PCOS. Then, we conducted protein-protein interaction (PPI) analysis and functional enrichment analyses and assessed the druggability of the identified protein biomarkers to prioritize the potential therapeutic targets. Additionally, we analyzed the mRNA levels of tier 1 and 2 therapeutic targets in human granulosa cells (GCs) and mouse ovaries in both PCOS and control groups. We investigated the EGLN1-HIF1α-ferroptosis axis’s role in PCOS induction using roxadustat, an approved EGLN1 inhibitor.
Figure 1.
Flowchart of the study design
Partially created with BioRender.com.
Study population and data source
The plasma pQTL data were retrieved from six large-scaled proteomic studies (Ferkingstad et al., 4,719 proteins11; Sun et al., 2,994 proteins12; Sun et al., 2,923 proteins13; Suhre et al., 1,124 proteins14; Folkersen et al., 90 proteins15; and Zhang et al., 4,657 proteins16). Each study provided comprehensive summary-level statistics, and detailed characteristics of each study are available in Table S1.
For the primary MR analysis, summary statistics of PCOS were obtained from a meta-analysis of seven previously published PCOS GWASs, including 113,238 people (GWAS Catalog: GCST007089, nPCOS individual = 10,074, nControl = 103,164).17 For external validation, summary statistics of PCOS were obtained from the UK Biobank (UK Biobank: 20002_1350, nPCOS individual = 436, nControl = 193,717)18 and the FinnGen research project (FinnGen: E4_PCOS, R9 release, nPCOS individual = 1,424, nControl = 200,581).19 In cases where SNP or effect allele frequency information was missed in the GWAS summary statistics, we referred to the matched 1000 Genomes reference panel to complete the dataset.
Local study population recruited individuals diagnosed with PCOS or without PCOS from the Reproductive Medicine Center of Women’s Hospital, School of Medicine, Zhejiang University from September 2018 to June 2020. The clinical characteristics of the individuals are shown in Table S2. PCOS was diagnosed according to the revised Rotterdam criteria.20 Women with any endocrine- or metabolism-related disorder were excluded, including congenital adrenal hyperplasia, androgen-secreting tumor, hypogonadotropic hypogonadism, thyroid dysfunction, Cushing syndrome, hyperprolactinemia, and premature ovarian failure. The control group comprised women without PCOS that attended the clinic owing to fallopian tube factors or male factors (including azoospermia, oligospermia, asthenozoospermia, and teratozoospermia) with regular menstrual menstruation, normal ovarian morphology, and no signs of hyperandrogenism and other systemic, gynecological or endocrine dysfunction. This study received approval from the Ethics Committee of Women’s Hospital, School of Medicine, Zhejiang University (no. IRB-20240097-R). The written informed consents were obtained from all participants.
Genetic instrument selection and primary mendelian randomization analysis
Genetic associations with plasma protein levels were extracted from the aforementioned six proteomic studies. Each protein was mapped to corresponding Uniprot ID using the bioDBnet database.21 Genomic coordinates for subsequent analyses were based on the human genome Build 37 (NCBI GRCh37) and processed using the “MungeSumstats” package.22 Instrumental variables (IVs) were selected based on the following criteria: (1) SNPs demonstrating genome-wide significant association with any protein were retained (p value <5 × 10−8), (2) SNPs located outside the major histocompatibility complex region were retained to avoid complex linkage disequilibrium (LD) structure, (3) LD clumping was conducted to identify independent pQTL (relative relaxed parameters [r2 < 0.1, clump_kb < 5,000] were used to enhance precision by incorporating more partial IVs9,23,24,25,26,27,28), and (4) the R2 and F-statistic (R2 = 2 × EAF × [1-EAF] × beta2; F = R2 × [N−2]/[1−R2]) were used to estimate the strength of IVs.29 R2 represented the proportion of variability in protein levels explained by each IV, while the F-statistic assesses the strength of the association between IVs and outcome. Only strong IVs with an F-statistic ≥10 were retained for each exposure of interest. In cases of duplicate proteins across studies, the protein with the highest sum of R2 was selected. Additionally, IVs were classified as cis or trans pQTL based on the following criteria: cis pQTL was defined when the leading SNP was located within ±1 megabase (Mb) region of the transcription start site of the protein-coding gene, whereas trans pQTL was defined when the leading SNP laid outside of this region.12 Ultimately, we identified a total of 74,693 cis pQTL and 2,590 unique plasma proteins as IVs for MR analysis.
We assessed the causality of circulating plasma proteins on PCOS by performing two-sample cis-MR using the “TwoSampleMR” package.8 In our study, the pQTL data served as IVs, and the GWAS of PCOS was utilized as the outcome trait data to investigate whether changes in plasma protein level have causal effects on PCOS. For proteins with only a single pQTL or with multiple pQTLs, we employed either the Wald ratio method or the inverse variance weighted MR (MR-IVW) to calculate MR estimates, respectively.30 The Wald ratio estimates were calculated as the regression coefficient for genetic association with the outcome divided by the regression coefficient for genetic association with circulating protein levels. The IVW estimates were calculated as the average of instrument ratio coefficients weighted by the inverse variance. The estimates obtained from MR analyses approximated odds ratios for an increased risk of PCOS per standard deviation (SD) increase in normalized protein levels. To account for multiple testing, we used the false discovery rate (FDR) at ɑ = 0.05 based on Benjamini-Hochberg method to prioritize the results for further analysis. Simultaneously, the MR-Egger, weighted median, simple mode, and weighted mode methods were used as supplementary analyses to enhance the robustness of our findings.31,32,33,34 There are partial sample overlaps between the proteomic study of Ferkingstad et al.11 and PCOS study of GCST007089,17 as well as the proteomic study of Sun et al.13 and PCOS study of ukb_20002_1350.18 To assess the impact introduced by sample overlap, we evaluated the bias and type 1 error incidence for sample overlap using an online calculator.35 Additionally, the MR-Pleiotropy Residual Sum and Outlier (MR-PRESSO) analysis detected and attempted to reduce horizontal pleiotropic SNPs and provided causal estimates after outlier correction.36
Bayesian colocalization analysis
To assess whether associations between protein and PCOS risk were driven by the confounding of LD, we performed Bayesian colocalization analysis using the “coloc” package.37 As described previously, Bayesian colocalization analysis assesses the probability that two traits share the same causal variant.37 It provides the posterior probability for different hypotheses: (1) neither the protein nor PCOS is associated with any variants in the genomic locus (H0); (2) there is one causal variant associated with protein only (H1); (3) there is one causal variant associated with PCOS only (H2); (4) there are two distinct causal variants associated with protein and PCOS separately (H3); and (5) there is a shared causal variant associated with protein and PCOS (H4). For each protein, we included SNPs within ±500 kb of the pQTL. When a protein had multiple pQTLs, colocalization analysis was performed based on each pQTL, respectively. The pQTL with the strongest evidence for colocalization was shown. We used the coloc.abf algorithm with default parameters (p1 at 1 × 10−4, p2 at 1 × 10−4, and p12 at 1 × 10−5) and defined a gene as having evidence of colocalization if the posterior probability for shared causal variants (PH4) exceeded 80%.
SMR and heterogeneity in dependent instruments tests
The SMR and the HEIDI test occupied the ability to distinguish a pleiotropic model from a linkage model. We adopted the SMR and the HEIDI test to provide further investigation for the causal relationships identified through the primary MR analyses.38 SMR analysis served as a supplementary method to examine for association between plasma protein level and PCOS, which may arise from a shared and potentially causal variant at a locus. The HEIDI test aimed to identify whether the observed association between proteins and PCOS was attributable to genetic LD of distinct causative variants.39 A significance threshold of p value <0.05 was set for the SMR analysis. In the HEIDI test, a p value less than 0.05 showed that the association was driven by LD between two distinct genetic variants.
PPI network and druggability evaluation
We conducted PPI network analysis of potential targeted plasma proteins associated with PCOS risk (FDR-corrected p value <0.05 at MR discovery stage). Our aim was to explore the interactions between those proteins identified in MR analysis and the targets for current medications of PCOS. Firstly, we obtained drugs and their corresponding targets for PCOS from the Opentargets database.40 Subsequently, we performed PPI network analysis using the Search Tool for the Retrieval of Interacting Genes (STRING) database.41 A minimum interaction score of 0.4 was set as the threshold for the PPI analysis.42 To assess the potential of identified proteins as potential therapeutic targets, we also extracted the druggable profile of candidate target proteins from the Drugbank.43
Functional enrichment analysis for therapeutic targets for PCOS
To assess the potential role of identified therapeutic targets for PCOS, we classified their function using the Gene Ontology (GO) and the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway database. The potential therapeutic targets for PCOS in primary MR analysis (p value <0.05) were used as the input for the GO and KEGG enrichment analysis. Functional enrichment analysis relative to the overall GO pattern and the KEGG pathway was performed using “clusterprofiler” R package with Fisher’s exact test (FDR <0.05).44
Sample collection
Ovarian GCs, follicular fluids, and peripheral blood samples were collected from both individuals with PCOS and controls. Follicular fluids were obtained from the largest follicle aspirated from each ovary, followed by centrifugation at 3,000 g for 10 min and subsequent storage at −80°C. GCs were isolated from the follicular fluid aspirated during oocyte retrieval by Ficoll density gradient centrifugation and then lysed in TRIzol for RNA extraction. Peripheral blood was collected on days 2~5 of a menstrual cycle. Venous blood was collected into heparin sodium tubes. The plasma was collected after centrifugation at 4°C and 3,000 g for 10 min and stored at −80°C for analysis.
Enzyme-linked immunosorbent assay
EGLN1 concentrations in human plasma and follicular fluid and mouse plasma were measured using enzyme-linked immunosorbent assay (ELISA) kits (MM-61907H1; MM-47886M1, Meimian, China). The ELISA procedure was conducted in accordance with the manufacturer’s recommendations.
Experimental animal care and ethics
Female Balb/c mice were housed in the laboratory animal center of Zhejiang University with free access to food and water and under a 12-h light/dark cycle. At 21 days of age, mice were randomly assigned to two groups as follows: the dehydroepiandrosterone (DHEA) group received subcutaneous injections of DHEA (HY-14650, USA) (6 mg/100 g body weight per day, dissolved in sesame oil) for 21 consecutive days to induce PCOS, while the control group received daily injections of the same volume of sesame oil. For roxadustat administration, mice were divided randomly into three groups from 21 days of age: the Oil + dimethyl sulfoxide (DMSO) group, the DHEA + DMSO group, and the DHEA + FG-4592 (FG) group. Roxadustat/FG-4592 (HY-13426, MCE) was dissolved in a mixed solvent containing 5% DMSO, 40% PEG-300, 5% Tween-80, and 50% N.S. Mice aged 21 days were intraperitoneally treated with roxadustat at a dosage of 10 mg/kg or an equal volume of vehicle once daily for 21 days as previously described.45 The Animal Care and Use Committee of Zhejiang University of Medicine approved all care and experimental procedures for mice (ID: ZJU20240116).
Estrous cycle determination
From the 10th day of modeling, vaginal smears were conducted daily at 09:00 for at least two estrous cycles. The stage of the estrus cycle was determined by microscopic examination of vaginal cytology using the Shorr method. Proestrus was characterized by round nucleated epithelial cells, estrus by keratinized squamous epithelial cells, metestrus by a mixture of epithelial cells and leukocytes, and diestrus by nucleated epithelial cells and a predominance of leukocytes.
Hematoxylin and eosin staining assay
The ovaries were initially fixed in 4% paraformaldehyde solution, then transitioned to 70% ethanol, and subsequently embedded in paraffin before being sectioned into 5 μm slices. Following deparaffinization, these sections were stained with hematoxylin and eosin (H&E). Observations, analysis, and photography of representative tissue sections were conducted under a light microscope.
Transmission electron microscopy
The ovaries were fixed with 2.5% glutaraldehyde solution in phosphate buffer overnight and washed with PBS three times for 10 min each. The ovaries were then immersed in 1% osmium tetroxide solution in phosphate buffer for 1 h and washed again with PBS three times for 10 min each. Thereafter, the ovaries were fixed in 2% uranyl acetate for 30 min; dehydrated with 50%, 70%, 90%, and finally 100% ethanol for 10 min each step; and rinsed twice with 100% acetone for 15 min each. Then, the ovaries were embedded in resin and sectioned. Observations and photography of the sections were carried out under an electron microscope (Thermo Scientific Talos L120C).
Protein isolation and western blot analysis
Cellular or tissue proteins were extracted by dissolving them in radioimmunoprecipitation assay (RIPA) lysis buffer containing protease and phosphatase inhibitors. Subsequently, total proteins were denatured in the 5× SDS-PAGE loading buffer by boiling at 95°C for 5 min. Equal amounts of proteins were separated using FuturePAGE 4%–20% gels (ET15412Gel, ACE Biotechnology) and then transferred onto nitrocellulose filter membranes. The membranes were blocked with 5% bovine serum albumin, incubated with primary antibodies overnight, and horseradish peroxidase-conjugated secondary antibodies at room temperature for an hour. Details regarding the antibodies and dilution ratio are listed in Table S3. The membrane was visualized using a BIO-RAD ChemiDoc MP Imaging System and analyzed with the Image Lab Software.
Quantitative polymerase chain reaction analysis
To measure the mRNA level of target genes, total RNA was extracted from cells or tissues using TRIzol reagent. Total RNA (1,000 ng) was reverse transcribed to complementary DNA (cDNA) using a DNase and reverse transcriptase reagent kit (R233, Vazyme). The quantitative polymerase chain reaction (qPCR) was carried out in a reaction mix containing SYBR Green (Q711-02, Vazyme) and cDNA on a StepOne Real-Time PCR System (Applied Biosystems) following the manufacturer’s instructions. To determine the relative expression levels of the target genes, the 2−ΔΔCt method was used. Ct values were normalized to the RPS18 and Rps18 (Ribosomal Protein S18) genes, respectively. The primer sequences used in this study were listed in Table S4.
GCs extraction and culture
For the culture of primary GCs, mice were sacrificed 46 h after intraperitoneal injection of 5IU of pregnant mare serum gonadotropin, as previously described.46 Ovarian antral follicles were punctured with a needle to release GCs. Cells were then cultured in DMEM/F12 supplemented with 5% fetal bovine serum and 1% penicillin and streptomycin at 37°C under a humidified atmosphere with 5% CO2.
FerroOrange assay
Mouse GCs were seeded into a 12-well cell culture plate and incubated overnight for detection. Cells were washed in PBS once and stained in 1 μM Ferroorange (F374, DOJINGO, Japan) in DMEM/F12 for 30 min. Then, images were captured by a confocal microscope (Leica DMi8). The fluorescence intensity of each individual cell was quantified using ImageJ2 software (version 2.9.0).
Statistical analysis
Statistical analyses were conducted using GraphPad Prism 9 software. Each experiment was conducted with a minimum of three independent samples and repeated at least three times. All data are presented as the mean ± standard error of the mean (SEM) derived from the specific number of experiments. Prior to subsequent analyses, the Shapiro-Wilk normality test was utilized to assess data distribution. For two-group comparisons, statistical significance (p value <0.05) was determined using unpaired Student’s t tests for parametric data, and Mann-Whitney U test for non-parametric data. For multiple-group comparisons, statistical significance (p value <0.05) was evaluated by one-way analysis of variance (ANOVA) test for parametric data and Kruskal-Wallis test for non-parametric data.
Results
Proteome-wide MR analysis identified 65 circulating proteins for PCOS
A total of 65 proteins exhibited significant associations with PCOS risk with FDR-corrected p value <0.05 using either the Wald ratio or IVW method (Figure 2; Table S5). Genetically predicted higher levels of 30 proteins were associated with an increased risk of PCOS, while the other 35 proteins were negatively associated with PCOS risk. In the replication stage, causal relationships for twelve proteins (FSHB, SIGLEC12, FUT10, NCR3LG1, NUCB2, C1QTNF9, EGLN1, MAPK9, SBSN, DBI, MRI1, and UROD) were successfully validated in independent datasets including the FinnGen or the UK Biobank dataset (p value <0.05) using the Wald ratio or IVW method (Figure 2; Table 1).18,19 Out of the 65 proteins identified, the majority showed consistent associations between circulating protein levels and higher PCOS risk across various MR methods (including MR-Egger, weighted median, simple mode, and weighted mode31,32,33,34) at both the discovery and replication stages, in comparison to the causalities obtained from the Wald ratio or IVW method (Table S6). In the MR-PRESSO analysis, there was insufficient evidence for horizontal pleiotropy in the association between the 65 circulating proteins and PCOS (global test p value >0.05; Table S7).
Figure 2.
Volcano plot showing effects of human plasma proteins on the risk of PCOS in the MR discovery stage
The red dots above the dashed horizontal black line correspond to the plasma proteins that are significant associated with the risk of PCOS (FDR-corrected p value <0.05).
Table 1.
Summary results from MR, colocalization, and SMR for proteome-wide candidate plasma proteins
| Protein | Protein full name |
MR |
Colocalization |
SMR |
Category | |||
|---|---|---|---|---|---|---|---|---|
| Pdiscovery | Preplication | PP4 > 0.80 | Beta | P | PHEIDI | |||
| FSHB | follicle-stimulating hormone subunit beta | 9.75E-09 | 2.93E-10 | yes | −1.43 | 1.21E-06 | 0.501 | tier 1 |
| SIGLEC12 | sialic acid binding ig like lectin 12 | 8.90E-06 | 3.35E-03 | yes | −0.111 | 0.005 | 0.564 | tier 1 |
| FUT10 | fucosyltransferase 10 | 5.99E-05 | 0.032 | yes | −0.096 | 0.042 | 0.601 | tier 1 |
| NUCB2 | nucleobindin 2 | 5.11E-04 | 0.044 | yes | −0.18 | 0.023 | 0.753 | tier 1 |
| NCR3LG1 | fatural killer cell cytotoxicity receptor 3 ligand 1 | 3.57E-04 | 0.047 | no | −0.208 | 0.019 | 0.905 | tier 2 |
| C1QTNF9 | C1q and TNF related 9 | 5.18E-06 | 0.029 | yes | −0.063 | 0.087 | 0.438 | tier 2 |
| EGLN1 | Egl-9 family hypoxia inducible factor 1 | 3.16E-05 | 0.016 | yes | 0.287 | 0.238 | 0.843 | tier 2 |
| MAPK9 | mitogen-activated protein kinase 9 | 1.79E-04 | 3.91E-03 | no | 0.126 | 0.271 | 0.848 | tier 2 |
| SBSN | suprabasin | 5.68E-04 | 4.43E-06 | yes | 0.089 | 0.149 | 0.217 | tier 2 |
| DBI | diazepam binding inhibitor | 7.04E-04 | 2.08E-03 | no | 0.079 | 0.294 | 0.751 | tier 2 |
| MRI1 | methylthioribose-1-phosphate isomerase 1 | 2.52E-04 | 8.66E-03 | yes | 0.199 | 0.056 | 0.365 | tier 2 |
| UROD | uroporphyrinogen decarboxylase | 1.15E-03 | 1.27E-03 | no | 0.102 | 0.021 | 0.964 | tier 2 |
To assess potential bias introduced by sample overlap between the exposure and outcome, we performed evaluation of the type 1 error rate and biases as previous study described.35 For each pair of causal association, the type 1 error rate was controlled under 0.05, and the biases due to sample overlap was less than 0.001 (Table S8). Hence, considerable weak instrument bias would not be expected, and the causalities between circulating proteins and PCOS were less likely to be biased by sample overlaps.
Colocalization analysis, SMR, and HEIDI tests supported the causality of circulating proteins with PCOS
Genetic colocalization analysis was conducted to mitigate potential confounding effect stemming from LD. Out of the 65 proteins identified as potential causal candidates through proteome-wide MR, 58 proteins exhibited evidence of genetic colocalization (PP4 > 80%) (Table S9). There was a substantial likelihood of a shared causal variant influencing both protein levels and PCOS risk.
To further assess the observed findings, we performed SMR and the HEIDI tests for the 65 proteins. Twenty-one of these proteins successfully passed the SMR test (p value <0.05) and aligned with the MR results in effect direction. Additionally, 63 proteins passed the HEIDI test (p value >0.05) (Table S10). Based on the above evidence, we categorized these proteins into three tiers. Four proteins (FSHB, SIGLEC12, FUT10, and NUCB2) successfully passed all tests and were classified into tier 1 (Table 1). Eight proteins that failed the colocalization analysis, SMR, or the HEIDI test (NCR3LG1, C1QTNF9, EGLN1, MAPK9, SBSN, DBI, MRI1, and UROD) were assigned to tier 2. The remaining 53 proteins, which failed in replication MR, were classified into tier 3.
PPI and druggability evaluation for candidate protein targets
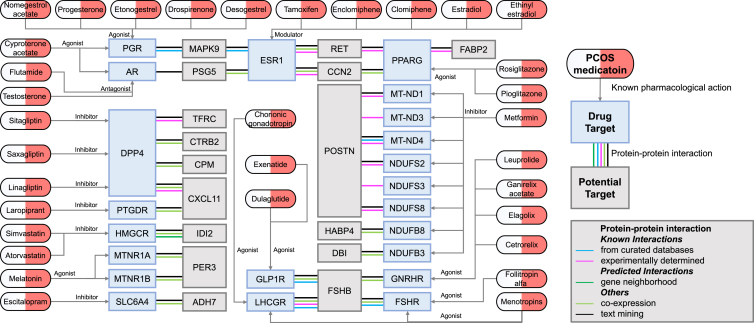
To evaluate the druggability of candidate targets, the PPI network analysis revealed interactions between prioritized proteins (ADH7, TFRC, CTRB2, CPM, CXCL11, DBI, PER3, FSHB, HABP4, MAPK9, PSG5, CCN2, RET, FABP2, IDI2, and POSTN) and the targets of current medications for PCOS (Figure 3; Tables S11 and S12). Utilizing STRING database,41 several interactions were determined as the most reliable interactions (known interactions), including FSHR-FSHB, NDUFS2-POSTN, ESR1-CCN2, DPP4-CXCL11, MT-ND4-POSTN, NDUFS8-POSTN, MT-ND1-POSTN, MT-ND3-POSTN, and NDUFS3-POSTN (Table S12). Specifically, FSHB was found to interact with FSHR, the target of LH/CG receptor agonist (menotropins) and FSHR agonist (follitropin alfa). CCN2 was associated with ESR1, the target of estrogen receptor alpha modulators (tamoxifen, enclomiphene, and clomiphene) and estrogen receptor alpha agonist (ethinyl estradiol and estradiol). CXCL11 was associated with DPP4, the target of dipeptidyl peptidase IV inhibitor (sitagliptin, saxagliptin, and linagliptin). Additionally, STRING revealed that POSTN interacted with a series of mitochondrially encoded NADH dehydrogenase (MT-ND1, MT-ND3, and MT-ND4) and NADH dehydrogenase (ubiquinone) Fe-S Protein (NDUFS2, NDUFS3, and NDUFS8).41 MT-ND1, MT-ND3, MT-ND4, NDUFS3, and NDUFS8 are all targets of metformin.
Figure 3.
Interaction between current PCOS medication targets and identified potential drug targets
To evaluate drug development activities of candidate targets, we searched for current medications targeting identified potential causal proteins in the Drugbank database43 (Table S13). We identified 80 medications targeting 20 proteins that may modify the disease, including antioxidant agent, estrogenic steroid, or drugs treating hypoestrogenism, acne, and autoimmune diseases. Overall, therapeutics targeting 65 proteins requires further evaluation among which 20 are druggable, and 45 represent future breakthrough points.
Validation and functional enrichment analysis for therapeutic targets of PCOS
To assess the impact of tier 1 and 2 therapeutic targets for PCOS on human follicle development, we analyzed the mRNA expression changes in human GCs. In comparison to the control group, PCOS individuals exhibited decreasing mRNA levels of FUT10 (MIM: 616931), NCR3LG1 (MIM: 613714), NUCB2 (MIM: 608020), and DBI (MIM: 125950), as well as increasing mRNA levels of EGLN1 (MIM: 606425) and MAPK9 (MIM: 602896) in GCs (Figure 4A). However, there were no significant changes in mRNA levels of SIGLEC12 (MIM: 606094), C1QTNF9 (MIM: 614285), SBSN (MIM: 609969), MRI1 (MIM: 615105), and UROD (MIM: 613521) in GCs between control group and PCOS individuals (Figure 4A). Furthermore, we utilized a DHEA-induced PCOS mouse model to detect the expression of therapeutic targets for PCOS. Compared with female control mice, DHEA mice exhibited significantly reduced mRNA levels of Fut10, Nucb2, C1qtnf9, and Dbi, as well as elevated mRNA levels of Egln1, Mapk9, Mri1, and Urod (Figure 4B). To determine the effect of therapeutic targets on the occurrence of PCOS, all potential therapeutic targets for PCOS were subjected to GO enrichment and KEGG pathway analysis to explore their putative biological function (Figures S1–S4). The KEGG pathway analysis identified the hypoxia-inducible factor 1 pathway among the various pathways related to the therapeutic targets for PCOS (Figure S4). EGLN1, identified as a tier 2 therapeutic target protein for PCOS, is known as prolyl hydroxylase domain-containing protein. In the presence of oxygen, EGLN1 hydroxylates HIF1α, resulting in a binding site for a ubiquitin ligase complex that includes von Hippel-Lindau (VHL), leading to HIF1α degradation.47 Based on the above analyses, there was substantial evidence supporting EGLN1 in the MR analysis and colocalization analysis, along with its consistent expression patterns observed in GCs of individuals with PCOS and ovaries of DHEA-induced PCOS mouse model.
Figure 4.
Validation for expression level of candidate proteins in individuals with PCOS and mouse model
(A) The mRNA level of tier 1 and 2 therapeutic targets in the GCs of individuals with PCOS and the control group according to the qPCR analyses.
(B) The mRNA level of tier 1 and 2 therapeutic targets in the ovarian tissue of DHEA-induced PCOS mouse model and the control group according to the qPCR analyses. All the results are expressed as the mean ± SEM; ∗p < 0.05 and ∗∗∗p < 0.001.
Roxadustat improves the onset of PCOS through targeting the EGLN1-HIF1α-ferroptosis axis
In both the follicular fluids and plasma of PCOS individuals, EGLN1 protein levels were significantly elevated compared with control group (Figures 5A and 5B). DHEA-induced PCOS mice also exhibited significantly increased plasma levels of EGLN1 compared with oil mouse controls (Figure 5C). Consequently, we delved further into the role of EGLN1-HIF1α signaling in the pathogenesis of PCOS and the potential of Roxadustat (FG-4592, an approving EGLN1 inhibitor for treating renal anemia) in intervening in PCOS. Compared with the control group (Oil + DMSO), the DHEA-induced PCOS mouse model (DHEA + DMSO) displayed typical polycystic ovaries and notable estrous acyclicity. After intraperitoneal administration of roxadustat, the DHEA + FG group reversed the disrupted estrous cycle and morphological changes in the ovary (Figures 5D–5F). Roxadustat-induced stabilization of HIF1α has been found to mitigate acute kidney injury by decreasing ferroptosis.48 Recent studies revealed that elevated miR-93-5p in GCs of PCOS individuals promoted ferroptosis, whereas metformin can inhibit ovarian ferroptosis through the SIRT3-AMPK-mTOR pathway to improve ovarian function in a PCOS mouse model.49,50 Transmission electron microscopy (TEM) was utilized to observe mitochondria ultrastructure in mouse GC cells before and after roxadustat administration (Figure 5G). Compared to the Oil + DMSO mice, we observed increased mitochondrial membrane density, reduced mitochondrial crista structures, and broken mitochondrial outer membrane in DHEA + DMSO mice, which was characteristic morphological features of ferroptosis reported previously.51 Relatively normal mitochondria ultrastructure in mouse GC cells was observed in the DHEA + FG group compared to the DHEA + DMSO group (Figure 5G). We also used the FerroOrange probe to detect intracellular Fe2+ levels in GCs. We observed enhanced intracellular Fe2+ levels in GCs of the DHEA + DMSO group and lowered Fe2+ levels after roxadustat treatment in the DHEA + FG group (Figure 5H).
Figure 5.
Roxadustat improves the onset of PCOS through the EGLN1-HIF1α-ferroptosis axis
(A) The follicular fluid levels of EGLN1 in PCOS and control groups.
(B) The plasma levels of EGLN1 in PCOS and control groups.
(C) The plasma levels of EGLN1 in control group mice (Oil) and DHEA-induced PCOS group mice (DHEA).
(D) Quantitative analysis of estrous cycles in vaginal smears of mice. M/D, metestrus/diestrus phase; P, proestrus; E, estrus. Scatterplot representing the percentage (%) of time spent in each estrous cycle.
(E) Representative diagram of estrous cycles.
(F) Representative ovarian morphology by H&E staining.
(G) Representative TEM images of GCs in ovarian tissues.
(H) Intracellular Fe2+ level in GCs was detected using FerroOrange probe by measuring the fluorescence in a confocal microscope. Quantification of FerroOrange fluorescence is shown on the right.
(I) The mRNA levels of ferroptosis regulation genes in ovarian tissues according to qPCR analyses.
(J) The protein accumulation of EGLN1-HIF1α-ferroptosis axis in ovarian tissues according to western blot.
(K) The mRNA levels of Fabp3 in ovarian tissues according to qPCR analyses. All the results are expressed as the mean ± SEM; ∗p value <0.05, ∗∗p value <0.01, ∗∗∗p value <0.001, and ∗∗∗∗p value <0.0001.
Next, we detected the expression of ferroptosis-related genes in ovarian tissue. Key genes involved in the positive regulation of ferroptosis, including Acsl4, Tfrc, Alox5, Alox12, Alox15, and Lpcat3, were markedly upregulated in the ovaries of the DHEA + DMSO group compared with those in the Oil + DMSO group (Figure 5I). Western blot analysis also showed higher protein accumulation of TFRC and ACSL4, as well as lower protein accumulation of GPX4 in the ovaries of the DHEA + DMSO group (Figure 5J). After administration with roxadustat, the expression of ferroptosis positive regulation genes (Acsl4, Tfrc, Alox5, Alox12, Alox15, Aloxe5, and Lpcat3) was lower than that in the DHEA + DMSO group (Figure 5I). Lower levels of TFRC and ACSL4, as well as higher level of GPX4, were also observed in the ovaries of the DHEA+FG group according to Western blot analysis (Figure 5J). Additionally, Roxadustat administration restored protein accumulation of HIF1α in the ovaries of the DHEA + FG group. Moreover, the mRNA expression of Fabp3 (fatty acid binding protein 3), a key HIF1α target genes responsible for fatty acid uptake and lipid storage,52 was restored by HIF1A activation (Figure 5K). Collectively, these findings confirmed that roxadustat treatment allows for HIF1α stabilization and antagonizes ferroptosis in the ovaries of the DHEA-induced mouse model.
Discussion
Principal findings
The current study systematically investigated circulating proteins associated with PCOS risk, particularly focusing on the impact of EGLN1 on onset of PCOS using an MR-Bayesian model averaging causal analyses, biological pathway enrichment analysis, and experimental approaches. A comprehensive pipeline comprising analytical techniques revealed causal associations between the decreased level of plasma proteins (FSHB, SIGLEC12, FUT10, NCR3LG1, NUCB2, and C1QTNF9) and a higher risk of PCOS. Causal associations were found between the elevation of plasma proteins (EGLN1, MAPK9, SBSN, DBI, MRI1, and UROD) and a higher risk of PCOS. In PPI analyses, FSHB, POSTN, CCN2, and CXCL11 ranked among the top proteins interacting with the targets of current PCOS medications. A list of 80 current drugs and 20 druggable proteins further supported the potential for drug development programs targeting PCOS. Additionally, KEGG functional enrichment analysis for therapeutic targets revealed involvement of the HIF1α pathway in PCOS. Using a DHEA-induced PCOS mouse model, we explored the EGLN1-HIF1α-ferroptosis axis in inducing PCOS and investigated the therapeutic potential of the EGLN1-targeting roxadustat.
Concordance of observational and causal associations for identified proteins
One of the key strengths of this study lies in comparing evidence between observational and MR analyses for candidate proteins. Given the heritability of PCOS, the MR method has recently gained traction in assessing causal associations between PCOS and etiological risk factors.53 Utilizing MR approaches, we have discovered drug targets having a causal association with an increased risk of PCOS. Several strategies were employed to enhance the biological credibility of these candidate proteins. Through our experimental approaches, we have partially confirmed the consistency between the expression levels of tier 1 and 2 targets (FUT10, NUCB2, EGLN1, and MAPK9) in ovaries and GCs, as well as the effect size of these targets in MR analyses. FUT10, a member of the α-1,3 fucosyltransferase family, mediates the α-1,3 fucosylation level of TNFR1, enhances its binding ability to TNF-α, and eventually accelerates the osteoarthritis process.54 In the ventricular zone of the embryonic brain, FUT10 activity is necessary to maintain the self-renewal of neurosphere-forming neural stem cells.55 While the regulation role of FUT10 in PCOS remains largely unknown, it is worth noting that core fucosylation catalyzed by fucosyltransferase 8 in GCs controls oocyte and follicular development via the FSH/FSHR pathway, loss of which contributes to poor ovary responders.56 Further investigation into the associations of fucosylation catalyzed by FUT10 and PCOS occurrence is warranted. NUCB2, initially identified in the nucleus of the hypothalamus, plays a role in diet and energy homeostasis.57 In a letrozole-induced rat PCOS model, plasma NUCB2 level was decreased compared to the control group, consistent with our findings.58 MAPK9, a member of the MAP kinase family, phosphorylated BCL2L11 upon activation, leading to its dissociation from BECN1 and subsequently participating in RIPK1-mediated activation of autophagy.59 Cultured GCs from women with PCOS exhibited increased levels of autophagy markers (ATG7, ATG5, BECN1, and LC3-II:LC3-I).60 Moreover, the mRNA level of MAPK9 was markedly elevated in human PCOS ovarian tissue compared to normal ovarian tissue,61 suggesting its involvement in autophagy activation in PCOS.
FSHB encodes the beta subunit of follicle-stimulating hormone, which is secreted from the pituitary gland. A recent study has revealed that FSH promotes glutamine synthesis in GCs, thereby maintaining follicle wall integrity and inhibiting GC apoptosis, which is crucial for initiating ovulation.62 Inadequate FSH levels are strongly associated with anovulation in PCOS. This finding aligns with our MR analysis, which suggests that FSHB is a protective factor for PCOS. In a GWAS of PCOS, rs11031006, an independent signal reach genome-wide significance for association with PCOS, is located near FSHB. The PCOS-susceptibility allele at rs11031006 is also robustly associated with lower circulating FSH concentrations.63,64 The allele rs11031006-G has been associated with higher serum FSH levels and a lower risk of PCOS.65
Implications for therapeutics targeting EGLN1-HIF1α-ferroptosis axis inducing PCOS
EGLN1 occupies hydroxylase activity to allow ubiquitin-mediated proteasomal degradation of HIF1α via VHL binding under physiological conditions.66 HIF1α is involved in glycolysis and lipid metabolism and regulates ferroptosis with varying effects among cell lines and animal models. In mouse models of diabetic renal tubular injury and testicular injury, HIF1α induces ferroptosis, resulting in damage to renal tubules, testicular Leydig, and Sertoli cells.67,68 In a cancer cell model and model of liver fibrosis, HIF1α antagonizes ferroptosis by upregulating lipid metabolism-related genes, including FABP3 and FABP7.69,70 In our DHEA-induced PCOS mouse model, we found that increased levels of ELGN1 were negatively associated with HIF1α stabilization, inducing ferroptosis of GCs in the ovary. Roxadustat, an EGLN1 inhibitor developed for treating anemia induced by chronic kidney disease or myelodysplastic syndromes, reversibly binds to and inhibits EGLN1.71 Roxadustat reverses the degradation of transcription factors in the HIF family under normal oxygen conditions. The role of roxadustat is cell background dependent. In some contexts, roxadustat amplifies HIF signaling and suppresses chemoresistant glioblastoma growth via inducing ferroptosis.72 Conversely, in other instances, HIF-1α stabilization by roxadustat has been found to mitigate acute kidney injury by decreasing ferroptosis.48 Additionally, roxadustat can also serve as a protective agent to reduce organ fibrosis and inflammation, correct metabolic disorders, reduce oxidative stress, improve mitochondrial function, and normalize angiogenesis.73 In our study, roxadustat improved the onset of PCOS by targeting the EGLN1-HIF1α-ferroptosis axis and reversed the disrupted estrous cycle and ovarian morphology. These findings suggest a considerable therapeutic potential for roxadustat in PCOS, providing an experimental basis before expanding its indication.
Strength and limitations
The strength of our investigation lies in the utilization of proteome-wide MR analyses to systematically estimate the causal effects of plasma proteins on PCOS. The MR design offers several advantages, including large-scale sample sizes, comprehensive proteome coverage, and the ability to minimize bias due to confounding and reverse causality. The replication MR analyses conducted across multiple datasets further validated the robustness of our findings. Additionally, colocalization analysis proved to be a powerful tool in revealing the pleiotropic effects of certain loci on multiple traits. Supplementary evidence from PPI networks, druggability evaluations, and experimental research provided insights into the potential pathogenic effect of druggable targets. Through this approach, we have discovered the HIF1α-ferroptosis axis as a relevant pathogenic pathway of PCOS in which the elevated level of EGLN1 plays a crucial role.
There are still several limitations in our analysis that merit consideration. Firstly, the plasma proteome data were obtained from European cohorts, which may limit the generalization of our conclusions to other populations.74,75 Secondly, there is a small percentage of sample overlap, which may result in potential bias and type 1 error inflation due to weak instrument bias. The F statistic quantifies the strength of the association between IVs and exposures. In our preliminary genetic instrument selection, we retained only IVs with an F statistic ≥10 to avoid weak instrument bias. We also calculated the type 1 error rate and found the estimated bias from current sample overlap rate was less than 0.1%. Recent studies have demonstrated the reliability and validity of two-sample MR using overlapping samples, under the premise of low expected analytic bias and minimal type 1 error.76,77,78,79 Therefore, the current degree of sample overlap does not significantly affect the robustness of causalities in our finding. Thirdly, in our experimental research, we detected expression levels of identified drug targets in GCs of PCOS but didn’t estimate their levels in other ovarian cells or organ tissues. Assessing protein levels from other ovarian cells or organ tissues in PCOS may provide further insights into pathogenesis and complication of PCOS. Fourthly, for candidate targets consistent across the observational validation and PCOS GWASs, our interpretation and hypothesis are limited by the lack of detailed understanding of pathogenesis and phenotypes of ovarian structure and function. Moreover, the MR analysis was based on the impact of circulating proteins, including cell-secreted and leaked proteins. The abundance of circulating proteins may differ from the abundance within cells and tissues; therefore, potential effects of cell- or tissue-specific protein abundance are not directly explored in our study.
Conclusions
In summary, this study identified numerous associations of circulating proteins with the risk of PCOS using integrated genetic approaches and experimental validation. FSHB, SIGLEC12, FUT10, NCR3LG1, NUCB2, C1QTNF9, EGLN1, MAPK9, SBSN, DBI, MRI1, and UROD were prioritized as potential drug targets for PCOS, pending verification in future trials. Building upon this, the EGLN1-HIF1α-ferroptosis axis was found to potentially mediate the pathogenesis of PCOS. Our findings underscore the value of population-scale genomic and proteomic analyses in identifying critical loci and pathways with causal implications for PCOS.
Data and code availability
Analyzing codes in this study are available on GitHub (https://github.com/Feidani/PCOS_Drug_targets).
Acknowledgments
We extend our gratitude to the participants and investigators of the FinnGen consortium and the UK Biobank database. This work was supported by grants from the National Key Research and Development Program of China (no. 2022YFC2703500, D.Z.), the National Natural Science Foundation of China (no. U23A20404, D.Z.; no. 82394424, D.Z.), the Key Research and Development Program of Zhejiang Province (no. 2021C03098, D.Z.), and the China Postdoctoral Science Foundation (no. 2023M743121, F.W.).
Author contributions
Conceptualization, F.N., F.W., and J.S.; methodology, F.N., F.W., J.S., X.L., and D.Z.; investigation, F.N., F.W., M.T., J.C., X.S., X.Y., R.C., Y.L., X.S., and J.C.; writing – original draft, F.N.; writing – review & editing, F.N., F.W., J.S., X.L., and D.Z.; funding acquisition, F.W. and D.Z.
Declaration of interests
The authors declare no competing interests.
Published: November 13, 2024
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.ajhg.2024.10.008.
Contributor Information
Xue Li, Email: [email protected].
Dan Zhang, Email: [email protected].
Web resources
BioDBnet, https://biodbnet-abcc.ncifcrf.gov/db/db2db.php.
Bias and Type 1 error rate calculator, https://sb452.shinyapps.io/overlap/
clusterProfiler, https://yulab-smu.top/biomedical-knowledge-mining-book/
coloc, https://github.com/chr1swallace/coloc
FinnGen, https://finngen.gitbook.io/documentation/v/r9
Gene Ontology, http://geneontology.org
GWAS Catalog, https://www.ebi.ac.uk/gwas/
Kyoto Encyclopedia of Genes and Genomes, http://www.kegg.jp/kegg
MendelianRandomization, https://cran.r-project.org/src/contrib/Archive/MendelianRandomization/
MungeSumstats, https://github.com/neurogenomics/MungeSumstats
Opentargets, https://www.opentargets.org
R software, https://www.r-project.org/
SMR, https://yanglab.westlake.edu.cn/software/smr/#Overview
TwoSampleMR, https://github.com/MRCIEU/TwoSampleMR
UK Biobank, https://www.nealelab.is/uk-biobank
Supplemental information
References
- 1.Dapas M., Dunaif A. Deconstructing a Syndrome: Genomic Insights Into PCOS Causal Mechanisms and Classification. Endocr. Rev. 2022;43:927–965. doi: 10.1210/endrev/bnac001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Teede H.J., Tay C.T., Laven J.J.E., Dokras A., Moran L.J., Piltonen T.T., Costello M.F., Boivin J., Redman L.M., Boyle J.A., et al. Recommendations From the 2023 International Evidence-based Guideline for the Assessment and Management of Polycystic Ovary Syndrome. J. Clin. Endocrinol. Metab. 2023;108:2447–2469. doi: 10.1210/clinem/dgad463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zheng J., Haberland V., Baird D., Walker V., Haycock P.C., Hurle M.R., Gutteridge A., Erola P., Liu Y., Luo S., et al. Phenome-wide Mendelian randomization mapping the influence of the plasma proteome on complex diseases. Nat. Genet. 2020;52:1122–1131. doi: 10.1038/s41588-020-0682-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Qu X., Donnelly R. Sex Hormone-Binding Globulin (SHBG) as an Early Biomarker and Therapeutic Target in Polycystic Ovary Syndrome. Int. J. Mol. Sci. 2020;21 doi: 10.3390/ijms21218191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Foda A.A., Foda E.A., El-Said Z.H. Serum irisin levels in polycystic ovary syndrome after ovarian drilling. Diabetes Metab. Syndr. 2019;13:1463–1468. doi: 10.1016/j.dsx.2019.02.019. [DOI] [PubMed] [Google Scholar]
- 6.Foda A.A., Foda E.A., El-Negeri M.A., El-Said Z.H. Serum chemerin levels in Polycystic Ovary Syndrome after metformin therapy. Diabetes Metab. Syndr. 2019;13:1309–1315. doi: 10.1016/j.dsx.2019.01.050. [DOI] [PubMed] [Google Scholar]
- 7.Panidis D., Tziomalos K., Koiou E., Kandaraki E.A., Tsourdi E., Delkos D., Kalaitzakis E., Katsikis I. The effects of obesity and polycystic ovary syndrome on serum lipocalin-2 levels: a cross-sectional study. Reprod. Biol. Endocrinol. 2010;8:151. doi: 10.1186/1477-7827-8-151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hemani G., Zheng J., Elsworth B., Wade K.H., Haberland V., Baird D., Laurin C., Burgess S., Bowden J., Langdon R., et al. The MR-Base platform supports systematic causal inference across the human phenome. Elife. 2018;7 doi: 10.7554/eLife.34408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Namba S., Konuma T., Wu K.H., Zhou W., Global Biobank Meta-analysis Initiative. Okada Y. A practical guideline of genomics-driven drug discovery in the era of global biobank meta-analysis. Cell Genom. 2022;2 doi: 10.1016/j.xgen.2022.100190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhao H., Rasheed H., Nøst T.H., Cho Y., Liu Y., Bhatta L., Bhattacharya A., Hemani G., Davey Smith G., Brumpton B.M., et al. Proteome-wide Mendelian randomization in global biobank meta-analysis reveals multi-ancestry drug targets for common diseases. Cell Genom. 2022;2 doi: 10.1016/j.xgen.2022.100195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ferkingstad E., Sulem P., Atlason B.A., Sveinbjornsson G., Magnusson M.I., Styrmisdottir E.L., Gunnarsdottir K., Helgason A., Oddsson A., Halldorsson B.V., et al. Large-scale integration of the plasma proteome with genetics and disease. Nat. Genet. 2021;53:1712–1721. doi: 10.1038/s41588-021-00978-w. [DOI] [PubMed] [Google Scholar]
- 12.Sun B.B., Maranville J.C., Peters J.E., Stacey D., Staley J.R., Blackshaw J., Burgess S., Jiang T., Paige E., Surendran P., et al. Genomic atlas of the human plasma proteome. Nature. 2018;558:73–79. doi: 10.1038/s41586-018-0175-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sun B.B., Chiou J., Traylor M., Benner C., Hsu Y.H., Richardson T.G., Surendran P., Mahajan A., Robins C., Vasquez-Grinnell S.G., et al. Plasma proteomic associations with genetics and health in the UK Biobank. Nature. 2023;622:329–338. doi: 10.1038/s41586-023-06592-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Suhre K., Arnold M., Bhagwat A.M., Cotton R.J., Engelke R., Raffler J., Sarwath H., Thareja G., Wahl A., DeLisle R.K., et al. Connecting genetic risk to disease end points through the human blood plasma proteome. Nat. Commun. 2017;8 doi: 10.1038/ncomms14357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Folkersen L., Fauman E., Sabater-Lleal M., Strawbridge R.J., Frånberg M., Sennblad B., Baldassarre D., Veglia F., Humphries S.E., Rauramaa R., et al. Mapping of 79 loci for 83 plasma protein biomarkers in cardiovascular disease. PLoS Genet. 2017;13 doi: 10.1371/journal.pgen.1006706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang J., Dutta D., Köttgen A., Tin A., Schlosser P., Grams M.E., Harvey B., CKDGen Consortium. Yu B., Boerwinkle E., et al. Plasma proteome analyses in individuals of European and African ancestry identify cis-pQTLs and models for proteome-wide association studies. Nat. Genet. 2022;54:593–602. doi: 10.1038/s41588-022-01051-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Day F., Karaderi T., Jones M.R., Meun C., He C., Drong A., Kraft P., Lin N., Huang H., Broer L., et al. Large-scale genome-wide meta-analysis of polycystic ovary syndrome suggests shared genetic architecture for different diagnosis criteria. PLoS Genet. 2018;14 doi: 10.1371/journal.pgen.1007813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sudlow C., Gallacher J., Allen N., Beral V., Burton P., Danesh J., Downey P., Elliott P., Green J., Landray M., et al. UK biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med. 2015;12 doi: 10.1371/journal.pmed.1001779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kurki M.I., Karjalainen J., Palta P., Sipilä T.P., Kristiansson K., Donner K.M., Reeve M.P., Laivuori H., Aavikko M., Kaunisto M.A., et al. FinnGen provides genetic insights from a well-phenotyped isolated population. Nature. 2023;613:508–518. doi: 10.1038/s41586-022-05473-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rotterdam ESHRE/ASRM-Sponsored PCOS Consensus Workshop Group Revised 2003 consensus on diagnostic criteria and long-term health risks related to polycystic ovary syndrome. Fertil. Steril. 2004;81:19–25. doi: 10.1016/j.fertnstert.2003.10.004. [DOI] [PubMed] [Google Scholar]
- 21.Mudunuri U., Che A., Yi M., Stephens R.M. bioDBnet: the biological database network. Bioinformatics. 2009;25:555–556. doi: 10.1093/bioinformatics/btn654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Murphy A.E., Schilder B.M., Skene N.G. MungeSumstats: a Bioconductor package for the standardization and quality control of many GWAS summary statistics. Bioinformatics. 2021;37:4593–4596. doi: 10.1093/bioinformatics/btab665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schmidt A.F., Finan C., Gordillo-Marañón M., Asselbergs F.W., Freitag D.F., Patel R.S., Tyl B., Chopade S., Faraway R., Zwierzyna M., Hingorani A.D. Genetic drug target validation using Mendelian randomisation. Nat. Commun. 2020;11:3255. doi: 10.1038/s41467-020-16969-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tang B., Wang Y., Jiang X., Thambisetty M., Ferrucci L., Johnell K., Hägg S. Genetic Variation in Targets of Antidiabetic Drugs and Alzheimer Disease Risk: A Mendelian Randomization Study. Neurology. 2022;99:e650–e659. doi: 10.1212/wnl.0000000000200771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Henry A., Gordillo-Marañón M., Finan C., Schmidt A.F., Ferreira J.P., Karra R., Sundström J., Lind L., Ärnlöv J., Zannad F., et al. Therapeutic Targets for Heart Failure Identified Using Proteomics and Mendelian Randomization. Circulation. 2022;145:1205–1217. doi: 10.1161/circulationaha.121.056663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Storm C.S., Kia D.A., Almramhi M.M., Bandres-Ciga S., Finan C., International Parkinson’s Disease Genomics Consortium IPDGC. Hingorani A.D., Wood N.W. Finding genetically-supported drug targets for Parkinson's disease using Mendelian randomization of the druggable genome. Nat. Commun. 2021;12:7342. doi: 10.1038/s41467-021-26280-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang Y., Xie J., Wen S., Cao P., Xiao W., Zhu J., Li S., Wang Z., Cen H., Zhu Z., et al. Evaluating the causal effect of circulating proteome on the risk of osteoarthritis-related traits. Ann. Rheum. Dis. 2023;82:1606–1617. doi: 10.1136/ard-2023-224459. [DOI] [PubMed] [Google Scholar]
- 28.Ghouse J., Tragante V., Ahlberg G., Rand S.A., Jespersen J.B., Leinøe E.B., Vissing C.R., Trudsø L., Jonsdottir I., Banasik K., et al. Genome-wide meta-analysis identifies 93 risk loci and enables risk prediction equivalent to monogenic forms of venous thromboembolism. Nat. Genet. 2023;55:399–409. doi: 10.1038/s41588-022-01286-7. [DOI] [PubMed] [Google Scholar]
- 29.Sun J., Zhao J., Jiang F., Wang L., Xiao Q., Han F., Chen J., Yuan S., Wei J., Larsson S.C., et al. Identification of novel protein biomarkers and drug targets for colorectal cancer by integrating human plasma proteome with genome. Genome Med. 2023;15:75. doi: 10.1186/s13073-023-01229-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Deng Y.T., Ou Y.N., Wu B.S., Yang Y.X., Jiang Y., Huang Y.Y., Liu Y., Tan L., Dong Q., Suckling J., et al. Identifying causal genes for depression via integration of the proteome and transcriptome from brain and blood. Mol. Psychiatry. 2022;27:2849–2857. doi: 10.1038/s41380-022-01507-9. [DOI] [PubMed] [Google Scholar]
- 31.Bowden J., Davey Smith G., Burgess S. Mendelian randomization with invalid instruments: effect estimation and bias detection through Egger regression. Int. J. Epidemiol. 2015;44:512–525. doi: 10.1093/ije/dyv080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Burgess S., Thompson S.G. Interpreting findings from Mendelian randomization using the MR-Egger method. Eur. J. Epidemiol. 2017;32:377–389. doi: 10.1007/s10654-017-0255-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bowden J., Davey Smith G., Haycock P.C., Burgess S. Consistent Estimation in Mendelian Randomization with Some Invalid Instruments Using a Weighted Median Estimator. Genet. Epidemiol. 2016;40:304–314. doi: 10.1002/gepi.21965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hartwig F.P., Davey Smith G., Bowden J. Robust inference in summary data Mendelian randomization via the zero modal pleiotropy assumption. Int. J. Epidemiol. 2017;46:1985–1998. doi: 10.1093/ije/dyx102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Burgess S., Davies N.M., Thompson S.G. Bias due to participant overlap in two-sample Mendelian randomization. Genet. Epidemiol. 2016;40:597–608. doi: 10.1002/gepi.21998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Verbanck M., Chen C.Y., Neale B., Do R. Detection of widespread horizontal pleiotropy in causal relationships inferred from Mendelian randomization between complex traits and diseases. Nat. Genet. 2018;50:693–698. doi: 10.1038/s41588-018-0099-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Giambartolomei C., Vukcevic D., Schadt E.E., Franke L., Hingorani A.D., Wallace C., Plagnol V. Bayesian test for colocalisation between pairs of genetic association studies using summary statistics. PLoS Genet. 2014;10 doi: 10.1371/journal.pgen.1004383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhu Z., Zhang F., Hu H., Bakshi A., Robinson M.R., Powell J.E., Montgomery G.W., Goddard M.E., Wray N.R., Visscher P.M., Yang J. Integration of summary data from GWAS and eQTL studies predicts complex trait gene targets. Nat. Genet. 2016;48:481–487. doi: 10.1038/ng.3538. [DOI] [PubMed] [Google Scholar]
- 39.Wu Y., Zeng J., Zhang F., Zhu Z., Qi T., Zheng Z., Lloyd-Jones L.R., Marioni R.E., Martin N.G., Montgomery G.W., et al. Integrative analysis of omics summary data reveals putative mechanisms underlying complex traits. Nat. Commun. 2018;9:918. doi: 10.1038/s41467-018-03371-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ochoa D., Hercules A., Carmona M., Suveges D., Baker J., Malangone C., Lopez I., Miranda A., Cruz-Castillo C., Fumis L., et al. The next-generation Open Targets Platform: reimagined, redesigned, rebuilt. Nucleic Acids Res. 2023;51:D1353–D1359. doi: 10.1093/nar/gkac1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Szklarczyk D., Kirsch R., Koutrouli M., Nastou K., Mehryary F., Hachilif R., Gable A.L., Fang T., Doncheva N.T., Pyysalo S., et al. The STRING database in 2023: protein-protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res. 2023;51:D638–D646. doi: 10.1093/nar/gkac1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lin J., Zhou J., Xu Y. Potential drug targets for multiple sclerosis identified through Mendelian randomization analysis. Brain. 2023;146:3364–3372. doi: 10.1093/brain/awad070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Knox C., Wilson M., Klinger C.M., Franklin M., Oler E., Wilson A., Pon A., Cox J., Chin N.E.L., Strawbridge S.A., et al. DrugBank 6.0: the DrugBank Knowledgebase for 2024. Nucleic Acids Res. 2024;52:D1265–D1275. doi: 10.1093/nar/gkad976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yu G., Wang L.G., Han Y., He Q.Y. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16:284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yan P., Li N., Ma M., Liu Z., Yang H., Li J., Wan C., Gao S., Li S., Zheng L., et al. Hypoxia-inducible factor upregulation by roxadustat attenuates drug reward by altering brain iron homoeostasis. Signal Transduct. Target. Ther. 2023;8:355. doi: 10.1038/s41392-023-01578-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jin J., Ma Y., Tong X., Yang W., Dai Y., Pan Y., Ren P., Liu L., Fan H.Y., Zhang Y., Zhang S. Metformin inhibits testosterone-induced endoplasmic reticulum stress in ovarian granulosa cells via inactivation of p38 MAPK. Hum. Reprod. 2020;35:1145–1158. doi: 10.1093/humrep/deaa077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Berra E., Benizri E., Ginouvès A., Volmat V., Roux D., Pouysségur J. HIF prolyl-hydroxylase 2 is the key oxygen sensor setting low steady-state levels of HIF-1alpha in normoxia. Embo j. 2003;22:4082–4090. doi: 10.1093/emboj/cdg392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li X., Zou Y., Xing J., Fu Y.Y., Wang K.Y., Wan P.Z., Zhai X.Y. Pretreatment with Roxadustat (FG-4592) Attenuates Folic Acid-Induced Kidney Injury through Antiferroptosis via Akt/GSK-3β/Nrf2 Pathway. Oxid. Med. Cell. Longev. 2020;2020 doi: 10.1155/2020/6286984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Peng Q., Chen X., Liang X., Ouyang J., Wang Q., Ren S., Xie H., Wang C., Sun Y., Wu X., et al. Metformin improves polycystic ovary syndrome in mice by inhibiting ovarian ferroptosis. Front. Endocrinol. 2023;14 doi: 10.3389/fendo.2023.1070264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tan W., Dai F., Yang D., Deng Z., Gu R., Zhao X., Cheng Y. MiR-93-5p promotes granulosa cell apoptosis and ferroptosis by the NF-kB signaling pathway in polycystic ovary syndrome. Front. Immunol. 2022;13 doi: 10.3389/fimmu.2022.967151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xie B.S., Wang Y.Q., Lin Y., Mao Q., Feng J.F., Gao G.Y., Jiang J.Y. Inhibition of ferroptosis attenuates tissue damage and improves long-term outcomes after traumatic brain injury in mice. CNS Neurosci. Ther. 2019;25:465–475. doi: 10.1111/cns.13069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bensaad K., Favaro E., Lewis C.A., Peck B., Lord S., Collins J.M., Pinnick K.E., Wigfield S., Buffa F.M., Li J.L., et al. Fatty acid uptake and lipid storage induced by HIF-1α contribute to cell growth and survival after hypoxia-reoxygenation. Cell Rep. 2014;9:349–365. doi: 10.1016/j.celrep.2014.08.056. [DOI] [PubMed] [Google Scholar]
- 53.Chen Y., Wang G., Chen J., Wang C., Dong X., Chang H.M., Yuan S., Zhao Y., Mu L. Genetic and Epigenetic Landscape for Drug Development in Polycystic Ovary Syndrome. Endocr. Rev. 2024;45:437–459. doi: 10.1210/endrev/bnae002. [DOI] [PubMed] [Google Scholar]
- 54.Yu H., Li M., Wen X., Yang J., Liang X., Li X., Bao X., Shu J., Ren X., Chen W., et al. Elevation of α-1,3 fucosylation promotes the binding ability of TNFR1 to TNF-α and contributes to osteoarthritic cartilage destruction and apoptosis. Arthritis Res. Ther. 2022;24:93. doi: 10.1186/s13075-022-02776-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kumar A., Torii T., Ishino Y., Muraoka D., Yoshimura T., Togayachi A., Narimatsu H., Ikenaka K., Hitoshi S. The Lewis X-related α1,3-fucosyltransferase, Fut10, is required for the maintenance of stem cell populations. J. Biol. Chem. 2013;288:28859–28868. doi: 10.1074/jbc.M113.469403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang T., Zhang Z., Qu C., Song W., Li M., Shao X., Fukuda T., Gu J., Taniguchi N., Li W. Core fucosylation regulates the ovarian response via FSH receptor during follicular development. J. Adv. Res. 2024 doi: 10.1016/j.jare.2024.01.025. [DOI] [PubMed] [Google Scholar]
- 57.Zhou Q., Liu Y., Feng R., Zhang W. NUCB2: roles in physiology and pathology. J. Physiol. Biochem. 2022;78:603–617. doi: 10.1007/s13105-022-00895-4. [DOI] [PubMed] [Google Scholar]
- 58.Xu Y., Zhang H., Li Q., Lao K., Wang Y. The role of nesfatin-1 expression in letrozole-induced polycystic ovaries in the rat. Gynecol. Endocrinol. 2017;33:438–441. doi: 10.1080/09513590.2017.1290068. [DOI] [PubMed] [Google Scholar]
- 59.Luan Q., Jin L., Jiang C.C., Tay K.H., Lai F., Liu X.Y., Liu Y.L., Guo S.T., Li C.Y., Yan X.G., et al. RIPK1 regulates survival of human melanoma cells upon endoplasmic reticulum stress through autophagy. Autophagy. 2015;11:975–994. doi: 10.1080/15548627.2015.1049800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li X., Qi J., Zhu Q., He Y., Wang Y., Lu Y., Wu H., Sun Y. The role of androgen in autophagy of granulosa cells from PCOS. Gynecol. Endocrinol. 2019;35:669–672. doi: 10.1080/09513590.2018.1540567. [DOI] [PubMed] [Google Scholar]
- 61.Li D., You Y., Bi F.F., Zhang T.N., Jiao J., Wang T.R., Zhou Y.M., Shen Z.Q., Wang X.X., Yang Q. Autophagy is activated in the ovarian tissue of polycystic ovary syndrome. Reproduction. 2018;155:85–92. doi: 10.1530/rep-17-0499. [DOI] [PubMed] [Google Scholar]
- 62.Zhang K.H., Zhang F.F., Zhang Z.L., Fang K.F., Sun W.X., Kong N., Wu M., Liu H.O., Liu Y., Li Z., et al. Follicle stimulating hormone controls granulosa cell glutamine synthesis to regulate ovulation. Protein Cell. 2024;15:512–529. doi: 10.1093/procel/pwad065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hayes M.G., Urbanek M., Ehrmann D.A., Armstrong L.L., Lee J.Y., Sisk R., Karaderi T., Barber T.M., McCarthy M.I., Franks S., et al. Genome-wide association of polycystic ovary syndrome implicates alterations in gonadotropin secretion in European ancestry populations. Nat. Commun. 2015;6:7502. doi: 10.1038/ncomms8502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Day F.R., Hinds D.A., Tung J.Y., Stolk L., Styrkarsdottir U., Saxena R., Bjonnes A., Broer L., Dunger D.B., Halldorsson B.V., et al. Causal mechanisms and balancing selection inferred from genetic associations with polycystic ovary syndrome. Nat. Commun. 2015;6:8464. doi: 10.1038/ncomms9464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mbarek H., Steinberg S., Nyholt D.R., Gordon S.D., Miller M.B., McRae A.F., Hottenga J.J., Day F.R., Willemsen G., de Geus E.J., et al. Identification of Common Genetic Variants Influencing Spontaneous Dizygotic Twinning and Female Fertility. Am. J. Hum. Genet. 2016;98:898–908. doi: 10.1016/j.ajhg.2016.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Semenza G.L. HIF-1 mediates metabolic responses to intratumoral hypoxia and oncogenic mutations. J. Clin. Invest. 2013;123:3664–3671. doi: 10.1172/jci67230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Feng X., Wang S., Sun Z., Dong H., Yu H., Huang M., Gao X. Ferroptosis Enhanced Diabetic Renal Tubular Injury via HIF-1α/HO-1 Pathway in db/db Mice. Front. Endocrinol. 2021;12 doi: 10.3389/fendo.2021.626390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wu Y., Wang J., Zhao T., Chen J., Kang L., Wei Y., Han L., Shen L., Long C., Wu S., Wei G. Di-(2-ethylhexyl) phthalate exposure leads to ferroptosis via the HIF-1α/HO-1 signaling pathway in mouse testes. J. Hazard Mater. 2022;426 doi: 10.1016/j.jhazmat.2021.127807. [DOI] [PubMed] [Google Scholar]
- 69.Yuan S., Wei C., Liu G., Zhang L., Li J., Li L., Cai S., Fang L. Sorafenib attenuates liver fibrosis by triggering hepatic stellate cell ferroptosis via HIF-1α/SLC7A11 pathway. Cell Prolif. 2022;55 doi: 10.1111/cpr.13158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yang M., Chen P., Liu J., Zhu S., Kroemer G., Klionsky D.J., Lotze M.T., Zeh H.J., Kang R., Tang D. Clockophagy is a novel selective autophagy process favoring ferroptosis. Sci. Adv. 2019;5 doi: 10.1126/sciadv.aaw2238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Dhillon S. Roxadustat: First Global Approval. Drugs. 2019;79:563–572. doi: 10.1007/s40265-019-01077-1. [DOI] [PubMed] [Google Scholar]
- 72.Su X., Xie Y., Zhang J., Li M., Zhang Q., Jin G., Liu F. HIF-α activation by the prolyl hydroxylase inhibitor roxadustat suppresses chemoresistant glioblastoma growth by inducing ferroptosis. Cell Death Dis. 2022;13:861. doi: 10.1038/s41419-022-05304-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhu X., Jiang L., Wei X., Long M., Du Y. Roxadustat: Not just for anemia. Front. Pharmacol. 2022;13 doi: 10.3389/fphar.2022.971795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Shi Y., Zhao H., Shi Y., Cao Y., Yang D., Li Z., Zhang B., Liang X., Li T., Chen J., et al. Genome-wide association study identifies eight new risk loci for polycystic ovary syndrome. Nat. Genet. 2012;44:1020–1025. doi: 10.1038/ng.2384. [DOI] [PubMed] [Google Scholar]
- 75.Chen Z.J., Zhao H., He L., Shi Y., Qin Y., Shi Y., Li Z., You L., Zhao J., Liu J., et al. Genome-wide association study identifies susceptibility loci for polycystic ovary syndrome on chromosome 2p16.3, 2p21 and 9q33.3. Nat. Genet. 2011;43:55–59. doi: 10.1038/ng.732. [DOI] [PubMed] [Google Scholar]
- 76.Xiong Z., Zhao L., Mei Y., Qiu D., Li X., Zhang P., Zhang M., Cao J., Wang Y. Proteome-wide Mendelian randomization identified potential drug targets for migraine. J. Headache Pain. 2024;25:148. doi: 10.1186/s10194-024-01853-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ou G., Wu J., Wang S., Jiang Y., Chen Y., Kong J., Xu H., Deng L., Zhao H., Chen X., Xu L. Dietary Factors and Risk of Gout: A Two-Sample Mendelian Randomization Study. Foods. 2024;13 doi: 10.3390/foods13081269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yang Y., Rao T., Wei S., Cheng J., Zhan Y., Lin T., Chen J., Zhong X., Jiang Y., Yang S. Role of inflammatory cytokines and the gut microbiome in vascular dementia: insights from Mendelian randomization analysis. Front. Microbiol. 2024;15 doi: 10.3389/fmicb.2024.1398618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hu J., Lu J., Lu Q., Weng W., Guan Z., Wang Z. Mendelian randomization and colocalization analyses reveal an association between short sleep duration or morning chronotype and altered leukocyte telomere length. Commun. Biol. 2023;6:1014. doi: 10.1038/s42003-023-05397-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Analyzing codes in this study are available on GitHub (https://github.com/Feidani/PCOS_Drug_targets).