Abstract
The recent global outbreak and spread of coronavirus disease (COVID-19) makes it an imperative to develop accurate and efficient diagnostic tools for the disease as medical resources are getting increasingly constrained. Artificial intelligence (AI)-aided tools have exhibited desirable potential; for example, chest computed tomography (CT) has been demonstrated to play a major role in the diagnosis and evaluation of COVID-19. However, developing a CT-based AI diagnostic system for the disease detection has faced considerable challenges, which is mainly due to the lack of adequate manually-delineated samples for training, as well as the requirement of sufficient sensitivity to subtle lesions in the early infection stages. In this study, we developed a dual-branch combination network (DCN) for COVID-19 diagnosis that can simultaneously achieve individual-level classification and lesion segmentation. To focus the classification branch more intensively on the lesion areas, a novel lesion attention module was developed to integrate the intermediate segmentation results. Furthermore, to manage the potential influence of different imaging parameters from individual facilities, a slice probability mapping method was proposed to learn the transformation from slice-level to individual-level classification. We conducted experiments on a large dataset of 1202 subjects from ten institutes in China. The results demonstrated that 1) the proposed DCN attained a classification accuracy of 96.74% on the internal dataset and 92.87% on the external validation dataset, thereby outperforming other models; 2) DCN obtained comparable performance with fewer samples and exhibited higher sensitivity, especially in subtle lesion detection; and 3) DCN provided good interpretability on the loci of infection compared to other deep models due to its classification guided by high-level semantic information. An online CT-based diagnostic platform for COVID-19 derived from our proposed framework is now available.
Keywords: COVID-19, Combined segmentation and classification, Attention, CT image
Graphic Abstract
1. Introduction
There has been a global outbreak and rapid spread of coronavirus disease (COVID-19) since the beginning of 2020. On March 1, 2020, the disease was declared a pandemic by the World Health Organization (WHO) (Roosa et al., 2020; Yan et al., 2020). According to real-time data published by WHO, more than 19 million people had been infected by the disease as at August 8, 2020, and over 716,000 victims had succumbed to it. Undoubtedly, the epidemic has become a severe challenge to the global human population. Therefore, accurate and efficient diagnosis of the disease is an imperative.
The reverse transcription-polymerase chain reaction (RT-PCR) test is regarded as the gold standard for COVID-19 diagnosis, but it is time-consuming and suffers from high false-negative rates (Ai et al., 2020; Chan et al., 2020; Fang et al., 2020). As a supplement, the chest computed tomography (CT) scan is more sensitive and efficient for COVID-19 diagnosis in practice and has been widely applied for early screening of the disease (Ai et al., 2020). Previous studies have shown that lesion size and severity can also be evaluated from chest CT images to facilitate the assessment of disease progression and subsequent treatment (Shi et al., 2020b). Thus, CT has been recognized as a COVID-19 diagnostic criterion in the Chinese “COVID-19 treatment plan (trial version 7)” (Chung et al., 2020; Huang et al., 2020a). However, manual evaluation of CT images typically takes several hours, which is not acceptable for COVID-19 clinical diagnosis given the efficiency demands of numerous suspected and confirmed cases. Therefore, it is critical to develop an AI-aided CT diagnostic system for rapid diagnosis and accurate evaluation of COVID-19 cases.
The past decade has witnessed the emergence of deep learning, which has proven relatively superior in computer vision and pattern recognition (LeCun et al., 2015). Classification models, such as AlexNet (Krizhevsky et al., 2012) and VGGNet (Simonyan and Zisserman, 2014), used a series of cascaded convolutional modules to extract features for image classification. ResNet (He et al., 2016) introduced shortcuts to convolutional neural network (CNN) and mitigated the vanishing gradient problem. DenseNet (Huang et al., 2017) utilized skip connections between every two layers and replaced summation with concatenation operation for easier information flow. In the field of image segmentation, Long et al. used a fully convolutional network to segment images and pioneered the application of deep learning in image segmentation tasks (Long et al., 2015). Several deep segmentation networks, such as DeepLab (Chen et al., 2018), PSPNet (Zhao et al., 2017b), and U-net (Ronneberger et al., 2015), were subsequently proposed and further improved image segmentation performance. Among them, U-net has been widely applied in medical image segmentation because of its simple and easy-to-train structure; hence, we adopted it in this study.
Deep learning methods are also widely used in medical image analysis (Chen et al., 2019; Huang et al., 2020b; Lei et al., 2020; Li et al., 2020b; Litjens et al., 2017; Shen et al., 2017). Recently, deep learning has been utilized in COVID-19 diagnosis and evaluation, and the results have been encouraging (Shi et al., 2020a). Several studies utilized end-to-end classification models for COVID-19 diagnosis. For example, Li et al. proposed a three-dimensional (3D) COVID-19 detection neural network (COVNet) to distinguish COVID-19 from community-acquired pneumonia and achieved an area under curve (AUC) score of 0.96 (Li et al., 2020a). Likewise, a 3D DeCoVNet was proposed for COVID-19 classification and achieved 90.7% sensitivity and 91.1% specificity (Zheng et al., 2020). However, the interpretability of the results was limited, thereby hindering its clinical application. In some other studies, lesion segmentation was accomplished first, and classification was performed based on the segmentation results. For instance, (Jin et al., 2020) proposed a three-stage model with U-net and 3D CNN for the diagnosis and evaluation of COVID-19. The model achieved a dice similarity coefficient (DSC) of 0.754, sensitivity of 97.2%, and specificity of 92.2% (Jin et al., 2020). Chen et al. used a Nested U-net to delineate the lesions and divided the results into quadrants for individual-level prediction (Chen et al., 2020). The accuracies at the slice level and individual level were 98.85% and 95.24%, respectively. Zhang et al. developed an AI system to differentiate COVID-19 from common pneumonia as well as normal controls and achieved a weighted accuracy of 92.49% (Zhang et al., 2020). The problem of this kind of method is that the classification results are highly dependent on the segmentation performance. Thus, useful information may be excluded from the CT images due to inaccurate segmentation, thereby worsening the classification performance.
To date, most of the studies have conducted the classification and segmentation processes separately. In fact, the two tasks can be combined to achieve better performance. Lesions in CT images are decisive in COVID-19 screening, but the lesion size is usually minor in the early stage of the disease and may be neglected by the classification network. However, the intermediate results from the segmentation network may help to focus the classification network more intensively on the lesion foci for accurate diagnosis through an attention mechanism (Fu et al., 2019; Hu et al., 2019; Oktay et al., 2018; Wang et al., 2017; Wang et al., 2018). Moreover, the attention maps can unveil regions that are crucial for classification, thus improving the interpretability of deep learning models and assisting in further assessment by clinicians. Hence, improved performance can be achieved by combining the classification and segmentation tasks.
In this study, we proposed a combined segmentation–classification framework that simultaneously accomplishes COVID-19 diagnosis and the segmentation of lesions based on chest CT images. A U-net-based lung segmentation was first performed to delineate the lung contours. Then, a proposed dual-branch combination network (DCN) was used to perform slice-level segmentation and classification. We proposed a lesion attention (LA) module in DCN to utilize the intermediate results of both segmentation and classification branches to improve the classification performance. Finally, a slice probability mapping strategy and a fully connected network (FCN) were adopted to obtain individual-level results from slice-level results, adapting our method to CT scans with different slice numbers. We compared the performance of DCN to other models and proved its efficacy in image classification. In addition, we found that the proposed method was more sensitive to the classification of images with minor lesions. This is extremely helpful for the early COVID-19 diagnosis as lesions in the early stage are usually subtle and difficult to detect (Macmahon et al., 2017).
More precisely, the contributions of this study are summarized as follows.
-
1.
COVID-19 segmentation and classification are simultaneously achieved using the proposed DCN, and a novel weighted Dice loss is proposed to ensure the trainability of the network.
-
2.
The sensitivity to COVID-19 is significantly improved, especially for subtle lesions.
-
3.
The intermediate attention maps produced by the proposed LA module provides interpretability for the classification.
2. Methods
2.1. Overall framework
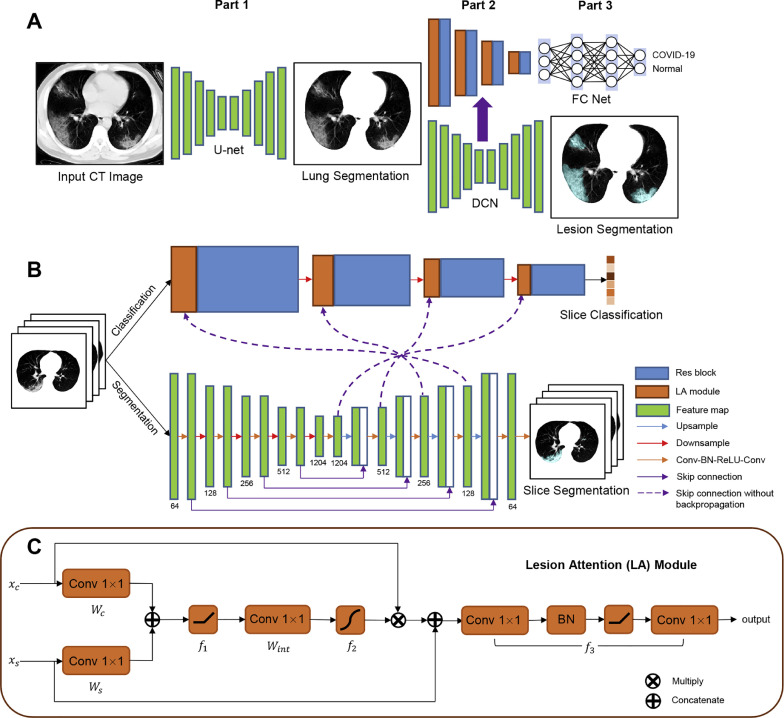
The overall framework of the proposed method (Fig. 1 (A)) can be divided into three parts. Part 1 is a lung segmentation network based on U-net to extract accurate lung regions. Part 2 is the proposed DCN (Fig. 1(B)), which can accomplish simultaneous slice-level classification and segmentation of CT images with the proposed LA module (Fig. 1(C)). In part 3, the slice results are integrated with a slice probability mapping method to obtain the classification results at individual level with a three-layer fully connected network.
Fig. 1.
A: The overall framework of our method, which consists of three parts: 1) lung segmentation using U-net; 2) slice-level combined segmentation and classification using the proposed dual-branch combination network (DCN); and 3) individual-level classification with a three-layer fully connected network. A slice probability mapping strategy is utilized to obtain individual-level results, considering inter-subject differences in slice number. B: Details of the DCN. The segmentation branch is based on U-net, and ResNet-50 with four residual blocks is utilized as the backbone of the classification branch. A lesion attention (LA) module is introduced before each residual block to combine classification features with segmentation features of corresponding scale using an attention mechanism. C: Internal structure of the LA module. f1, f2 represent rectified linear unit (ReLU) and sigmoid activation, respectively, and f3 is a series of convolution, batch normalization, and ReLU operations.
2.2. Lung segmentation
The images require preprocessing to eliminate interference and obtain the region of interest, that is, the lung. Thresholding methods based on Hounsfield unit (HU) values are widely used for chest CT image preprocessing (Iii and Sensakovic, 2004). However, these thresholding methods are not accurate enough in practice, especially for CT images of patients with COVID-19. A possible explanation is that the HU values of the lesions in patients are relatively high, and it is difficult to distinguish them from other organs using thresholding methods; thus, the subsequent analysis is affected. Therefore, we trained a lung segmentation model based on U-net (Ronneberger et al., 2015) to achieve better lung segmentation results. The lung segmentation model has the same architecture as the segmentation branch of DCN, which is described in Section 2.3.1.
2.3. Dual-branch combination network
2.3.1. Model structure
We proposed DCN to accomplish simultaneous classification and segmentation of CT images. The network consists of a classification branch and segmentation branch, corresponding to the classification and segmentation tasks, respectively. The backbone of the classification branch is ResNet-50 (Wang et al., 2017), including four residual blocks. The backbone of the segmentation branch is U-net and comprises an encoder and a decoder. The five blocks of the encoder consist of 64, 128, 256, 512, and 1024 channels respectively. Four 2 × 2 max-pooling layers and four 2 × 2 up-sample layers are used for down-sampling and up-sampling. Each convolution block consists of a 3 × 3 convolution (Conv) layer, a batch normalization (BN) layer (Ioffe and Szegedy, 2015), a rectified linear unit (ReLU) (Nair and Hinton, 2010), and a second 3 × 3 Conv layer. The outputs of the encoding blocks are concatenated with the corresponding decoding blocks using skip connections (Huang et al., 2017). The intermediate results of the two branches are combined with the proposed LA modules. Backpropagation between the two branches is cut off to ensure the trainability of the model. DCN receives the segmented lung images obtained from Section 2.2 as inputs, and outputs the slice-level classification and segmentation results.
2.3.2. Lesion attention module
To better integrate the information of the two branches and improve the classification performance, we proposed the LA module. The inputs of the LA module contain two parts: xc from the classification branch and xs from the segmentation branch. The attention mechanism is utilized to focus the classification branch more on lesions. The formulations of the LA module are as follows:
| (1) |
| (2) |
where is the channel-level concatenation; , , and are weights of 1 × 1 Conv layers; bc, bs, and bint are the corresponding biases; Fc and Fs refer to input channel sizes of the classification and segmentation branches, respectively; and Fint represents the output channel size of the corresponding Conv layers. Functions and correspond to ReLU and sigmoid activation function, respectively. The attention map is then normalized to [0, 1]. The final output of the LA module can be written as:
| (3) |
where f 3 comprises a series of units including two 1 × 1 Conv layers (,), BN, and a ReLU.
2.4. Slice probability mapping
DCN handles the classification of each slice. We then need to incorporate the slice results to achieve individual-level classification and determine whether the subject is infected by COVID-19. However, the slice numbers vary in different subjects owing to the diverse slice thicknesses, fields of view, or volumes of lungs. Some studies utilized max-pooling or average-pooling on fully connected layers to eliminate the effects of this problem (Li et al., 2020a). However, this may lead to loss of information as the approach only saves the max or average signals of all slices. To maximize the information from each slice, we proposed a slice probability mapping strategy based on resampling. Specifically, we sorted the results of slices (that is, the probability of being infected) in descending order and fitted the curve with a bilinear interpolation approach (Li and Orchard, 2001). We then acquired 100 values from the curve in identical intervals and obtained consecutive probabilities in descending order. A simple three-layer FCN was then applied to the classification of individuals with the derived 100 values as input. The numbers of nodes in the two hidden layers are 256 and 128, respectively.
2.5. Loss function
The proposed DCN is a slice-level end-to-end network composed of a classification branch and a segmentation branch. Its loss function also comprises two parts: classification and segmentation losses. Similar to ResNet, we used cross-entropy loss (Zhao et al., 2017a) for the slice-level classification:
| (4) |
where y denotes the true label of the sample, and refers to the predicted label.
The original U-net used binary cross-entropy (BCE) loss (Ronneberger et al., 2015; Zhao et al., 2017a), which performed poorly on our dataset. CT images of patients with COVID-19 are extremely imbalanced data for segmentation because the region of lesions is usually much smaller compared with the normal region and background; and BCE loss is not suitable for this circumstance (Milletari et al., 2016; Sudre et al., 2017).
To deal with this problem, we used Dice loss (Milletari et al., 2016), which is an objective function that directly optimizes the network on the evaluation metric (Dice similarity coefficient (DSC)). The slice-level Dice loss can be written as:
| (5) |
where X is the ground truth; Y is the predicted result; and pi, gi represent the value of the ith pixel of the predicted result and ground truth, respectively. The smooth parameter s was used to prevent division by 0 and was set to 1 in this paper.
Samples from normal subjects are necessary to train the classification branch. However, for the segmentation task, images of normal subjects are all negative samples. This can exacerbate the imbalance of samples, which will affect the training of the segmentation branch. To solve the problem, we proposed a novel weighted Dice loss for the segmentation branch:
| (6) |
| (7) |
where w is the loss weight determined by the label of samples. The weights of slices with/without annotated lesions are set to 1/0, which means only slices with annotated lesions participate in the backpropagation of the segmentation branch. The total loss function can be written as:
| (8) |
where λ is the trade-off parameter for the two losses, and we set in this study experimentally.
We used Dice and BCE losses for the lung segmentation network and FCN, respectively.
3. Experiments and results
3.1. Materials
3.1.1. Subjects
A total of 1918 CT scans from 1202 subjects (704 patients versus 498 controls, 210,395 slices) collected in ten hospitals were enrolled in the study. The data were divided into an internal training set (48 patients versus 75 controls, 6130 slices) from the First Hospital of Yueyang and an external validation set (656 patients versus 423 controls, 204,265 slices) from nine other hospitals. Detailed information of the data source can be found in Table 1 . The internal training set was used for training and testing with a five-fold cross-validation strategy. The external validation set was used to evaluate the generalization performance of the model. All patients were laboratory-confirmed COVID-19 cases by RT-PCR test. The Institutional Review Board of Third Xiangya Hospital approved our study and waived the informed consent of patients based on the retrospective nature of the study. The personal information of the patients was removed in this study.
Table 1.
Data composition and sources.
| COVID-19 |
Normal |
|||||
|---|---|---|---|---|---|---|
| patients | scans | slices | patients | scans | slices | |
| Internal | 48 | 48 | 2371 | 75 | 75 | 3759 |
| External | 656 | 1372 | 166937 | 423 | 423 | 37328 |
| Independent cohorts | ||||||
| Yueyang | 48 | 48 | 2371 | 75 | 75 | 3759 |
| Changsha 1 | 39 | 110 | 8976 | 423 | 423 | 37328 |
| Changsha 2 | 201 | 578 | 46898 | - | - | - |
| Wuhan | 190 | 190 | 12199 | - | - | - |
| Changde | 76 | 133 | 50668 | - | - | - |
| Xiangtan | 39 | 106 | 24270 | - | - | - |
| Shaoyang | 62 | 144 | 8085 | - | - | - |
| Hengyang | 11 | 35 | 11704 | - | - | - |
| Loudi | 32 | 70 | 3966 | - | - | - |
| Yiyang | 6 | 6 | 171 | - | - | - |
3.1.2. Image acquisition and preprocessing
All subjects in the internal training set underwent a thick-section CT scan (Anke ANATOM 16 HD, First Hospital of Yueyang, China). The CT protocol was as follows: tube voltage, 120 kV; automatic tube current, 120 mA–240 mA; iterative reconstruction; 64 mm detector; slice thickness, 5 mm–6 mm; pitch, 1; matrix, 512 × 512; field of view, 360 × 360; and breath-hold at full inspiration. The scan parameters of the external testing set can be found in Table S1.
Dicom files were converted into images using the Pydicom toolkit (Mason, 2011). The pixel values of the images represent HU values within the window of -900 HU–100 HU. They were further normalized into 8-bit grayscale (0–255).
3.1.3. Data annotation
Although the thresholding methods are inaccurate for severely infected lungs, they can still be utilized to reduce the pressure of manual annotation by manual supervision and correction. We first used a threshold-based lung CT preprocessing approach to extract the lung areas (Iii and Sensakovic, 2004). A series of morphological processes, such as dilation and erosion, were then performed to obtain better results. We checked each slice and selected slices with a good shape as the ground truth for lung segmentation. Slices with unsatisfactory results were manually re-delineated.
Furthermore, we asked six experienced radiologists to annotate the CT images of patients with COVID-19 in the internal dataset at pixel level. In the segmentation task, each pixel was annotated as a lesion of COVID-19 or background (labeled as 1 or 0). A total of 2371 slices from patients were annotated manually, and each slice was annotated by one radiologist. We asked three radiologists to annotate the same CT images from part of patients as a comparison between the segmentation performance of DCN and radiologists. For each slice of patients in the classification task, we considered it as a positive sample if lesions were marked by radiologists and set the slice label to 1. Otherwise, we considered the slice as a negative sample and set the label to 0. Slices from healthy controls were labeled as 0. Given the large amount of data in the external dataset and the lack of annotation experts, we did not annotate the external dataset at slice level.
3.2. Parameters and metrics
3.2.1. Training details
All training and testing processes were performed using Pytorch (Steiner et al., 2019) on a server with NVIDIA Tesla P100 GPUs. The lung segmentation, DCN, and FCN models were trained separately. The lung segmentation model was trained in 50 epochs with a batch size of 16. Likewise, DCN, VGGNet, ResNets, and DenseNet were trained in 100 epochs with a batch size of 8. The FCN model was trained in 20 epochs with a batch size of 16. All the models were optimized using Adam optimizer (Kingma and Ba, 2015) with an initial learning rate of 0.001 and a learning decay rate of 0.95 per epoch. Five-fold cross-validation was utilized in the internal training stage. For the external validation stage, the model was pre-trained using all samples of the internal dataset and tested on the external dataset.
To deal with the problem of imbalanced data sizes in the training stage, an under-sampling approach (Buda et al., 2018) was adopted for negative samples. Precisely, all positive samples and an equivalent number of randomly selected negative samples were used for training in an epoch, and negative samples were re-sampled in the next epoch.
3.2.2. Evaluation metrics
In this study, we adopted a commonly used metric, DSC, to evaluate segmentation performance; precision and recall were also calculated at a threshold of 0.5:
| (9) |
| (10) |
| (11) |
where N represents the number of pixels; subscripted T/F means the pixel is correctly/incorrectly predicted; and subscripted P/N refers to whether the pixel is a positive/negative sample.
Accuracy (Acc), sensitivity (Sen), and specificity (Spc) were utilized to evaluate the classification performance. Accuracy is used to describe the performance on the whole dataset, whereas sensitivity and specificity represent the classification results for patients and normal controls, respectively:
| (12) |
| (13) |
| (14) |
where TP, FP, TN, and FN refer to the numbers of true-positive, false-positive, true-negative, and false-negative samples, respectively. The average accuracy (AA) was also introduced to eliminate the interference of data imbalance:
| (15) |
The receiver operating characteristic (ROC) curve and AUC were used to evaluate the network segmentation and classification performances.
3.3. Segmentation results
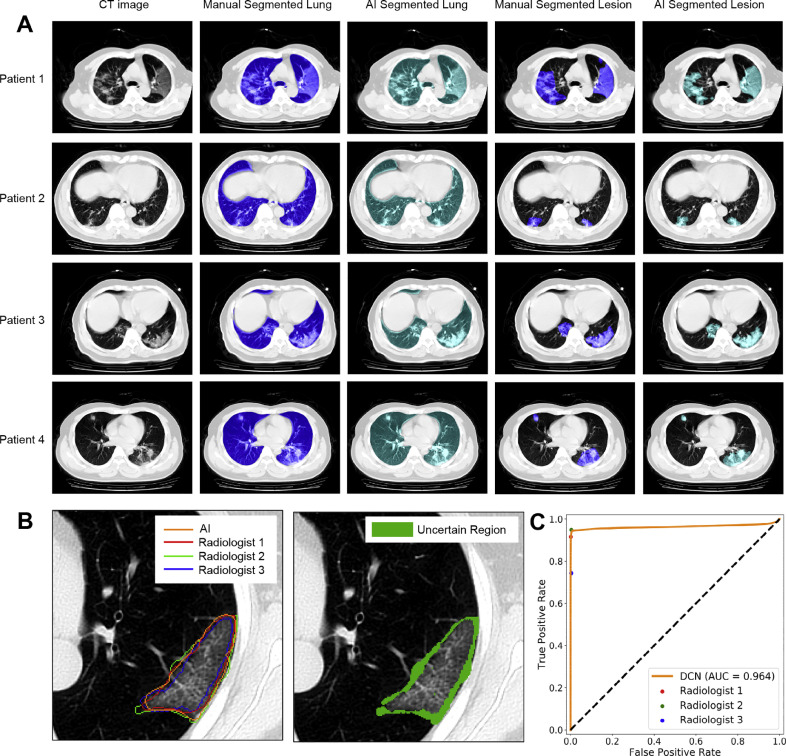
The DSC of lung segmentation was 99.11% (Table 2 ). A comparison between manual annotation and U-net-based lung segmentation is shown in Fig. 2 (A). It can be observed that the segmentation of U-net is highly consistent with the ground truth, which provides a strong guarantee for subsequent analysis.
Table 2.
Segmentation performance of lung and lesion on the internal dataset with five-fold cross-validation.
| DSC | Precision | Recall | |
|---|---|---|---|
| Lung | 99.11% | 99.33% | 98.89% |
| Lesion | 83.51% | 83.46% | 83.55% |
Fig. 2.
A: Manual and AI-based segmentation of lung and lesions in CT images from four patients. B: The left figure shows the segmentation results of three radiologists and the automatic results of DCN. The right figure shows the uncertain region without the consensus of all three radiologists. C: Pixel level ROC curve of DCN and performance of radiologists.
For the segmentation of lesions, we achieved a DSC of 83.51%. The segmentation results are shown in Fig. 2(A). To better evaluate the performance of the proposed segmentation method, a comparison between the proposed DCN and segmentation of three radiologists was performed, and the results are shown in Fig. 2(B) and (C). Annotated lesions without the consensus of all three radiologists are labeled as uncertain regions. A pixel-level ROC curve is shown in Fig. 2(C); our method reached an AUC of 0.964. The results of the three radiologists are also shown in the diagram. The results show that the performance of our method is comparable with an average of three radiologists, which indicates that our segmentation algorithm is comparable to human-level annotation and capable of COVID-19 auxiliary diagnosis.
3.4. Classification results
3.4.1. Internal dataset
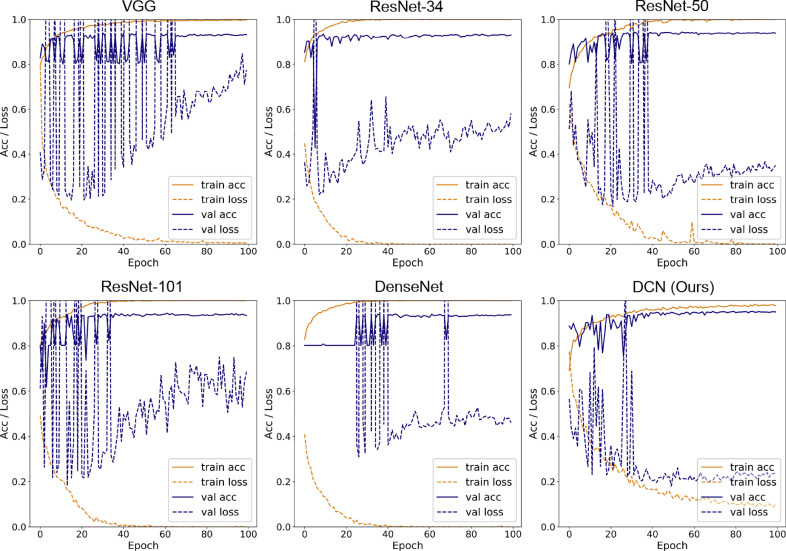
As we used a slice-based strategy, two results would be obtained on both the slice level and individual level. Five other deep learning models (VGG-16, ResNet-34, ResNet50, ResNet101, and DenseNet-121) were also used for comparison with DCN, and the other parts of the framework (lung segmentation and FCN) were kept for fair comparisons. The slice-level training and testing performances of fold 1 are shown in Fig. 3 . We found that the training process of DCN was more stable compared to that of other networks. All the other five models suffered from overfitting according to the significant difference between the training and testing performances. Using our model, the gap between the training and testing stages was smaller, which means DCN is more resistant to overfitting. This is probably due to the extra input in each LA module from the segmentation branch and the benefits from the attention mechanism.
Fig. 3.
Training and testing performance of six models (VGG-16, ResNet-34, ResNet-50, ResNet-101, DenseNet-121, and the proposed DCN) on the internal dataset (fold 1). Solid lines refer to training and testing accuracies, and dashed lines denote corresponding losses.
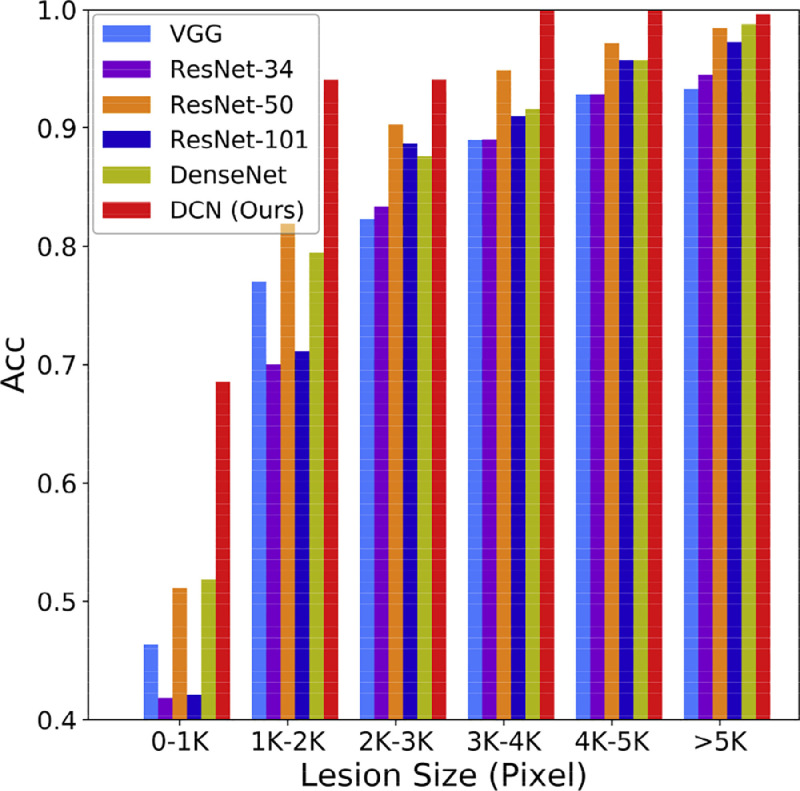
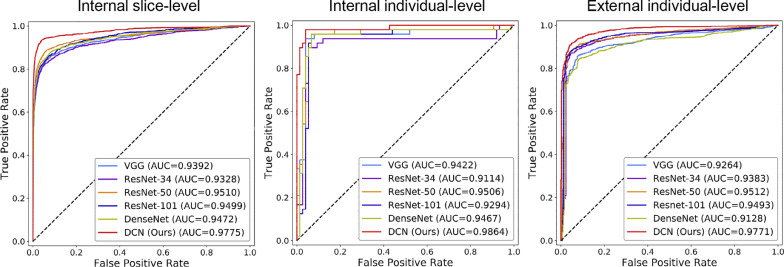
For all patients and healthy controls, we achieved a slice-level accuracy of 95.99% and an individual-level accuracy of 96.74%, which are significantly higher than the results of other models. The ROC curves are shown in Fig. 3. The proposed DCN also achieved the best performance with a slice-level AUC of 0.9755 and an individual-level AUC of 0.9864. The detailed results are presented in Table 3 . We further divided the slices with lesions into six groups (0–1 k, 1–2 k, 2–3 k, 3–4–k, 4–5 k, ≥5 k) according to the number of pixels of the lesion regions and calculated the accuracy of each group. As shown in Fig. 5, the proposed method outperformed other methods in all six groups and significantly improved the classification accuracy of small-lesion slices, which is vital for the early diagnosis of COVID-19.
Table 3.
Classification performance. Slice-level and individual-level results of the internal dataset and individual-level results of the external dataset are illustrated. Slice-level results of the external dataset are unavailable due to the lack of slice-level annotation. Acc, AA, Sen, Spc represent accuracy, average accuracy, sensitivity, and specificity, respectively.
| Slice-level |
Individual-level |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Cohort | Method | Acc (%) | AA (%) | Sen (%) | Spc (%) | AUC | Acc (%) | AA (%) | Sen (%) | Spc (%) | AUC |
| Internal validation | VGG16 | 92.68 | 86.46 | 74.89 | 98.02 | 0.9392 | 93.49 | 93.54 | 93.75 | 93.33 | 0.9422 |
| ResNet-34 | 92.25 | 85.37 | 72.57 | 98.17 | 0.9328 | 91.87 | 92.13 | 89.58 | 94.67 | 0.9114 | |
| ResNet-50 | 93.96 | 89.15 | 80.20 | 98.09 | 0.9510 | 94.31 | 94.58 | 95.83 | 93.33 | 0.9506 | |
| ResNet-101 | 93.05 | 86.57 | 74.51 | 98.62 | 0.9499 | 94.31 | 94.58 | 95.83 | 93.33 | 0.9294 | |
| DenseNet-121 | 93.50 | 88.51 | 79.20 | 97.81 | 0.9472 | 93.49 | 93.54 | 93.75 | 93.33 | 0.9467 | |
| DCN (ours) | 95.99 | 93.59 | 89.14 | 98.04 | 0.9755 | 96.74 | 96.95 | 97.91 | 96.00 | 0.9864 | |
| External validation | VGG16 | - | - | - | - | - | 87.58 | 87.87 | 87.32 | 88.42 | 0.9264 |
| ResNet-34 | - | - | - | - | - | 90.03 | 89.47 | 90.52 | 88.42 | 0.9383 | |
| ResNet-50 | - | - | - | - | - | 90.92 | 90.14 | 91.62 | 88.65 | 0.9512 | |
| ResNet-101 | - | - | - | - | - | 90.58 | 90.74 | 90.45 | 91.02 | 0.9493 | |
| DenseNet-121 | - | - | - | - | - | 86.41 | 85.09 | 87.68 | 82.51 | 0.9128 | |
| DCN (ours) | - | - | - | - | - | 92.87 | 92.89 | 92.86 | 92.91 | 0.9771 | |
Fig. 5.
Classification results on slices of different lesion sizes. The annotated images were divided into 6 groups according to lesion size, and we calculated the sensitivity of each group.
3.4.2. External dataset
Different CT scanning equipment and parameters may cause variations in CT data. To verify the generalization performance of our method, we tested the model on the external dataset from nine different hospitals scanned with different equipment and parameters. The external dataset included 1795 CT scans from 656 patients and 423 normal controls. The slice thickness varied from 0.6 mm to 10 mm. The models were pre-trained on the internal dataset and tested on the external dataset. The proposed DCN achieved 92.87% accuracy, 92.86% sensitivity, and 92.91% specificity at the individual level, which significantly outperformed those of other models. The ROC curves are shown in Fig. 4 , and the proposed method achieved the best AUC of 0.9771.
Fig. 4.
Slice-level and individual-level ROC curves on the internal dataset and individual-level ROC curve on the external dataset.
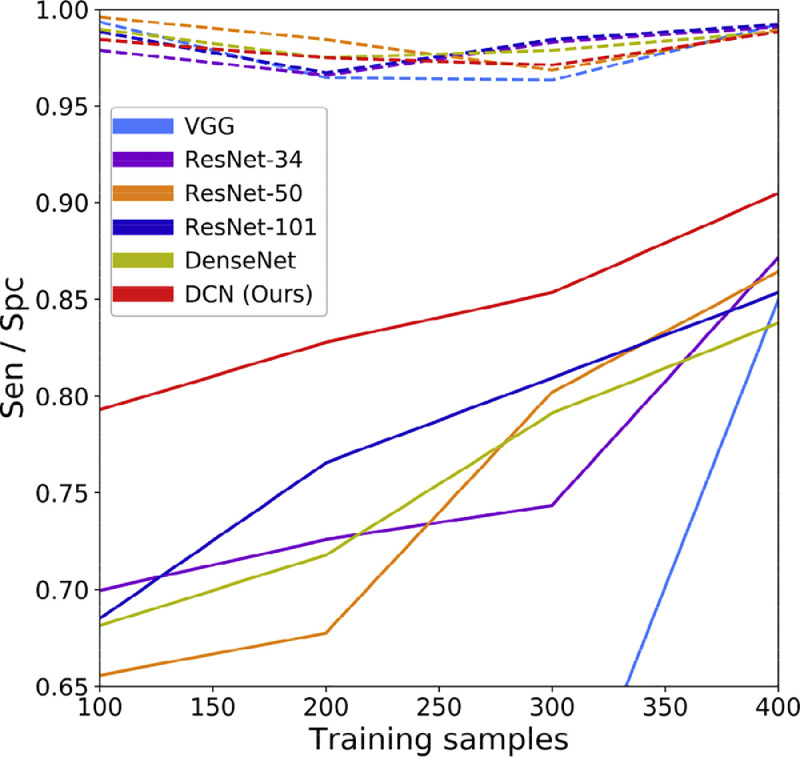
3.4.3. Training with small samples
Training with small samples was also performed to evaluate the generalization performance of the models. The models were trained with different sample sizes and tested on a balanced dataset with 1000 images. The sensitivity (solid lines) and specificity (dashed lines) are shown in Fig. 6 . The specificity of all six models maintained a relatively high level (over 95%), and the increase in the model performance was mainly due to the increase in sensitivity; this means the increment of the training samples enhanced the ability of the networks to detect lesions. The proposed DCN achieved significant progress in sensitivity on the small training samples.
Fig. 6.
Model performance with small-size training datasets. Solid lines refer to sensitivity on slices with lesions while dashed lines represent specificity on normal slices. The models were tested on a selected dataset with 500 positive samples and 500 negative samples.
3.4.4. Comparison to other COVID-19 study
To better evaluate our method, we compared it with other methods designed for COVID-19 classification. COVNet (Li et al., 2020a) and 3D-ResNet (Zhang et al., 2020) were implemented on our dataset, and the results are shown in Table 4 . The results demonstrate the superiority of our method. We also observed the significant drop in performance of COVNet and 3D-ResNet on the external dataset. It is maybe due to data heterogeneity because the external dataset was scanned using different parameters with the internal dataset. In comparison, our DCN has better compatibility with data heterogeneity.
Table 4.
Comparison of DCN with other COVID-19 classification methods. COVNet and 3D-ResNet were tested, and the individual-level results of internal and external datasets were obtained. DCNs with different modules were tested in an ablation study to evaluate the effectiveness of the modules. LA refers to lesion attention; SPM represents slice probability mapping.
| Slice-level |
Individual-level |
External validation |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Method | LA module | SPM module | Acc (%) | AA (%) | Sen (%) | Spc (%) | Acc (%) | AA (%) | Sen (%) | Spc (%) | Acc (%) | AA (%) | Sen (%) | Spc (%) |
| COVNet | - | - | - | - | 88.61 | 87.65 | 83.33 | 92.00 | 77.58 | 79.83 | 75.72 | 83.94 | ||
| 3D-ResNet | - | - | - | - | 92.68 | 91.75 | 87.50 | 96.00 | 77.64 | 81.27 | 74.64 | 87.90 | ||
| DCN(base) | 94.40 | 92.93 | 90.19 | 95.67 | 91.87 | 91.50 | 89.58 | 93.33 | 77.64 | 84.85 | 71.67 | 98.02 | ||
| DCN | ✓ | 94.40 | 92.93 | 90.19 | 95.67 | 94.31 | 94.58 | 95.83 | 93.33 | 89.44 | 90.81 | 88.29 | 93.33 | |
| DCN | ✓ | 95.99 | 93.59 | 89.14 | 98.04 | 94.31 | 94.21 | 94.67 | 93.75 | 87.48 | 89.90 | 85.47 | 94.32 | |
| DCN | ✓ | ✓ | 95.99 | 95.59 | 89.14 | 98.04 | 96.74 | 96.95 | 97.91 | 96.00 | 92.87 | 92.89 | 92.86 | 92.91 |
Moreover, we conducted an ablation study on DCN to measure the effects of the LA module and slice probability mapping. In the base model of DCN, the LA module was replaced with a 1 × 1 Conv layer, and the slice probability mapping was replaced with the max-pooling of the features derived from the last residual block. We observed that the LA module significantly improved the slice-level classification accuracy, which emphasizes the effectiveness of the attention mechanism for COVID-19 classification. Moreover, the slice probability mapping improved the individual-level accuracy, especially for the external dataset, which proved that slice probability mapping improved the generalization of the model.
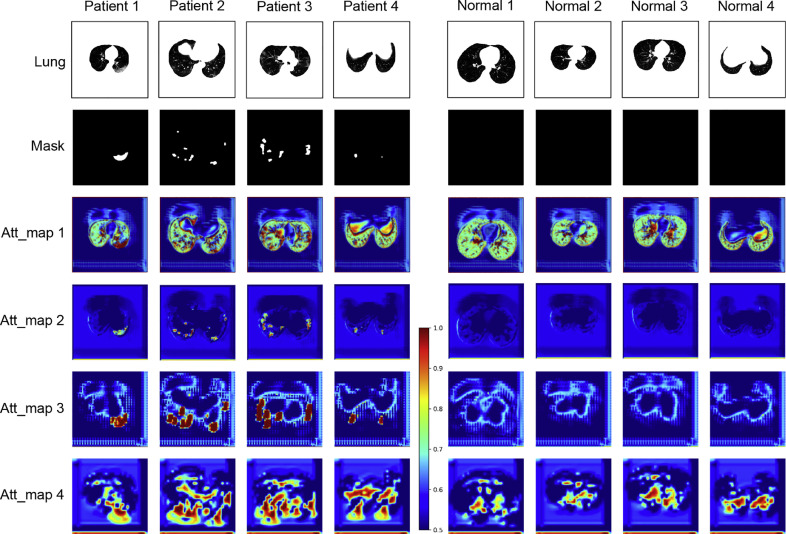
3.4.5. Attention maps
In further analyzing the proposed DCN, the attention maps derived from the testing stage are shown in Fig. 7 , including four patients and four controls. The images of the six rows represent original testing images, lesion masks, and attention maps from four LA modules, respectively. It can be observed that the consistency between lesion masks and attention maps is very high, especially for LA modules 2 and 3. In other words, the module enables the network to focus on areas with lesions. The attention maps reveal the emphasized areas for classification and promote interpretability for the classification results. We also found that some activated areas in the first and last attention maps were inconsistent with the lesion masks. This is probably because the classification input of the first LA module and the segmentation input of the last LA module come from the shallow layer of the network and contain more shallow semantic information. Quantitative analysis was also performed by calculating the DSC of the generated masks. We resized the generated masks into the same size of input images and calculated the DSC of the resized masks. DSCs of 0.28, 0.56, 0.46, and 0.17 were achieved for four LA modules, respectively, which are consistent with the analysis above.
Fig. 7.
Attention maps of internal dataset produced by LA modules. The first row shows the lung segmentation images, and the second row shows the manually annotated lesion masks. Att_maps 1-4 refer to attention maps of LA modules 1-4, respectively.
3.5. Online platform
Based on the high accuracy of the proposed method, we built a cloud platform for COVID-19 auxiliary diagnosis and lesion segmentation (http://218.77.58.164:8808/index). The system can process data in batches and provide feedback on the risks of being infected and possible lesion regions in a few seconds. The platform provides COVID-19 diagnostic and segmentation assistance to doctors and others worldwide, thereby relieving their burden and providing support for the global fight against the COVID-19 epidemic.
4. Discussion
CT imaging has proven to be an effective tool for the diagnosis and quantification of COVID-19, but the image reading is time-consuming. AI-based auxiliary diagnoses of CT scans are crucial for the early screening of COVID-19. In this study, we proposed a combined segmentation–classification framework for the segmentation of lesions and diagnosis of COVID-19 based on chest CT images. The method achieved an accuracy of 96.74% and AUC of 0.9864 on the internal dataset with five-fold cross-validation. The generalization performance of the proposed method was confirmed on a large multi-site external dataset with an accuracy of 92.87%. The experiments demonstrated that DCN outperformed five other commonly used classification models on both internal and external datasets. Furthermore, we compared DCN with two other COVID-19 classification methods, and DCN achieved superior performance. This is probably because we trained the models on a relatively small dataset, and our slice-based method is easier to be trained than the individual-based methods that require more training data. The proposed DCN achieved a lung segmentation DSC of 99.11% and a lesion segmentation DSC of 83.51%. Although it is difficult to compare DCN with other COVID-19 segmentation methods due to their different datasets and annotation quality, we compared our results with segmentation results of radiologists and demonstrated the reliability of our lesion segmentation results.
An LA module was proposed to fuse the intermediate results of the segmentation and classification branches for better performance. The LA module was inspired by the attention mechanism (Fu et al., 2019; Oktay et al., 2018). The intermediate results from two branches were concatenated and produced the attention maps for image classification. The classification branch could then concentrate more on the infected loci. The ablation study in Section 3.4.4 demonstrated the effectiveness of the LA module as it improved the accuracy significantly (1.59% for slice level, 2.43% for individual level, and 3.43% for external validation). The high degree of consistency between the manually annotated and attention masks (Fig. 7) verified the effectiveness of the LA module. Based on accurate attention maps of LA modules 2 and 3, our method can provide good interpretations of the classification results.
Another advantage of our method is its sensitivity in processing images with small lesions. As shown in Fig. 5, the proposed DCN achieved an average promotion of over 20% for images with lesion sizes of less than 1000 pixels, compared with other models. This is mainly due to the attention mechanism provided by the proposed LA module, which allows the network to focus on the infected loci. Considering that lesions are subtle at the early stage of COVID-19, our method is highly applicable to early screening of the disease. Moreover, DCN also achieved significant progress in the case of small training samples, especially for sensitivity. Thus, DCN would prove invaluable in the absence of sufficient samples, such as in the early stage of the COVID-19 epidemic or other similar situations.
We proposed some other techniques in this study to ensure the efficacy of our model. A weighted Dice loss function was proposed to handle the different requirements of the training data and different optimization goals between the classification and segmentation branches. The loss function also facilitates the training of the segmentation branch by reducing the sample imbalance. The difference in slice numbers caused by the diversity of scanning machines and parameters raised another technical challenge for the slice-based methods. Hence, to utilize the information in every slice, we proposed a slice probability mapping strategy, with which we can derive features with the same dimensions in each scan case for subsequent calculations. The slice probability mapping enables the analyses of scans with different slice numbers, thereby facilitating the implementation of our method on diverse datasets. Moreover, the results of ablation study, especially the results on external dataset, has proved the effectiveness of the slice probability mapping.
The proposed DCN has several limitations. First, the precision of the attention masks partly depends on the accuracy of the segmentation branch. The segmentation branch learns from manual annotation (in which quality is not guaranteed), and inconsistencies between different radiologists may introduce biases. Semi-supervised or unsupervised methods may provide new perspectives for resolving this problem. Second, due to the large data size, the external dataset was not labeled at slice level. Hence, we could not analyze the slice-level performance at the external validation stage. Human-in-the-loop methods may be useful for further analysis.
5. Conclusion
The proposed combined segmentation–classification network for the diagnosis of COVID-19 outperformed commonly used classification models on both internal and external validation datasets. Further, the proposed LA module enables the network to focus on infected loci and significantly improves the detection of small lesions for early screening of COVID-19. Moreover, the attention maps aid the identification of lesion loci, thereby improving the interpretation of classification.
In the future, we will continue to improve the network performance and extend DCN to a wider range of applications such as lung nodule classification and tumor detection.
CRediT authorship contribution statement
Kai Gao: Methodology, Software, Writing - original draft. Jianpo Su: Software, Visualization, Writing - review & editing. Zhongbiao Jiang: Investigation, Resources, Data curation. Ling-Li Zeng: Supervision, Validation. Zhichao Feng: Investigation, Writing - review & editing. Hui Shen: Conceptualization, Methodology, Project administration. Pengfei Rong: Conceptualization, Data curation, Project administration. Xin Xu: Supervision. Jian Qin: Methodology. Yuexiang Yang: Software, Resources. Wei Wang: Resources, Data curation. Dewen Hu: Conceptualization, Funding acquisition.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This work was supported by the National Key Research and Development Program (No. 2018YFB1305101) and the National Natural Science Foundation of China (NSFC 61773391, 31773319, 61722313, 62036013).
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.media.2020.101836.
Appendix. Supplementary materials
References
- Ai T., Yang Z., Hou H., Zhan C., Chen C., Lv W., Tao Q., Sun Z., Xia L. Correlation of chest CT and RT-PCR testing in coronavirus disease 2019 (COVID-19) in China: a report of 1014 cases. Radiology. 2020 doi: 10.1148/radiol.2020200642. 200642-200642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buda M., Maki A., Mazurowski M.A. A systematic study of the class imbalance problem in convolutional neural networks. Neural Netw. 2018;106:249–259. doi: 10.1016/j.neunet.2018.07.011. [DOI] [PubMed] [Google Scholar]
- Chan J.F.W., Yuan S., Kok K., To K.K.W., Chu H., Yang J., Xing F., Liu J., Yip C.C., Poon R.W.S. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: a study of a family cluster. Lancet North Am. Ed. 2020;395:514–523. doi: 10.1016/S0140-6736(20)30154-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Wu L., Zhang J., Zhang L., Gong D., Zhao Y., Hu S., Wang Y., Hu X., Zheng B. Deep learning-based model for detecting 2019 novel coronavirus pneumonia on high-resolution computed tomography: a prospective study. MedRxiv. 2020 doi: 10.1101/2020.02.25.20021568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Papandreou G., Kokkinos I., Murphy K., Yuille A.L. DeepLab: semantic image segmentation with deep convolutional nets, atrous convolution, and fully connected CRFs. IEEE Trans. Pattern Anal. Mach. Intell. 2018;40:834–848. doi: 10.1109/TPAMI.2017.2699184. [DOI] [PubMed] [Google Scholar]
- Chen M., Fang L., Zhuang Q., Liu H. Deep learning assessment of myocardial infarction from MR image sequences. IEEE Access. 2019;7:5438–5446. doi: 10.1088/1361-6560/ab3103. [DOI] [Google Scholar]
- Chung M., Bernheim A., Mei X., Zhang N., Huang M., Zeng X., Cui J., Xu W., Yang Y., Fayad Z.A. CT imaging features of 2019 novel coronavirus (2019-nCoV) Radiology. 2020;295:202–207. doi: 10.1148/radiol.2020200230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang Y., Zhang H., Xie J., Lin M., Ying L., Pang P., Ji W. Sensitivity of chest CT for COVID-19: comparison to RT-PCR. Radiology. 2020 doi: 10.1148/radiol.2020200432. 200432–200432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu J., Liu J., Tian H., Li Y., Bao Y., Fang Z., Lu H. Proceedings of the IEEE International Conference on Computer Vision and Pattern Recognition (CVPR) 2019. Dual attention network for scene segmentation; pp. 3146–3154. [DOI] [Google Scholar]
- He K., Zhang X., Ren S., Sun J. Proceedings of the IEEE International Conference on Computer Vision and Pattern Recognition (CVPR) 2016. Deep residual learning for image recognition; pp. 770–778. [DOI] [Google Scholar]
- Hu J., Shen L., Albanie S., Sun G., Wu E. Squeeze-and-Excitation Networks. IEEE Trans. Pattern Anal. Mach. Intell. 2019 doi: 10.1109/TPAMI.2019.2913372. 1–1. [DOI] [PubMed] [Google Scholar]
- Huang C., Wang Y., Li X., Ren L., Cao B. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang G., Liu Z., Der Maaten L.V., Weinberger K.Q. Proceedings of the IEEE International Conference on Computer Vision and Pattern Recognition (CVPR) 2017. Densely connected convolutional networks; pp. 2261–2269. [DOI] [Google Scholar]
- Huang W., Yan H., Wang C., Li J., Chen H. Perception-to-Image: reconstructing natural images from the brain activity of visual perception. Ann. Biomed. Eng. 2020 doi: 10.1007/s10439-020-02502-3. [DOI] [PubMed] [Google Scholar]
- Iii S.G.A., Sensakovic W.F. Automated lung segmentation for thoracic CT: impact on computer-aided diagnosis1. Acad. Radiol. 2004;11:1011–1021. doi: 10.1016/j.acra.2004.06.005. [DOI] [PubMed] [Google Scholar]
- Ioffe S., Szegedy C. International Conference on Machine Learning. 2015. Batch Normalization: accelerating deep network training by reducing internal covariate shift; pp. 448–456. [Google Scholar]
- Jin S., Wang b., Xu H., Luo C., Wei L., Zhao W., Hou X. AI-assisted CT imaging analysis for COVID-19 screening: Building and deploying a medical AI system in four weeks. MedRxiv. 2020 doi: 10.1101/2020.03.19.20039354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingma D.P., Ba J. International Conference on Learning Representations (ICLR) 2015. Adam: a method for stochastic optimization. [Google Scholar]
- Krizhevsky A., Sutskever I., Hinton G. ImageNet classification with deep convolutional neural networks. Adv. Neural Inf. Process. Syst. 2012;25:1097–1105. [Google Scholar]
- LeCun Y., Bengio Y., Hinton G. Deep learning. Nature. 2015;521:436–444. doi: 10.1038/nature14539. [DOI] [PubMed] [Google Scholar]
- Lei Y., Tian Y., Shan H., Zhang J., Wang G., Kalra M.K. Shape and margin-aware lung nodule classification in low-dose CT images via soft activation mapping. Med. Image Anal. 2020;60 doi: 10.1016/j.media.2019.101628. [DOI] [PubMed] [Google Scholar]
- Li L., Qin L., Xu Z., Yin Y., Wang X., Kong B., Bai J., Lu Y., Fang Z., Song Q. Artificial intelligence distinguishes COVID-19 from community acquired pneumonia on chest CT. Radiology. 2020 doi: 10.1148/radiol.2020200905. 200905–200905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Wu F., Yang G., Xu L., Wong T., Mohiaddin R.H., Firmin D.N., Keegan J., Zhuang X. Atrial scar quantification via multi-scale CNN in the graph-cuts framework. Med. Image Anal. 2020;60 doi: 10.1016/j.media.2019.101595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Orchard M.T. New edge-directed interpolation. IEEE Trans. Image Process. 2001;10:1521–1527. doi: 10.1109/83.951537. [DOI] [PubMed] [Google Scholar]
- Litjens G., Kooi T., Bejnordi B.E., Setio A.A.A., Ciompi F., Ghafoorian M., van der Laak J.A.W.M., van Ginneken B., Sanchez C.I. A survey on deep learning in medical image analysis. Med. Image Anal. 2017;42:60–88. doi: 10.1016/j.media.2017.07.005. [DOI] [PubMed] [Google Scholar]
- Long J., Shelhamer E., Darrell T. Computer Vision and Pattern Recognition. 2015. Fully convolutional networks for semantic segmentation; pp. 3431–3440. [DOI] [PubMed] [Google Scholar]
- Macmahon H., Naidich D.P., Goo J.M., Lee K.S., Leung A.N., Mayo J.R., Mehta A.C., Ohno Y., Powell C.A., Prokop M. Guidelines for management of incidental pulmonary nodules detected on CT images: from the Fleischner Society 2017. Radiology. 2017;284:228–243. doi: 10.1148/radiol.2017161659. [DOI] [PubMed] [Google Scholar]
- Mason D. SU‐E‐T‐33: Pydicom: an open source DICOM library. Med. Phys. 2011;38 doi: 10.1118/1.3611983. 3493–3493. [DOI] [Google Scholar]
- Milletari F., Navab N., Ahmadi S. International Conference on 3d Vision. 2016. V-Net: fully convolutional neural networks for volumetric medical image segmentation; pp. 565–571. [DOI] [Google Scholar]
- Nair V., Hinton G.E. Proceedings of the 27th International Conference on Machine Learning (ICML) 2010. Rectified linear units improve restricted boltzmann machines. [Google Scholar]
- Oktay, O., Schlemper, J., Folgoc, L.L., Lee, M.C.H., Heinrich, M.P., Misawa, K., Mori, K., Mcdonagh, S., Hammerla, N., Kainz, B., 2018. Attention U-Net: learning where to look for the pancreas. arXiv preprint arXiv: 1804.03999.
- Ronneberger O., Fischer P., Brox T. Medical Image Computing and Computer Assisted Intervention (MICCAI) 2015. U-Net: convolutional networks for biomedical image segmentation; pp. 234–241. [DOI] [Google Scholar]
- Roosa K., Lee Y., Luo R., Kirpich A., Rothenberg R., Hyman J.M., Yan P., Chowell G. Real-time forecasts of the COVID-19 epidemic in China from February 5th to February 24th, 2020. Infect. Dis. Model. 2020;5:256–263. doi: 10.1016/j.idm.2020.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen D., Wu G., Suk H. Deep learning in medical image analysis. Annu. Rev. Biomed. Eng. 2017;19:221–248. doi: 10.1146/annurev-bioeng-071516-044442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi F., Wang J., Shi J., Wu Z., Wang Q., Tang Z., He K., Shi Y., Shen D. Review of artificial intelligence techniques in imaging data acquisition, segmentation and diagnosis for COVID-19. IEEE Rev. Biomed. Eng. 2020 doi: 10.1109/RBME.2020.2987975. [DOI] [PubMed] [Google Scholar]
- Shi H., Han X., Jiang N., Cao Y., Alwalid O., Gu J., Fan Y., Zheng C. Radiological findings from 81 patients with COVID-19 pneumonia in Wuhan, China: a descriptive study. Lancet Infect. Dis. 2020;20:425–434. doi: 10.1016/S1473-3099(20)30086-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonyan, K., Zisserman, A., 2014. Very deep convolutional networks for large-scale image recognition. In: arXiv perprint arXiv: 1409.1556.
- Steiner B., Devito Z., Chintala S., Gross S., Paszke A., Massa F., Lerer A., Chanan G., Lin Z., Yang E. PyTorch: an imperative style, high-performance deep learning library. Adv. Neural Inf. Process. Syst. 2019:8026–8037. [Google Scholar]
- Sudre, C.H., Li, W., Vercauteren, T., Ourselin, S., Cardoso, M.J., 2017. Generalised Dice overlap as a deep learning loss function for highly unbalanced segmentations. arXiv preprint arXiv:1707.03237. 10.1007/978-3-319-67558-9_28 [DOI] [PMC free article] [PubMed]
- Wang F., Jiang M., Qian C., Yang S., Li C., Zhang H., Wang X., Tang X. Proceedings of the IEEE International Conference on Computer Vision and Pattern Recognition (CVPR) 2017. Residual attention network for image classification; pp. 6450–6458. [DOI] [Google Scholar]
- Wang X., Girshick R., Gupta A., He K. Proceedings of the IEEE International Conference on Computer Vision and Pattern Recognition (CVPR) 2018. Non-local neural networks; pp. 7794–7803. [DOI] [Google Scholar]
- Yan L., Zhang H., Xiao Y., Wang M., Sun C., Liang J., Li S., Zhang M., Guo Y., Xiao Y. Prediction of criticality in patients with severe Covid-19 infection using three clinical features: a machine learning-based prognostic model with clinical data in Wuhan. MedRxiv. 2020 doi: 10.1101/2020.02.27.20028027. [DOI] [Google Scholar]
- Zhang K., Liu X., Shen J., Li Z., Sang Y., Wu X., Zha Y., Liang W., Wang C., Wang K., Ye L., Gao M., Zhou Z., Li L., Wang J., Yang Z., Cai H., Xu J., Yang L., Cai W., Xu W., Wu S., Zhang W., Jiang S., Zheng L., Zhang X., Wang L., Lu L., Li J., Yin H., Wang W., Li O., Zhang C., Liang L., Wu T., Deng R., Wei K., Zhou Y., Chen T., Lau J.Y.-N., Fok M., He J., Lin T., Li W., Wang G. Clinically applicable AI system for accurate diagnosis, quantitative measurements, and prognosis of COVID-19 pneumonia using computed tomography. Cell. 2020;181:1423–1433. doi: 10.1016/j.cell.2020.04.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H., Gallo O., Frosio I., Kautz J. Loss functions for image restoration with neural networks. IEEE Trans. Comput. Imaging. 2017;3:47–57. doi: 10.1109/TCI.2016.2644865. [DOI] [Google Scholar]
- Zhao H., Shi J., Qi X., Wang X., Jia J. Proceedings of the IEEE International Conference on Computer Vision and Pattern Recognition (CVPR) 2017. Pyramid scene parsing network; pp. 6230–6239. [DOI] [Google Scholar]
- Zheng C., Deng X., Fu Q., Zhou Q., Feng J., Ma H., Liu W., Wang X. Deep learning-based detection for COVID-19 from chest CT using weak label. MedRxiv. 2020 doi: 10.1101/2020.03.12.20027185. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.