Abstract
Cardiac MRI is used to diagnose and treat patients with a multitude of cardiovascular diseases. Despite the growth of clinical cardiac MRI, complicated image prescriptions and long acquisition protocols limit the specialty and restrain its impact on the practice of medicine. Artificial intelligence (AI)—the ability to mimic human intelligence in learning and performing tasks—will impact nearly all aspects of MRI. Deep learning (DL) primarily uses an artificial neural network to learn a specific task from example data sets. Self-driving scanners are increasingly available, where AI automatically controls cardiac image prescriptions. These scanners offer faster image collection with higher spatial and temporal resolution, eliminating the need for cardiac triggering or breath holding. In the future, fully automated inline image analysis will most likely provide all contour drawings and initial measurements to the reader. Advanced analysis using radiomic or DL features may provide new insights and information not typically extracted in the current analysis workflow. AI may further help integrate these features with clinical, genetic, wearable-device, and “omics” data to improve patient outcomes. This article presents an overview of AI and its application in cardiac MRI, including in image acquisition, reconstruction, and processing, and opportunities for more personalized cardiovascular care through extraction of novel imaging markers.
© RSNA, 2024
Supplemental material is available for this article.
See also the review “How AI May Transform Musculoskeletal Imaging” by Guermazi et al in this issue.
Summary
Artificial intelligence is increasingly used to make cardiac MRI faster and easier, with the potential to improve patient care through learning and with the imager role shifting to one of oversight.
Essentials
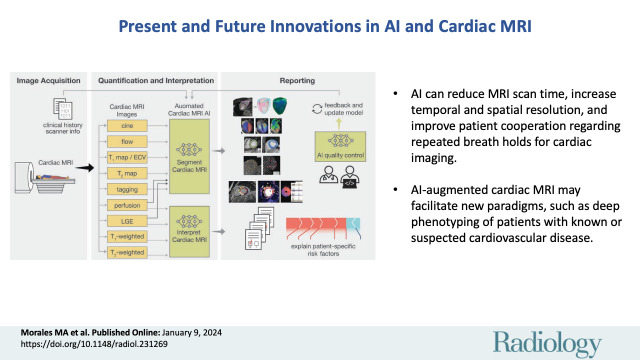
■ Advances in rapid imaging with artificial intelligence (AI) can reduce MRI scan time, increase temporal and spatial resolution, and improve patient cooperation regarding repeated breath holds.
■ Automated AI postprocessing applications are starting to become available, reducing the analysis burden on cardiac imagers and shifting their role from one of drawing regions of interest to quality control and clinical image interpretation.
■ The use of AI in cardiac MRI may provide a new paradigm for gaining insights into images; combined with clinical, genetic, wearable-device, and “omics” data, AI will enable deep phenotyping of patients with known or suspected cardiovascular disease.
Introduction
Cardiac MRI plays a prominent role in managing patients with various cardiovascular diseases (1,2). Artificial intelligence (AI) is transforming the entire field of cardiac imaging and is ubiquitous in cardiac MRI (3) (Fig 1). Of 520 AI medical algorithms cleared by the U.S. Food and Drug Administration as of October 2023, 454 were in radiology and cardiology (4). AI is already integrated into daily cardiac MRI workflow in terms of facilitating image analysis. The use of AI can improve cardiac MRI, including patient selection, protocoling, scanning, accelerated data acquisition and reconstruction, and automated and inline image analysis. It can also improve the integration of cardiac MRI data with other clinical, genetic, “omics” (genomics, transcriptomics, etc), and biomarker data to improve patient management and thus outcomes. AI could also change how image readers extract relevant information from cardiac MRI data by learning from images to automatically extract the most important information instead of requiring human input to select the parameters beforehand. AI techniques based on machine learning (ML) and deep learning (DL) are frequently used in cardiac MRI with great success. Herein, we present an in-depth review of the current and future impact of AI on cardiac MRI.
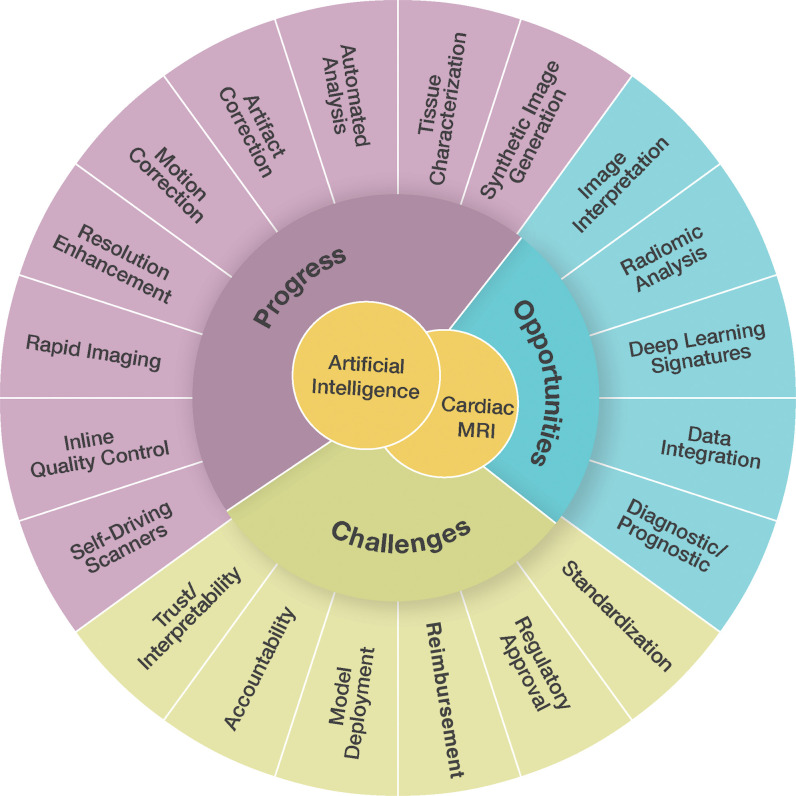
Figure 1:
Diagram shows the progress, opportunities, and current challenges in the use of artificial intelligence in cardiac MRI.
AI Fundamentals
AI models require training on a large data set to “learn” a relationship of interest and then generalize this relationship to new data. The architecture defines the computational steps between inputs and outputs, while the learning strategy and loss function set the criteria used to penalize mistakes during training (Fig 2). In AI, particularly DL, the choice of architecture and learning strategy and the availability of data are among the most critical aspects of developing a successful model. Several architectures have been used in cardiac MRI, including convolutional neural networks (CNNs), recurrent neural networks, U-Net (a type of CNN) (5), generative adversarial networks (6), and transformers (7). Many factors impact the optimal choice of network architecture, including the specific task (eg, reconstruction, segmentation, or prediction), data set size and structure (eg, numerical, static image, time series, or k-space), computational needs, and practical clinical deployment. The strategies to train AI models include supervised, semisupervised, transfer, and ensemble learning strategies (Fig 3). Common successful applications use multiple approaches combined together in various ways. A supervised strategy depends on ground-truth data with well-defined input and output pairs. It is the main strategy if enough unbiased ground-truth data exist. A semisupervised model is a hybrid model based on a desired output that is auxiliary to the main task or only partly known. Transfer learning (8) offers a way to adapt a trained network to new but related data, settings, or tasks. For instance, models can be pretrained with publicly available cardiac MRI data sets, such as those listed in Table S1, with further training based on images relevant for specific applications. Ensemble learning (9) combines the outputs from many models with various designs to improve overall performance. Irrespective of the strategy, AI training involves an iterative optimization of model parameters using a prespecified loss function that directly assesses the model performance for a specific task. Thus, selecting the loss function is important in training robust, high-performance AI models (10).
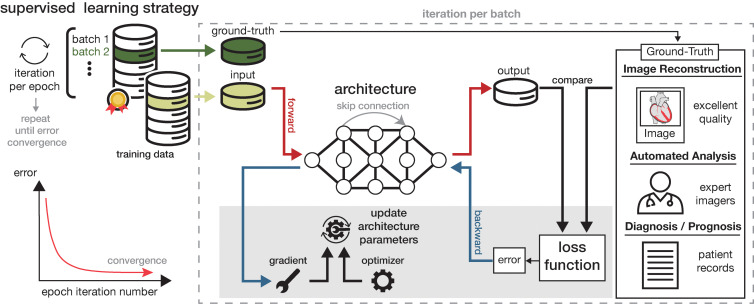
Figure 2:
Diagram of supervised learning strategy for an artificial intelligence model. An architectural layer performs computational operations on data passing through it, with skip connections linking nonsequential layers. A forward pass transforms input into output. The output is compared with an ideal or correct ground truth. The task of the model (eg, image reconstruction, automated analysis, or clinical diagnosis) dictates the form of “ground truth,” which could be high-quality images, expert annotations, or patient records. The loss function quantifies the discrepancy between the output and the ground truth, converting the disagreement into an error signal. The backward pass propagates the error back through the model. This is accomplished by calculating gradients, which represent the direction and rate of change of the error, and selecting an optimizer, which determines how the architecture parameters are updated based on these gradients. This process reduces the error over time. An epoch is one full pass over the training data set, with the architecture trained across many epochs until error convergence is achieved.
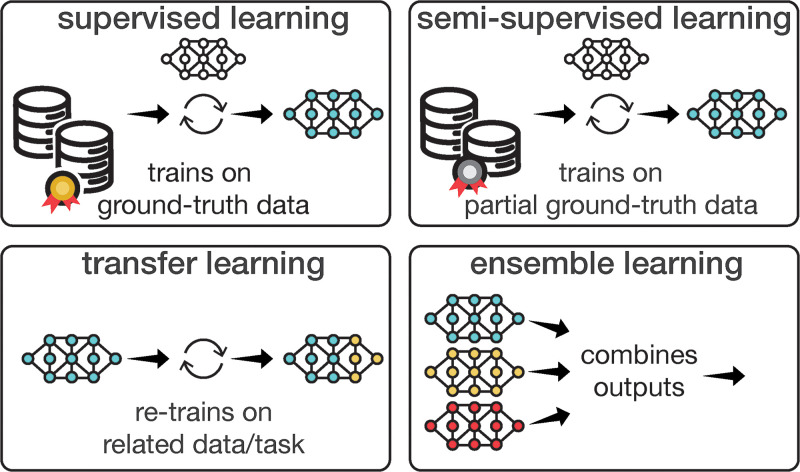
Figure 3:
Diagram of common learning strategies for artificial intelligence models. Supervised learning trains models on input-output relationships using “ground truth” as a reference to guide the learning process. Semisupervised learning, with partial or imperfect ground truth, is effective where only partial supervision is feasible. Transfer learning refines a preexisting model using related data or tasks, saving time and computational resources compared with starting from scratch. Ensemble learning combines outputs from multiple uniquely trained models, reducing error risks and enhancing model robustness for more reliable, accurate predictions.
One of the crucial aspects of building a robust AI model is data availability for development (ie, training and testing) and rigorous validation. Development data sets include all data used to train and optimize the model. The data used for development should represent the setting where the AI model will ultimately be deployed. Independent validation (also called “external testing”) data sets are essential to investigate model performance and robustness. There are different levels of “independence,” involving location, imaging equipment, patient population, and timing. Ideally, the development data set should include a broad spectrum of scenarios that the model will encounter during independent testing. However, this is not always possible, resulting in diminished model performance (11). Additionally, several other factors may impact model performance. For example, expert labeling does not always represent truth, and “experts” from different institutions do not always agree, even when measuring simple cardiac MRI parameters such as myocardial wall thickness (12). These “uncertainties” will impact model performance and robustness. With the limited training data available in cardiac MRI, AI models are more susceptible to race, sex, ethnicity, body size, and referral bias. Furthermore, many current AI models for cardiac MRI segmentation are based on images collected in population health–based studies, which do not represent clinical patient populations, diminishing their clinical robustness. Therefore, when evaluating the generalizability and robustness of an AI model, one should consider the characteristics of the patient and imaging data used for model development, how the model was trained, and its robustness in independent data sets. Guidelines and recommendations will be needed to highlight best practices for model development and evaluation, including data collection and curation documentation.
Applications of AI in Cardiac MRI
Self-driving Scanning
Despite the rapid growth of clinical cardiac MRI over the past 2 decades, its availability remains largely limited to major academic centers. One of the challenges in making this imaging available in smaller health care systems is complicated image prescription. There have been efforts to develop automated or semiautomated image prescription in cardiac MRI (13). However, these approaches need to be more clinically robust. For instance, DL-based prescriptions eliminate manual contouring and do not require the collection of extra images beyond what is already included in the clinical protocol (14). Therefore, a DL framework can automate the entire scanning procedure, with automated section prescription and parameter optimization, shifting the role of the MRI technologists to one of quality control (14). A DL model can calculate imaging parameters automatically to automate the scan further (15) or annotate anatomic landmarks (16). AI-assisted scanning could also improve throughput, yielding shorter scan times, higher consistency, and enhanced image quality (17). Nevertheless, there will always be a need for active supervision and correction strategists, as AI-assisted scanning will be prone to errors. With advances in AI, growth in cardiac MRI, and the economic pressure for improved throughput, scanning will undoubtedly be nearly fully automated in the future. However, how technologists will adapt to such automation for routine clinical scanning remains to be determined.
Image Reconstruction
Cardiac MRI protocols are long, with a 45–60-minute examination time requiring repeated breath holds. Therefore, reducing scan time and breath holding is desirable. AI-based image reconstruction has been proposed to improve imaging efficiency (18). It can be generally classified into four strategies: (a) image reconstruction, (b) image enhancement, (c) denoising, and (d) quantitative myocardial tissue characterization.
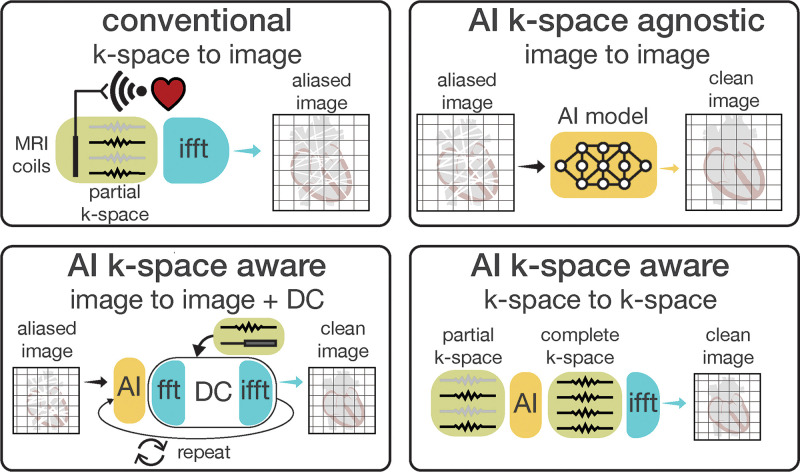
Image reconstruction.—In MRI, k-space data are the raw data obtained from the MRI signal, with “fully sampled” data adhering to the Nyquist theorem for complete image information. “Undersampling” occurs when the data points collected are fewer than what is prescribed with this theorem. Undersampling reduces the signal-to-noise ratio (SNR) and results in “aliasing artifacts,” where the final images display distortions. Using AI-accelerated cardiac MRI techniques can remove such artifacts. These techniques are broadly divided into k-space-agnostic and -aware approaches (Fig 4). In k-space-agnostic models, an image-to-image network, such as a U-Net CNN (19–23), is trained to convert images reconstructed from undersampled k-space data to match images created from fully sampled k-space. Only reconstructed images are used during training for k-space-agnostic models, but in some contexts generating realistic input-output image pairs could require k-space data and access to reconstruction pipelines. K-space-aware models (24–27) include prior knowledge of the acquisition process and k-space consistency terms. For example, traditional regularization functions, which prevent overfitting or enforce prior knowledge, are learned using CNNs. In variational networks (28), iterative steps of a reconstruction algorithm are unrolled and replaced with CNNs, although unrolling is not always used (29). K-space-aware approaches offer some guarantees about the reconstructed images because of the mathematical formulation and the enforced k-space data consistencies. However, a direct head-to-head comparison of different AI-accelerated cardiac MRI techniques is challenging, and it is unlikely that one method will always be superior to the other. Many factors should be considered, including the training set size, k-space sampling trajectory (Fig 5), availability of a fully sampled data set, practical deployment, and reconstruction latency.
Figure 4:
Diagram of accelerated cardiac MRI reconstruction techniques. Accelerated imaging shortens scan time by collecting only partial k-space data instead of a complete set. The traditional reconstruction uses inverse fast Fourier transform (ifft) for k-space-to-image transformation, causing aliasing artifacts in accelerated imaging. K-space-agnostic artificial intelligence (AI) methods transform aliased images (input) into clean, high-quality images (output) without detailed knowledge of the acquisition process. Conversely, k-space-aware AI methods use information of the acquisition process as prior knowledge, integrating image-to-image models with alternating k-space data consistency (DC) steps to better handle incomplete k-space data. Alternatively, AI models can convert partial k-space inputs into a complete set, aiding in generating cleaner, higher-quality images. fft = fast Fourier transform.
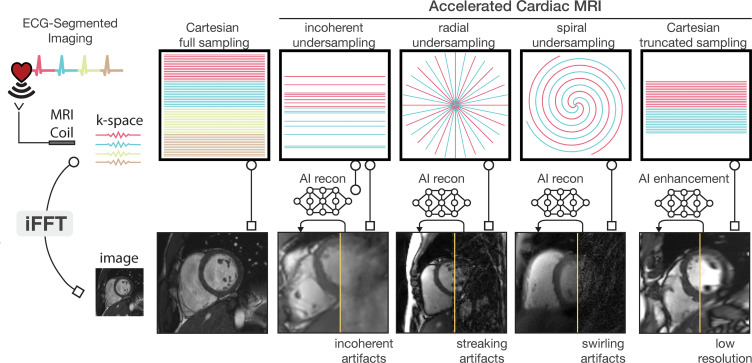
Figure 5:
Diagram of artificial intelligence (AI)–based image reconstruction (recon) and enhancement. The sampling trajectory dictates the k-space data collection path; a Cartesian trajectory is a grid-like path. Reconstruction latency is the delay from data collection to viewing the image. K-space data are collected over multiple heartbeats via electrocardiogram (ECG)–segmented imaging, usually using a fully sampled Cartesian trajectory, which aids in functional imaging and minimizes motion artifacts at the cost of longer scan time. Accelerated imaging, employing undersampled Cartesian, radial, or spiral sampling, reduces scan time by acquiring partial k-space data. However, utilizing a simple inverse fast Fourier transform (iFFT) for frequency (ie, k-space)–to-image domain conversion causes artifacts like incoherence, streaking, and swirling in images. Various AI reconstruction models mitigate these artifacts, improving image clarity and quality. Another method to reduce scan time is truncating the k-space data, which yields lower-resolution images without aliasing artifacts. AI enhancement is used in this context to restore the lost spatial resolution.
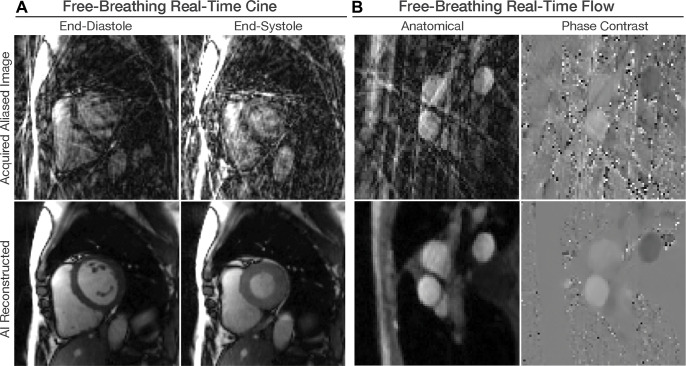
Most published AI literature in rapid cardiac MRI focuses on cine imaging using k-space-agnostic (19,22,23) or k-space-aware approaches (26,27,30). U-Net models (Fig 6A) trained using relatively modest training sets with typical sizes of 250–500 healthy subjects and patients have been widely used (19,23). Images reconstructed with compressed sensing have been used as a reference standard when only undersampled data are available (27). Although this approach can reduce reconstruction time, it limits data quality to that of current algorithms. The k-space-aware models have been trained on smaller cohorts, typically fewer than 100 healthy subjects and patients (26,27). The motion-compensation step may also be recast as a DL approach and combined with unrolled reconstruction networks (31). Numerous recent studies have applied DL to accelerate cardiac MRI. These studies include U-Net models that have also been proposed for rapid MRI angiography (32). AI-accelerated coronary MRI has shown good quality and good diagnostic performance (33), while AI-accelerated two-dimensional or four-dimensional phase-contrast MRI using U-Net (Fig 6B) or unrolled networks (24,25,34) is also available. Despite the need to accelerate myocardial perfusion and late gadolinium enhancement (LGE) image acquisition, few studies have focused on AI acceleration of these sequences (20,21,35,36).
Figure 6:
Artificial intelligence (AI)–based image reconstruction. Free-breathing electrocardiogram-free real-time cardiac MRI scans collected with highly accelerated imaging sequences have substantial aliasing artifacts. Convolutional U-Net models are trained using paired high-quality and aliased images to remove such artifacts. (A) A single U-Net model can remove artifacts from balanced steady-state free precession real-time cine images, as shown in these diastolic and systolic cardiac phase MRI scans in the short-axis plane. (B) Gradient-recalled echo real-time phase-contrast flow imaging acquires velocity-compensated and velocity-encoded images, as shown in these aortic outflow tract plane MRI scans. Two separate U-Net models were used to remove the artifacts while preserving the phase-contrast information.
Image enhancement.—In image resolution enhancement or restoration (also called superresolution techniques), AI models are trained to “recover” the spatial resolution of images acquired with a lower resolution (37,38). Acquiring images with lower resolution reduces scan time since fewer data are acquired. In that case, the acquired k-space data become truncated or cropped relative to higher-resolution images. Synthetic lower-resolution images for training can be generated by retrospectively cropping the raw k-space data before applying the inverse fast Fourier transform. In generative adversarial network–based models for resolution enhancement, the discriminator training can be modified. Specifically, the discriminator estimates the probability that an image based on complete k-space data appears “more realistic” than the resolution-enhanced image produced by the generator. This generative adversarial network approach aims to achieve more than just training the discriminator to differentiate between the two images. Instead, it aims to produce generated images that mimic those from the target data set and possess sharper details and edges, bringing them closer in quality to true high-resolution images. In addition to the adversarial loss function, there is a perceptual loss function for resolution enhancement that compares the features extracted from true high-resolution and resolution-enhanced images. This enables the production of resolution-enhanced images that are perceptually similar to those acquired with higher resolutions. Resolution enhancement models can reduce scan time in various sequences, including cine (37,39,40), coronary (41), and cardiac diffusion (42).
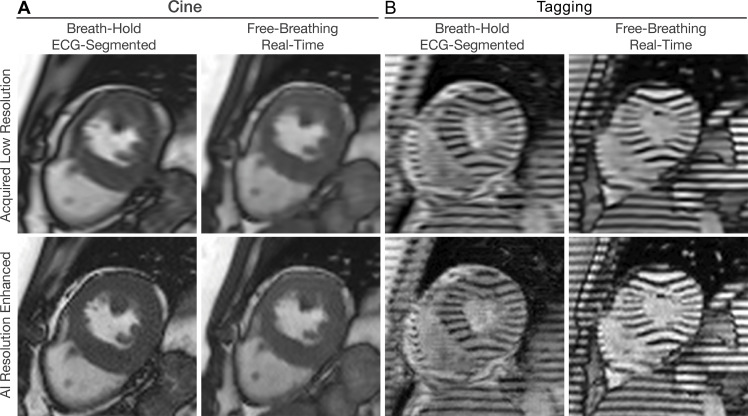
The advantages of resolution enhancement techniques include ease of deployment, flexibility for different sequences, and there being no need for sequence modifications. For instance, a recently proposed inline resolution enhancement technique pretrained with cine data (40) (Fig 7A) can be readily used without any modifications to reconstruct real-time tagging images (Fig 7B) or to reduce the scan time of LGE imaging (Fig 8). However, there are concerns that such models can hallucinate and create realistic-looking images that are not true. A certain threshold of k-space data and spatial resolution is likely essential to guarantee the capture of crucial anatomic details in low-resolution images, where the function of the AI model is primarily to refine the sharpness of existing structures. Thus, rigorous evaluation is necessary before the clinical adoption of these models.
Figure 7:
Artificial intelligence (AI)–based resolution enhancement. Collecting balanced steady-state free precession cardiac MRI scans with lower resolution reduces scan time since fewer data are acquired. Acceleration reduces the breath-hold time needed in breath-hold electrocardiogram (ECG)–gated sequences or enables free-breathing real-time imaging with high temporal resolution. (A) Cine images in the short-axis plane show enhancement of spatial resolution using a generative adversarial network. (B) Tagging images in the short-axis plane show enhancement of spatial resolution using a pretrained model based on cine images.
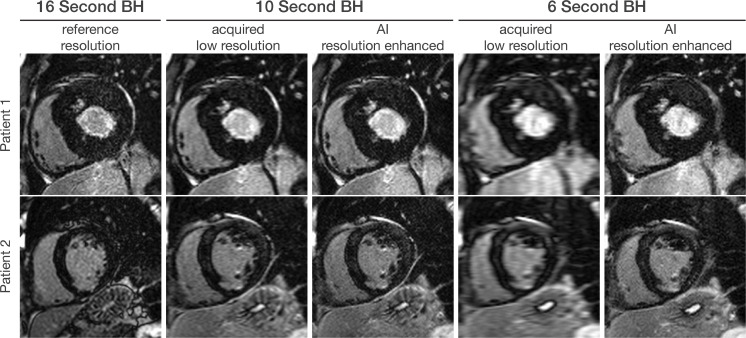
Figure 8:
Artificial intelligence (AI)–based resolution enhancement. A pretrained AI-based resolution enhancement model based on balanced steady-state free precession cardiac MRI cine data can be used to accelerate inversion recovery late gadolinium enhancement imaging, as shown in images in the short-axis plane. For instance, current existing sequences require a 16-second breath hold (BH) for imaging, with one breath hold for each section. The imaging time can be reduced to 10 seconds or even 6 seconds at the expense of diminished spatial resolution. The AI model is used to enhance the spatial resolution in images with shorter imaging time.
Image denoising.—Image denoising in MRI can improve the SNR and is frequently accomplished using image filtering during image reconstruction. However, traditional denoising methods could also result in image blurring. In recent years, there have been substantial improvements in natural image denoising using AI (43), which could be adopted in MRI (44,45). AI denoising could achieve higher denoising capability with reduced image blurring (46). In supervised learning image denoising, a pair of noisy and noise-free images are used to train a DL model, such as a CNN (47) or a generative adversarial network (45). A trained model can generate the denoised image from acquired images. However, in vivo acquisition of noise-free MRI scans is not feasible; therefore, simulated noise is often added to generate synthetic paired images for training. The denoising technique has several applications in cardiac MRI. It can enhance image SNR when images have intrinsically low SNR, such as when they are acquired with a low magnetic field or in low-SNR sequences (eg, LGE). It can also be combined with imaging acceleration to recover the SNR intrinsically lost because of the imaging acceleration. Image denoising could also improve the precision of myocardial tissue characterization techniques. Finally, AI-based denoising algorithms can also reduce the number of signal averages. Signal averaging is a technique commonly used in cardiac diffusion imaging that improves the SNR by running multiple acquisitions (48).
Despite the progress, there are many practical challenges in AI-based denoising of cardiac MRI scans. The current pipeline for data collection using multicoil arrays and nonlinear image reconstruction impacts the noise characteristics of images. Removing spatially varying noise is more complicated than denoising natural images. Furthermore, each cardiac MRI examination generates thousands of images; therefore, the speed of image denoising will be an important factor in the clinical feasibility of AI-based denoising in cardiac MRI. Further developments and rigorous evaluation are still needed to demonstrate the clinical utility of AI-based denoising of cardiac MRI scans.
Quantitative myocardial tissue characterization.—In myocardial tissue characterization sequences (eg, T1 mapping), images with different weighting across multiple heartbeats are collected to estimate the tissue parameters (eg, native T1) using pixel-wise numerical fitting. Therefore, reducing the scan time of each image does not reduce the total scan time, which is governed by the number of collected images and the recovery time needed between images. AI-based models can reduce scan time by estimating quantitative values (eg, T1) from fewer images collected with different contrasts (49,50). A model can then be trained to estimate the tissue parameters using a simple neural network to replace numerical fitting based on the Bloch equation (49). The use of CNNs can exploit spatial information in images to improve the precision of tissue parameters (51). Further refinements, including training using simulated data or scanner information, have improved the robustness of such models (52,53). Cardiac MR fingerprinting–based myocardial tissue characterization, in which images acquired with different weighting are used to estimate different tissue properties simultaneously, can also benefit from using AI to speed up dictionary generation, matching, and image reconstruction (54).
Despite considerable growth in research in AI-based accelerated cardiac MRI techniques, many challenges remain. Irrespective of which architecture or imaging acceleration strategy, the quality of the training data and how they are synthesized will have an immense impact on the performance of imaging acceleration. Researchers must rely on carefully synthesized training data from original single-coil raw k-space data, mimicking rapid imaging. In addition, loss functions and metrics used to quantitatively evaluate the performance of image reconstruction methods, such as mean square error or structural similarity index, do not always reflect diagnostic image quality. AI reconstruction methods that do not consider these issues will provide overly optimistic results that will have little practical value. Therefore, there is always a need for rigorous subjective assessment by clinical imaging readers to evaluate the performance of AI-based cardiac MRI reconstruction algorithms tested in rigorous prospectively collected imaging experiments.
Image Analysis
AI can reduce the analysis burden on imagers at multiple stages (Fig 9). For instance, image segmentation remains one of the most time-consuming and least desired tasks in cardiac MRI. Research has steadily increased in automated AI-based cardiac MRI segmentation tools (55), which has resulted in improved performance compared with traditional non-DL approaches (56). U-Net has been a widely used DL architecture for cardiac MRI segmentation (Fig 10). A model based on U-Net achieved the highest performance in a competition of automated segmentation of short-axis cine images (57). In a similar competition, all proposed models were based on U-Net architectures (58). U-Net models also achieved the highest performance in a recent challenge in segmentation of the right ventricle across multiple anatomic views (59). Indeed, the majority of the models in the challenge were based on a self-configuring method that selected the optimal U-Net configuration based on the specific biomedical image segmentation task (60).
Figure 9:
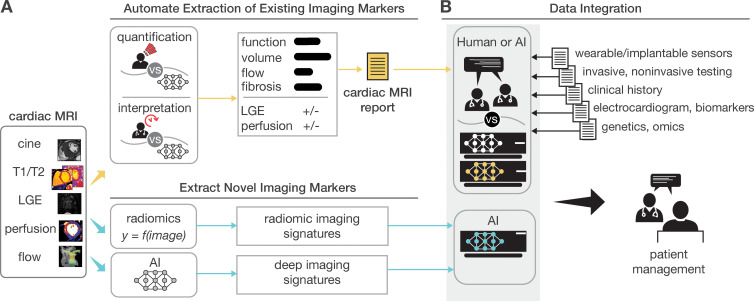
Diagram shows artificial intelligence (AI)–enabled cardiac MRI analysis, interpretation, and reporting. In cardiac MRI, images are collected to characterize function (ie, cine and tagging images), flow, tissue properties and fibrosis (ie, T1 and T2 maps and T1-weighted, T2-weighted, and extracellular volume [ECV] images), and scarring (ie, late gadolinium enhancement [LGE] images). AI can substantially impact the analysis workflow after imaging. The clinical reading includes quantification (eg, function on cine images) and clinical interpretation of individual sequences (eg, presence and location of scarring on LGE images). AI can assist in both the analysis and interpretation of clinical readings and can change the role of imagers to oversight.
Figure 10:
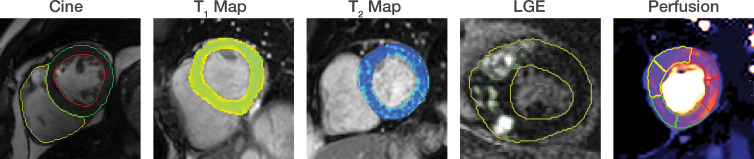
Automated segmentation using convolutional U-Net convolutional neural networks. Automated segmentation based on U-Net architectures has been demonstrated across many cardiac MRI sequences, including cine, T1 and T2 mapping, late gadolinium enhancement (LGE), and perfusion images.
The Sørensen-Dice coefficient is the most used metric in validating medical segmentations (61). However, the accuracy of relevant clinical parameters, such as structural and functional parameters measured on cine images, should also be considered. For instance, automated AI cine segmentation software can provide functional and anatomic quantitative parameters with good correlation with parameters acquired using manual methods. However, in many cases, there can be substantial errors, and the ability of automated measurements to classify functions for treatment decision-making is limited (62). Therefore, the value of AI segmentation algorithms for diagnosis or prognostication should also be considered and arguably is more clinically relevant than segmentation metrics. Indeed, while AI-based segmentation has been relatively successful, there is often a need for manual adjustments and quality control (63), and such human adjustments can improve AI model segmentation performance and robustness (64).
Nearly all vendors provide AI-based solutions for calculating functional and volumetric parameters from cine images. A fully automated AI model outperformed human readers in speed and precision in measuring left ventricular structure and function (65). Automating image analysis for cardiac MRI sequences beyond cine has been more challenging due to lower data availability and lower contrast-to-noise ratio and SNR. Recent studies have investigated automating the processing of four-dimensional flow data (66–68), a very time-consuming process hindering the clinical application of four-dimensional flow MRI. Automated inline AI segmentation of myocardial perfusion images for pixel-wise quantification has been shown to provide measures of myocardial blood flow comparable to manually obtained measures (69). Automated analysis of T1 mapping also provides T1 measures with good correlation with manual analysis (70). Ensemble learning techniques for on-the-fly quality control of T1 mapping can minimize the need for manual adjustments (71). Further, pretrained AI models for T1 mapping along with transfer learning techniques can be leveraged for T2 quantification (72). LGE imaging is a cornerstone of cardiac MRI examinations and is part of most clinical cardiac MRI protocols. However, LGE scar quantification remains challenging, and evaluation remains mostly subjective, limited to assessing presence and location. Automating LGE quantification using DL techniques such as two-dimensional or three-dimensional U-Net is promising; however, such models are currently not commercially available (73–76).
There are many remaining challenges in robust cardiac MRI segmentation, particularly access to large training data sets from diverse patient populations, imaging equipment, and sequences. Integration of automated AI analysis algorithms into commercial analysis software remains challenging, and inline implementation could be limited to academic research centers. In addition, adjustments and quality control of inline segmentation tools are challenging; there is also no standardized format for contours in MRI, resulting in the incompatibility of contours among different analysis systems. These practical but important issues hinder the potential use of AI-based tools in cardiac MRI analysis. Due to the increasing use of cardiac MRI for clinical purposes, automated analysis will be the most desired and impactful application of AI in cardiac MRI in the immediate future.
AI and Precision Cardiovascular Medicine
In cardiac MRI, researchers are moving from imaging to clinical end points impacting patient diagnosis, prognosis, and management. The bar is substantially higher than in other applications to demonstrate “AI value” and demands rigorous study design (77). Despite the successful application of AI in cardiac MRI scan acquisition and analysis, researchers are only in the early phase of developing AI tools that could impact care for an individual patient. AI could impact diagnostic and prognostic accuracy in cardiac MRI in two different ways, through (a) the extraction of new imaging features that image readers do not typically extract or that are not apparent to even expert readers and (b) the ability to combine existing imaging, clinical, genetics, and omics data (Fig 11). In this section, we will review the potential use of AI in diagnosis and prognosis of patients undergoing cardiac MRI.
Figure 11:
The impact of artificial intelligence (AI)–enabled cardiac MRI on the patient care pipeline. The diagram shows how AI impacts information extraction and flow at different steps. (A) AI facilitates and improves the analysis and interpretation of images by extracting standard cardiac MRI parameters. This reduces the analysis burden and improves the accuracy, precision, and reproducibility of analysis and interpretation. AI may also provide a new paradigm for gaining insights into cardiac MRI scans by extracting radiomic or deep imaging signatures of cardiac disease not currently being extracted. (B) AI enables the efficient combination of clinical, imaging, wearable device, biomarker, genetics, and “omics” data to provide clinically actionable information to improve patient care. LGE = late gadolinium enhancement.
In current practice, imaging measurements (eg, functional left ventricular ejection fraction), collected manually or automatically, are selected based on a prior assumption of their clinical relevance. A simple application of AI could be to automate image analysis to extract these imaging-derived parameters. Alternatively, AI could challenge this presumption by providing knowledge and insight that would otherwise not be possible using traditional image interpretation. There are two approaches to extracting novel imaging markers, involving radiomic signatures and DL feature extraction.
Radiomic Signatures
With radiomic signatures (78), one extracts and analyzes many quantitative predefined image features (eg, myocardial texture) with high throughput. The hypothesis is that mining many quantitative features yields useful features with diagnostic or prognostic value not commonly obtained with conventional image analysis. A comprehensive review of radiomic analysis methods is available (79). Texture analysis of nonenhanced cine MRI enables the diagnosis of subacute and chronic myocardial infarction with high accuracy (80). Radiomic analysis of native T1 images can discriminate between hypertrophic and hypertensive cardiomyopathies (81). Radiomic features of LGE images distinguish between myocardial infarction and myocarditis (82). Radiomic features of LGE and cine images, relevant clinical information, and conventional MRI parameters can help predict major adverse cardiac events in patients with ST-segment elevation myocardial infarction (83). Texture analysis of myocardial T1 and T2 maps delivers quantitative imaging parameters for diagnosing acute or chronic heart failure–like myocarditis (84). Radiomic analysis of extracellular volume could help discriminate reversible from irreversible myocardial injury (85). Radiomic analysis of noncontrast MRI scans can also provide hints about the presence of scarring (80,86–88). Radiomic analysis of LGE images also provides prognostic value beyond traditional scar burden assessment (89).
While there is growing interest in applying radiomics in cardiac MRI, the field is in its infancy. There are many challenges regarding appropriate methodology, reproducibility, sensitivity, and generalizability that need addressing before using radiomic analysis in a clinical setting. Radiomic features are calculated using predefined mathematical operations, making the extraction step reproducible. However, the extracted features will vary based on the input images. For example, shape features depend highly on the region of interest where the radiomic features are calculated; therefore, if there is variability in the drawing of the region of interest, this will impact radiomic features. Similarly, imaging filters applied to original images impact the radiomic features. Radiomic features extracted from cardiac MRI scans acquired with different contrast weightings (eg, T1 or T2 weighting) have different sensitivity and reproducibility (90). Furthermore, sequence parameters and processing impact radiomic feature values (91,92).
However, these issues are not fundamentally different from any quantification in cardiac MRI. For example, it is well established that myocardial T1 imaging is sensitive to field strength and sequence. Similarly, radiomic features extracted from different sequences, with different field strengths, or on equipment from different vendors will differ. These limitations should be considered when interpreting radiomic studies. Additional efforts are warranted to develop standardized approaches to implementing radiomic analysis and promoting accurate reporting of methodology and more insightful interpretation.
DL Feature Extraction
Unlike radiomics-based analysis, DL models automatically extract features from input images such that the prediction of an outcome is successfully achieved. DL models for extracting deep imaging features are mainly based on CNNs. The hypothesis is that deep features, extracted after applying a large number of convolutional layers, have incremental diagnostic or prognostic value beyond established clinical and imaging parameters. DL models applied to medical imaging data have been widely used in other diseases and imaging modalities (93). However, fewer studies have explored their potential in cardiac MRI. A DL analysis of cardiac cine motion can predict survival in patients with pulmonary arterial hypertension (94). A DL analysis of LGE images from patients with ischemic heart disease can also predict survival (95). In both studies (94,95), the input and output images of a U-Net CNN were set to be identical. In such approaches, the imaging features are obtained from the halfway “bottleneck” features between the encoder and the decoder, rather than from the output of the decoder. Transfer learning has also been used for diagnosis (96). In one study, a semisupervised model was trained on the auxiliary task of classifying image type, and transfer learning was then applied to fine-tune the model for diagnosing cardiac amyloidosis (96).
Comparing Radiomic and DL Models and Their Real-World Potential
While both radiomic and DL models can extract features from images, to our knowledge a direct head-to-head comparison of these two methods in cardiac MRI has not been conducted. The jury is still out on whether there has been a rigorous demonstration of the potential of radiomic or DL models to extract novel imaging features beyond current metrics. In addition, it is necessary to develop approaches to understand what features contribute to model performance. While there are existing techniques for explaining model behavior (97), such as saliency maps, their practical utility and trustworthiness have recently been questioned (98). One could argue that cardiologists sometimes use medications for which the underlying mechanism of action is not completely understood and that researchers should treat AI models similarly. But AI researchers need to acknowledge that the efficacy of these medications has been investigated in rigorous prospective clinical trials. AI models should likewise be tested in rigorous multicenter prospective clinical trials to demonstrate accuracy and efficacy and thereby gain trust.
Statistical Modeling versus ML to Describe Data
After extracting all relevant clinical and imaging markers from patient data, the next step is to use these rich data sets to answer the relevant clinical question using statistical modeling or ML, each with its strengths and weaknesses. There are ongoing misconceptions and controversies in distinguishing statistical modeling from ML. In statistical modeling, the aim is to derive a mathematical model to describe the data. Statistical models are interpretable and can be readily used to investigate the effect of imaging or clinical variables and their uncertainty on outcomes of interest. With statistical modeling, estimating the effects of predictors on the outcome requires that the number of patients or observations be larger than the number of predictors. In ML, the machine learns the association from the presented data. The goal in ML is to learn the prediction by studying the data pattern, not necessarily to learn the effect of variables on the outcome; ML does not provide information about causality. An ML model typically results in a “black box” system and requires a much larger data set for the training process (99), which focuses on optimizing objective criteria rather than being based on statistical assumptions. ML models can handle high-dimensional data, as applies to cardiac disease involving multimodality images, clinical risk markers, serum biomarkers, genetic data, and omics data. However, one should not always assume that ML models outperform statistical modeling in cardiac care and should consider their strengths and limitations with caution. When dealing with numeric data such as cardiac MRI measurements or clinical variables, there are many advantages to using conventional statistical modeling, including interpretability and generalizability. In recent years, ongoing efforts have focused on improving the interpretability of ML models to enhance their clinical adoption (97,100). These “explainable” models could provide confidence in ML modeling but will not completely overcome its limitations (100).
Conclusion and Future Perspectives
Advanced artificial intelligence (AI) methods will increasingly permeate all aspects of the cardiac MRI workspace, shortening scan acquisition, image reconstruction, and data analysis times. Unique insights from advanced analysis methods will likely lead to paradigm shifts in the diagnosis and management of a large spectrum of cardiovascular diseases. Just as in the case of adaptive cruise control and self-parking features in cars, which take some burdens off drivers, imagers should not fear these advances. Troubleshooting and analysis oversight will continue to be needed, and the knowledge of those trained in the “pre-AI” era will be increasingly valuable. Researchers should also recognize that AI models should learn cause and effect, and that learning only association and prediction will not be sufficient in cardiovascular care. In cardiovascular disease, clinicians will need AI models that enable deep phenotyping and understanding of patients such that the models can provide information about causation and actionable information for individual patients. We are embarking on the era of artificial general intelligence that uses and makes sense of the vast information from cardiac imaging, clinical examination, wearable devices, genetic testing, and omics studies to improve patient care.
Disclosures of conflicts of interest: M.A.M. No relevant relationships. W.J.M. Research agreement with Siemens Healthineers; prior research agreement with Philips Medical Systems; payment for expert testimony, but not in the area related to this article; and participation on a data and safety monitoring board for the Jackson Heart Study. R.N. Funding from the National Institutes of Health; patents issued but unrelated to this review article; and research agreement with Siemens Healthineers.
Abbreviations:
- AI
- artificial intelligence
- CNN
- convolutional neural network
- DL
- deep learning
- LGE
- late gadolinium enhancement
- ML
- machine learning
- SNR
- signal-to-noise ratio
References
- 1. Guo R , Weingärtner S , Šiurytė P , et al . Emerging techniques in cardiac magnetic resonance imaging . J Magn Reson Imaging 2022. ; 55 ( 4 ): 1043 – 1059 . [DOI] [PubMed] [Google Scholar]
- 2. Rajiah PS , François CJ , Leiner T . Cardiac MRI: state of the art . Radiology 2023. ; 307 ( 3 ): e223008 . [DOI] [PubMed] [Google Scholar]
- 3. Leiner T , Rueckert D , Suinesiaputra A , et al . Machine learning in cardiovascular magnetic resonance: basic concepts and applications . J Cardiovasc Magn Reson 2019. ; 21 ( 1 ): 61 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Artificial intelligence and machine learning (AI/ML)-enabled medical devices . U.S. Food and Drug Administration . https://www.fda.gov/medical-devices/software-medical-device-samd/artificial-intelligence-and-machine-learning-aiml-enabled-medical-devices. Updated October 19, 2023. Accessed October 19, 2023 .
- 5. Ronneberger O , Fischer P , Brox T . U-Net: convolutional networks for biomedical image segmentation . In: Navab N , Hornegger J , Wells WM , Frangi AF , eds. Medical image computing and computer-assisted intervention—MICCAI 2015. Part III . Cham: : Springer International Publishing; , 2015. ; 234 – 241 . [Google Scholar]
- 6. Goodfellow I , Pouget-Abadie J , Mirza M , et al . Generative adversarial networks . Commun ACM 2020. ; 63 ( 11 ): 139 – 144 . [Google Scholar]
- 7. Vaswani A , Shazeer N , Parmar N , et al . Attention is all you need . In: Guyon I , Von Luxburg U , Bengio S , et al. , editors. Advances in Neural Information Processing Systems 30 (NIPS 2017) . Red Hook, NY: : Curran Associates; , 2017. . [Google Scholar]
- 8. Yang Q , Zhang Y , Dai W , Pan SJ . Transfer learning . Cambridge: : Cambridge University Press; , 2020. . [Google Scholar]
- 9. Murphy KP . Probabilistic machine learning: an introduction . Cambridge: : MIT Press; , 2022. . [Google Scholar]
- 10. Zhao H , Gallo O , Frosio I , Kautz J . Loss functions for image restoration with neural networks . IEEE Trans Comput Imaging 2016. ; 3 ( 1 ): 47 – 57 . [Google Scholar]
- 11. Yu AC , Mohajer B , Eng J . External validation of deep learning algorithms for radiologic diagnosis: a systematic review . Radiol Artif Intell 2022. ; 4 ( 3 ): e210064 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Captur G , Manisty CH , Raman B , et al . Maximal wall thickness measurement in hypertrophic cardiomyopathy: biomarker variability and its impact on clinical care . JACC Cardiovasc Imaging 2021. ; 14 ( 11 ): 2123 – 2134 . [DOI] [PubMed] [Google Scholar]
- 13. Lelieveldt BP , van der Geest RJ , Lamb HJ , Kayser HW , Reiber JH . Automated observer-independent acquisition of cardiac short-axis MR images: a pilot study . Radiology 2001. ; 221 ( 2 ): 537 – 542 . [DOI] [PubMed] [Google Scholar]
- 14. Blansit K , Retson T , Masutani E , Bahrami N , Hsiao A . Deep learning-based prescription of cardiac MRI planes . Radiol Artif Intell 2019. ; 1 ( 6 ): e180069 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ohta Y , Tateishi E , Morita Y , et al . Optimization of null point in Look-Locker images for myocardial late gadolinium enhancement imaging using deep learning and a smartphone . Eur Radiol 2023. ; 33 ( 7 ): 4688 – 4697 . [DOI] [PubMed] [Google Scholar]
- 16. Xue H , Artico J , Fontana M , Moon JC , Davies RH , Kellman P . Landmark detection in cardiac MRI by using a convolutional neural network . Radiol Artif Intell 2021. ; 3 ( 5 ): e200197 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kwong RY , Jerosch-Herold M , Hainer JM , et al . Artificial intelligence-guided cardiac magnetic resonance imaging as a clinical routine procedure leads to substantial reduction of scan time and improvement of imaging quality. Comparative results of 1,147 patient studies from a single US center . J Am Coll Cardiol 2023. ; 81 ( 8 Supplement ): 1363 . [Google Scholar]
- 18. Chen Y , Schönlieb C-B , Lio P , et al . AI-based reconstruction for fast MRI—a systematic review and meta-analysis . In: Proceedings of the IEEE . 2022. ; 110 ( 2 ): 224 – 245 . [Google Scholar]
- 19. Hauptmann A , Arridge S , Lucka F , Muthurangu V , Steeden JA . Real-time cardiovascular MR with spatio-temporal artifact suppression using deep learning-proof of concept in congenital heart disease . Magn Reson Med 2019. ; 81 ( 2 ): 1143 – 1156 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wang J , Weller DS , Kramer CM , Salerno M . DEep learning-based rapid Spiral Image REconstruction (DESIRE) for high-resolution spiral first-pass myocardial perfusion imaging . NMR Biomed 2022. ; 35 ( 5 ): e4661 . [DOI] [PubMed] [Google Scholar]
- 21. Fan L , Shen D , Haji-Valizadeh H , et al . Rapid dealiasing of undersampled, non-Cartesian cardiac perfusion images using U-net . NMR Biomed 2020. ; 33 ( 5 ): e4239 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Shen D , Ghosh S , Haji-Valizadeh H , et al . Rapid reconstruction of highly undersampled, non-Cartesian real-time cine k-space data using a perceptual complex neural network (PCNN) . NMR Biomed 2021. ; 34 ( 1 ): e4405 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Morales MA , Assana S , Cai X , et al . An inline deep learning based free-breathing ECG-free cine for exercise cardiovascular magnetic resonance . J Cardiovasc Magn Reson 2022. ; 24 ( 1 ): 47 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Oscanoa JA , Middione MJ , Syed AB , Sandino CM , Vasanawala SS , Ennis DB . Accelerated two-dimensional phase-contrast for cardiovascular MRI using deep learning-based reconstruction with complex difference estimation . Magn Reson Med 2023. ; 89 ( 1 ): 356 – 369 . [DOI] [PubMed] [Google Scholar]
- 25. Vishnevskiy V , Walheim J , Kozerke S . Deep variational network for rapid 4D flow MRI reconstruction . Nat Mach Intell 2020. ; 2 ( 4 ): 228 – 235 . [Google Scholar]
- 26. Sandino CM , Lai P , Vasanawala SS , Cheng JY . Accelerating cardiac cine MRI using a deep learning-based ESPIRiT reconstruction . Magn Reson Med 2021. ; 85 ( 1 ): 152 – 167 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. El-Rewaidy H , Fahmy AS , Pashakhanloo F , et al . Multi-domain convolutional neural network (MD-CNN) for radial reconstruction of dynamic cardiac MRI . Magn Reson Med 2021. ; 85 ( 3 ): 1195 – 1208 . [DOI] [PubMed] [Google Scholar]
- 28. Hammernik K , Klatzer T , Kobler E , et al . Learning a variational network for reconstruction of accelerated MRI data . Magn Reson Med 2018. ; 79 ( 6 ): 3055 – 3071 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Qin C , Schlemper J , Caballero J , Price AN , Hajnal JV , Rueckert D . Convolutional recurrent neural networks for dynamic MR image reconstruction . IEEE Trans Med Imaging 2019. ; 38 ( 1 ): 280 – 290 . [DOI] [PubMed] [Google Scholar]
- 30. Küstner T , Fuin N , Hammernik K , et al . CINENet: deep learning-based 3D cardiac CINE MRI reconstruction with multi-coil complex-valued 4D spatio-temporal convolutions . Sci Rep 2020. ; 10 ( 1 ): 13710 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Qi H , Hajhosseiny R , Cruz G , et al . End-to-end deep learning nonrigid motion-corrected reconstruction for highly accelerated free-breathing coronary MRA . Magn Reson Med 2021. ; 86 ( 4 ): 1983 – 1996 . [DOI] [PubMed] [Google Scholar]
- 32. Haji-Valizadeh H , Shen D , Avery RJ , et al . Rapid reconstruction of four-dimensional MR angiography of the thoracic aorta using a convolutional neural network . Radiol Cardiothorac Imaging 2020. ; 2 ( 3 ): e190205 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wu X , Deng L , Li W , et al . Deep learning-based acceleration of compressed sensing for noncontrast-enhanced coronary magnetic resonance angiography in patients with suspected coronary artery disease . J Magn Reson Imaging 2023. ; 58 ( 5 ): 1521 – 1530 . [DOI] [PubMed] [Google Scholar]
- 34. Haji-Valizadeh H , Guo R , Kucukseymen S , et al . Highly accelerated free-breathing real-time phase contrast cardiovascular MRI via complex-difference deep learning . Magn Reson Med 2021. ; 86 ( 2 ): 804 – 819 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. El-Rewaidy H , Neisius U , Mancio J , et al . Deep complex convolutional network for fast reconstruction of 3D late gadolinium enhancement cardiac MRI . NMR Biomed 2020. ; 33 ( 7 ): e4312 . [DOI] [PubMed] [Google Scholar]
- 36. Le J , Tian Y , Mendes J , et al . Deep learning for radial SMS myocardial perfusion reconstruction using the 3D residual booster U-net . Magn Reson Imaging 2021. ; 83 : 178 – 188 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Masutani EM , Bahrami N , Hsiao A . Deep learning single-frame and multiframe super-resolution for cardiac MRI . Radiology 2020. ; 295 ( 3 ): 552 – 561 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Steeden JA , Quail M , Gotschy A , et al . Rapid whole-heart CMR with single volume super-resolution . J Cardiovasc Magn Reson 2020. ; 22 ( 1 ): 56 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Berggren K , Ryd D , Heiberg E , Aletras AH , Hedström E . Super-resolution cine image enhancement for fetal cardiac magnetic resonance imaging . J Magn Reson Imaging 2022. ; 56 ( 1 ): 223 – 231 . [DOI] [PubMed] [Google Scholar]
- 40. Yoon S , Nakamori S , Amyar A , et al . Accelerated cardiac MRI cine with use of resolution enhancement generative adversarial inline neural network . Radiology 2023. ; 307 ( 5 ): e222878 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Küstner T , Munoz C , Psenicny A , et al . Deep-learning based super-resolution for 3D isotropic coronary MR angiography in less than a minute . Magn Reson Med 2021. ; 86 ( 5 ): 2837 – 2852 . [DOI] [PubMed] [Google Scholar]
- 42. Teh I , McClymont D , Carruth E , Omens J , McCulloch A , Schneider JE . Improved compressed sensing and super-resolution of cardiac diffusion MRI with structure-guided total variation . Magn Reson Med 2020. ; 84 ( 4 ): 1868 – 1880 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Tian C , Fei L , Zheng W , Xu Y , Zuo W , Lin CW . Deep learning on image denoising: an overview . Neural Netw 2020. ; 131 : 251 – 275 . [DOI] [PubMed] [Google Scholar]
- 44. Ran M , Hu J , Chen Y , et al . Denoising of 3D magnetic resonance images using a residual encoder-decoder Wasserstein generative adversarial network . Med Image Anal 2019. ; 55 : 165 – 180 . [DOI] [PubMed] [Google Scholar]
- 45. Jiang D , Dou W , Vosters L , Xu X , Sun Y , Tan T . Denoising of 3D magnetic resonance images with multi-channel residual learning of convolutional neural network . Jpn J Radiol 2018. ; 36 ( 9 ): 566 – 574 . [DOI] [PubMed] [Google Scholar]
- 46. Fan L , Zhang F , Fan H , Zhang C . Brief review of image denoising techniques . Vis Comput Ind Biomed Art 2019. ; 2 ( 1 ): 7 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Zhang K , Zuo W , Chen Y , Meng D , Zhang L . Beyond a Gaussian denoiser: residual learning of deep CNN for image denoising . IEEE Trans Image Process 2017. ; 26 ( 7 ): 3142 – 3155 . [DOI] [PubMed] [Google Scholar]
- 48. Phipps K , van de Boomen M , Eder R , et al . Accelerated in vivo cardiac diffusion-tensor MRI using residual deep learning-based denoising in participants with obesity . Radiol Cardiothorac Imaging 2021. ; 3 ( 3 ): e200580 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Guo R , El-Rewaidy H , Assana S , et al . Accelerated cardiac T1 mapping in four heartbeats with inline MyoMapNet: a deep learning-based T1 estimation approach . J Cardiovasc Magn Reson 2022. ; 24 ( 1 ): 6 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Le JV , Mendes JK , McKibben N , et al . Accelerated cardiac T1 mapping with recurrent networks and cyclic, model-based loss . Med Phys 2022. ; 49 ( 11 ): 6986 – 7000 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Amyar A , Guo R , Cai X , et al . Impact of deep learning architectures on accelerated cardiac T1 mapping using MyoMapNet . NMR Biomed 2022. ; 35 ( 11 ): e4794 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Amyar A , Fahmy AS , Guo R , et al . Scanner-independent myoMapNet for accelerated cardiac MRI T1 mapping across vendors and field strengths . J Magn Reson Imaging . 10.1002/jmri.28739. Published online April 13, 2023 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Guo R , Chen Z , Amyar A , et al . Improving accuracy of myocardial T1 estimation in MyoMapNet . Magn Reson Med 2022. ; 88 ( 6 ): 2573 – 2582 . [DOI] [PubMed] [Google Scholar]
- 54. Velasco C , Fletcher TJ , Botnar RM , Prieto C . Artificial intelligence in cardiac magnetic resonance fingerprinting . Front Cardiovasc Med 2022. ; 9 : 1009131 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Litjens G , Ciompi F , Wolterink JM , et al . State-of-the-art deep learning in cardiovascular image analysis . JACC Cardiovasc Imaging 2019. ; 12 ( 8 Pt 1 ): 1549 – 1565 . [DOI] [PubMed] [Google Scholar]
- 56. Galati F , Ourselin S , Zuluaga MA . From accuracy to reliability and robustness in cardiac magnetic resonance image segmentation: a review . Appl Sci (Basel) 2022. ; 12 ( 8 ): 3936 . [Google Scholar]
- 57. Bernard O , Lalande A , Zotti C , et al . Deep learning techniques for automatic MRI cardiac multi-structures segmentation and diagnosis: is the problem solved? IEEE Trans Med Imaging 2018. ; 37 ( 11 ): 2514 – 2525 . [DOI] [PubMed] [Google Scholar]
- 58. Campello VM , Gkontra P , Izquierdo C , et al . Multi-centre, multi-vendor and multi-disease cardiac segmentation: the M&Ms Challenge . IEEE Trans Med Imaging 2021. ; 40 ( 12 ): 3543 – 3554 . [DOI] [PubMed] [Google Scholar]
- 59. Martin-Isla C , Campello VM , Izquierdo C , et al . Deep learning segmentation of the right ventricle in cardiac MRI: the M&Ms challenge . IEEE J Biomed Health Inform 2023. ; 27 ( 7 ): 3302 – 3313 . [DOI] [PubMed] [Google Scholar]
- 60. Isensee F , Jaeger PF , Kohl SAA , Petersen J , Maier-Hein KH . nnU-Net: a self-configuring method for deep learning-based biomedical image segmentation . Nat Methods 2021. ; 18 ( 2 ): 203 – 211 . [DOI] [PubMed] [Google Scholar]
- 61. Taha AA , Hanbury A . Metrics for evaluating 3D medical image segmentation: analysis, selection, and tool . BMC Med Imaging 2015. ; 15 ( 1 ): 29 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Wang S , Patel H , Miller T , et al . AI based CMR assessment of biventricular function: clinical significance of intervendor variability and measurement errors . JACC Cardiovasc Imaging 2022. ; 15 ( 3 ): 413 – 427 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Tao Q , Yan W , Wang Y , et al . Deep learning-based method for fully automatic quantification of left ventricle function from cine MR images: a multivendor, multicenter study . Radiology 2019. ; 290 ( 1 ): 81 – 88 . [DOI] [PubMed] [Google Scholar]
- 64. Alabed S , Alandejani F , Dwivedi K , et al . Validation of artificial intelligence cardiac MRI measurements: relationship to heart catheterization and mortality prediction . Radiology 2022. ; 305 ( 1 ): 68 – 79 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Davies RH , Augusto JB , Bhuva A , et al . Precision measurement of cardiac structure and function in cardiovascular magnetic resonance using machine learning . J Cardiovasc Magn Reson 2022. ; 24 ( 1 ): 16 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Corrado PA , Wentland AL , Starekova J , Dhyani A , Goss KN , Wieben O . Fully automated intracardiac 4D flow MRI post-processing using deep learning for biventricular segmentation . Eur Radiol 2022. ; 32 ( 8 ): 5669 – 5678 . [DOI] [PubMed] [Google Scholar]
- 67. Bustamante M , Viola F , Engvall J , Carlhäll CJ , Ebbers T . Automatic time-resolved cardiovascular segmentation of 4D flow MRI using deep learning . J Magn Reson Imaging 2023. ; 57 ( 1 ): 191 – 203 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. You S , Masutani EM , Alley MT , et al . Deep learning automated background phase error correction for abdominopelvic 4D flow MRI . Radiology 2022. ; 302 ( 3 ): 584 – 592 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Xue H , Davies RH , Brown LAE , et al . Automated inline analysis of myocardial perfusion MRI with deep learning . Radiol Artif Intell 2020. ; 2 ( 6 ): e200009 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Fahmy AS , El-Rewaidy H , Nezafat M , Nakamori S , Nezafat R . Automated analysis of cardiovascular magnetic resonance myocardial native T1 mapping images using fully convolutional neural networks . J Cardiovasc Magn Reson 2019. ; 21 ( 1 ): 7 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Hann E , Popescu IA , Zhang Q , et al . Deep neural network ensemble for on-the-fly quality control-driven segmentation of cardiac MRI T1 mapping . Med Image Anal 2021. ; 71 : 102029 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Zhu Y , Fahmy AS , Duan C , Nakamori S , Nezafat R . Automated myocardial T2 and extracellular volume quantification in cardiac MRI using transfer learning-based myocardium segmentation . Radiol Artif Intell 2020. ; 2 ( 1 ): e190034 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Fahmy AS , Neisius U , Chan RH , et al . Three-dimensional deep convolutional neural networks for automated myocardial scar quantification in hypertrophic cardiomyopathy: a multicenter multivendor study . Radiology 2020. ; 294 ( 1 ): 52 – 60 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Fahmy AS , Rausch J , Neisius U , et al . Automated cardiac MR scar quantification in hypertrophic cardiomyopathy using deep convolutional neural networks . JACC Cardiovasc Imaging 2018. ; 11 ( 12 ): 1917 – 1918 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Zabihollahy F , White JA , Ukwatta E . Convolutional neural network-based approach for segmentation of left ventricle myocardial scar from 3D late gadolinium enhancement MR images . Med Phys 2019. ; 46 ( 4 ): 1740 – 1751 . [DOI] [PubMed] [Google Scholar]
- 76. Fahmy AS , Rowin EJ , Chan RH , Manning WJ , Maron MS , Nezafat R . Improved quantification of myocardium scar in late gadolinium enhancement images: deep learning based image fusion approach . J Magn Reson Imaging 2021. ; 54 ( 1 ): 303 – 312 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Quer G , Arnaout R , Henne M , Arnaout R . Machine learning and the future of cardiovascular care: JACC state-of-the-art review . J Am Coll Cardiol 2021. ; 77 ( 3 ): 300 – 313 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Gillies RJ , Kinahan PE , Hricak H . Radiomics: images are more than pictures, they are data . Radiology 2016. ; 278 ( 2 ): 563 – 577 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Hassani C , Saremi F , Varghese BA , Duddalwar V . Myocardial radiomics in cardiac MRI . AJR Am J Roentgenol 2020. ; 214 ( 3 ): 536 – 545 . [DOI] [PubMed] [Google Scholar]
- 80. Baessler B , Mannil M , Oebel S , Maintz D , Alkadhi H , Manka R . Subacute and chronic left ventricular myocardial scar: accuracy of texture analysis on nonenhanced cine MR images . Radiology 2018. ; 286 ( 1 ): 103 – 112 . [DOI] [PubMed] [Google Scholar]
- 81. Neisius U , El-Rewaidy H , Nakamori S , Rodriguez J , Manning WJ , Nezafat R . Radiomic analysis of myocardial native T1 imaging discriminates between hypertensive heart disease and hypertrophic cardiomyopathy . JACC Cardiovasc Imaging 2019. ; 12 ( 10 ): 1946 – 1954 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Di Noto T , von Spiczak J , Mannil M , et al . Radiomics for distinguishing myocardial infarction from myocarditis at late gadolinium enhancement at MRI: comparison with subjective visual analysis . Radiol Cardiothorac Imaging 2019. ; 1 ( 5 ): e180026 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Durmaz ES , Karabacak M , Ozkara BB , et al . Radiomics-based machine learning models in STEMI: a promising tool for the prediction of major adverse cardiac events . Eur Radiol 2023. ; 33 ( 7 ): 4611 – 4620 . [DOI] [PubMed] [Google Scholar]
- 84. Baessler B , Luecke C , Lurz J , et al . Cardiac MRI and texture analysis of myocardial T1 and T2 maps in myocarditis with acute versus chronic symptoms of heart failure . Radiology 2019. ; 292 ( 3 ): 608 – 617 . [DOI] [PubMed] [Google Scholar]
- 85. Chen BH , An DA , He J , et al . Myocardial extracellular volume fraction radiomics analysis for differentiation of reversible versus irreversible myocardial damage and prediction of left ventricular adverse remodeling after ST-elevation myocardial infarction . Eur Radiol 2021. ; 31 ( 1 ): 504 – 514 . [DOI] [PubMed] [Google Scholar]
- 86. Fahmy AS , Rowin EJ , Arafati A , Al-Otaibi T , Maron MS , Nezafat R . Radiomics and deep learning for myocardial scar screening in hypertrophic cardiomyopathy . J Cardiovasc Magn Reson 2022. ; 24 ( 1 ): 40 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Neisius U , El-Rewaidy H , Kucukseymen S , et al . Texture signatures of native myocardial T1 as novel imaging markers for identification of hypertrophic cardiomyopathy patients without scar . J Magn Reson Imaging 2020. ; 52 ( 3 ): 906 – 919 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Mancio J , Pashakhanloo F , El-Rewaidy H , et al . Machine learning phenotyping of scarred myocardium from cine in hypertrophic cardiomyopathy . Eur Heart J Cardiovasc Imaging 2022. ; 23 ( 4 ): 532 – 542 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Fahmy AS , Rowin EJ , Jaafar N , et al . Radiomics of late gadolinium enhancement reveals prognostic value of myocardial scar heterogeneity in hypertrophic cardiomyopathy . JACC Cardiovasc Imaging . 10.1016/j.jcmg.2023.05.003. Published online June 8, 2023 . [DOI] [PubMed] [Google Scholar]
- 90. Jang J , Ngo LH , Mancio J , et al . Reproducibility of segmentation-based myocardial radiomic features with cardiac MRI . Radiol Cardiothorac Imaging 2020. ; 2 ( 3 ): e190216 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Jang J , El-Rewaidy H , Ngo LH , et al . Sensitivity of myocardial radiomic features to imaging parameters in cardiac MR imaging . J Magn Reson Imaging 2021. ; 54 ( 3 ): 787 – 794 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Wichtmann BD , Harder FN , Weiss K , et al . Influence of image processing on radiomic features from magnetic resonance imaging . Invest Radiol 2023. ; 58 ( 3 ): 199 – 208 . [DOI] [PubMed] [Google Scholar]
- 93. Aggarwal R , Sounderajah V , Martin G , et al . Diagnostic accuracy of deep learning in medical imaging: a systematic review and meta-analysis . NPJ Digit Med 2021. ; 4 ( 1 ): 65 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Bello GA , Dawes TJW , Duan J , et al . Deep learning cardiac motion analysis for human survival prediction . Nat Mach Intell 2019. ; 1 ( 2 ): 95 – 104 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Popescu DM , Shade JK , Lai C , et al . Arrhythmic sudden death survival prediction using deep learning analysis of scarring in the heart . Nat Cardiovasc Res 2022. ; 1 ( 4 ): 334 – 343 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Martini N , Aimo A , Barison A , et al . Deep learning to diagnose cardiac amyloidosis from cardiovascular magnetic resonance . J Cardiovasc Magn Reson 2020. ; 22 ( 1 ): 84 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Reyes M , Meier R , Pereira S , et al . On the interpretability of artificial intelligence in radiology: challenges and opportunities . Radiol Artif Intell 2020. ; 2 ( 3 ): e190043 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Arun N , Gaw N , Singh P , et al . Assessing the trustworthiness of saliency maps for localizing abnormalities in medical imaging . Radiol Artif Intell 2021. ; 3 ( 6 ): e200267 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Bzdok D , Altman N , Krzywinski M . Statistics versus machine learning . Nat Methods 2018. ; 15 ( 4 ): 233 – 234 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Salih A , Boscolo Galazzo I , Gkontra P , et al . Explainable artificial intelligence and cardiac imaging: toward more interpretable models . Circ Cardiovasc Imaging 2023. ; 16 ( 4 ): e014519 . [DOI] [PubMed] [Google Scholar]



![Diagram shows artificial intelligence (AI)–enabled cardiac MRI analysis, interpretation, and reporting. In cardiac MRI, images are collected to characterize function (ie, cine and tagging images), flow, tissue properties and fibrosis (ie, T1 and T2 maps and T1-weighted, T2-weighted, and extracellular volume [ECV] images), and scarring (ie, late gadolinium enhancement [LGE] images). AI can substantially impact the analysis workflow after imaging. The clinical reading includes quantification (eg, function on cine images) and clinical interpretation of individual sequences (eg, presence and location of scarring on LGE images). AI can assist in both the analysis and interpretation of clinical readings and can change the role of imagers to oversight.](https://cdn.ncbi.nlm.nih.gov/pmc/blobs/3fb5/10831479/b2236a2ea533/radiol.231269.fig9.jpg)