Abstract
Purpose
To explore specific conditions and types of genetic variants that specialists in genetics recommend should be returned as incidental findings in clinical sequencing.
Methods
Sixteen specialists in clinical genetics and/or molecular medicine selected variants in 99 common conditions to return to the ordering physician if discovered incidentally through whole genome sequencing. For most conditions, the specialists independently considered 3 molecular scenarios for both adults and minor children: a known pathogenic mutation, a truncating variant presumed pathogenic (where other truncating variants were known to be pathogenic), or a missense variant predicted in silico to be pathogenic.
Results
On average, for adults and children respectively, each specialist selected 83.5 and 79.0 conditions or genes out of 99 in the known pathogenic mutation categories, 57.0 and 53.5 out of 72 in the truncating variant categories, and 33.4 and 29.7 out of 72 in the missense variant categories. Concordance in favor of disclosure within the adult/known pathogenic mutation category was 100% for 21 conditions or genes and 80% or higher for 64 conditions or genes.
Conclusion
Specialists were highly concordant for the return of findings in 64 conditions or genes if discovered incidentally during whole exome or whole genome sequencing.
Keywords: whole genome sequencing, incidental findings
INTRODUCTION
There is an increasing consensus that whole exome sequencing (WES) and whole genome sequencing (WGS) will continue to improve in accuracy and decline in price, and that the use of these technologies will eventually become an integral part of clinical medicine.1–7 Several recent reports have highlighted the use of WES/WGS in the diagnosis or treatment of patients,8–12 and there is rapid expansion of CLIA approved molecular laboratories now providing, or soon planning to provide these services to clinicians.13–15 One of the greatest impediments to the immediate application of sequence data to clinical medicine relates to whether, and to what degree, molecular laboratories and clinicians should seek out, interpret and communicate incidental or secondary (i.e. unrelated to reasons for ordering) genetic findings.
Whatever the clinical indication for WES/WGS, there is considerable potential in each patient to discover large numbers of variants associated with human disease.16 Currently, there are no guidelines for return of incidental findings from clinical sequencing, although there have been proposals for lower and higher risk categories or “bins” based upon clinical validity and actionability.17 Yet, no one has asked whether well-meaning specialists would agree upon incidental findings that would be appropriate to disclose. To explore concordance or discordance that such efforts might face, 16 specialists in genetics independently evaluated 99 common genetic conditions and individual genes and selected those they would recommend reporting back to the patient’s physician as incidental findings after whole genome sequencing.
MATERIALS AND METHODS
A list was generated of all diseases for which testing is clinically available as registered on the GeneTests website.18 Genes associated with the same disease were combined. The top 88 conditions or genes based on the frequency of laboratory testing were supplemented by adding hereditary breast and ovarian cancer, a condition that was not frequent among laboratories due to patented genes, along with a number of common chromosomal conditions and deletion syndromes currently diagnosed through cytogenetic analysis (Table 1).
Table 1.
Number (%) of Specialists Selecting Each Incidental Genetic Finding for Return
| Condition/Gene | Known mutation | Truncating variant | Missense variant | |||
|---|---|---|---|---|---|---|
| Adult | Child | Adult | Child | Adult | Child | |
| Cancer | ||||||
| Hereditary Breast and Ovarian Cancer | 16 (100.0%) | 12 (75.0%) | 15 (93.8%) | 11 (68.8%) | 9 (56.3%) | 4 (25.0%) |
| Li-Fraumeni Syndrome | 16 (100.0%) | 14 (87.5%) | 16 (100.0%) | 13 (81.3%) | 9 (56.3%) | 5 (31.3%) |
| Lynch Syndrome | 16 (100.0%) | 12 (75.0%) | 16 (100.0%) | 11 (68.8%) | 9 (56.3%) | 4 (25.0%) |
| APC-Associated Polyposis | 16 (100.0%) | 15 (93.8%) | 16 (100.0%) | 14 (87.5%) | 9 (56.3%) | 7 (43.8%) |
| MUTYH Polyposis | 16 (100.0%) | 13 (81.3%) | 15 (93.8%) | 13 (81.3%) | 9 (56.3%) | 5 (31.3%) |
| Von Hippel-Lindau | 16 (100.0%) | 16 (100.0%) | 16 (100.0%) | 15 (93.8%) | 10 (62.5%) | 7 (43.8%) |
| MEN 1 | 16 (100.0%) | 15 (93.8%) | 16 (100.0%) | 14 (87.5%) | 9 (56.3%) | 7 (43.8%) |
| MEN 2 | 16 (100.0%) | 15 (93.8%) | 15 (93.8%) | 13 (81.3%) | 9 (56.3%) | 7 (43.8%) |
| PTEN Hamartoma Tumor Syndrome | 16 (100.0%) | 16 (100.0%) | 16 (100.0%) | 15 (93.8%) | 8 (50.0%) | 5 (31.3%) |
| Neurofibromatosis 1 | 15 (93.8%) | 15 (93.8%) | 15 (93.8%) | 15 (93.8%) | 7 (43.8%) | 6 (37.5%) |
| Retinoblastoma | 16 (100.0%) | 16 (100.0%) | 16 (100.0%) | 15 (93.8%) | 8 (50.0%) | 7 (43.8%) |
| Metabolic/Storage | ||||||
| MCAD Deficiency | 15 (93.8%) | 15 (93.8%) | 14 (87.5%) | 14 (87.5%) | 9 (56.3%) | 8 (50.0%) |
| Gaucher Disease | 16 (100.0%) | 15 (93.8%) | 15 (93.8%) | 14 (87.5%) | 10 (62.5%) | 9 (56.3%) |
| Tay-Sachs Disease | 15 (93.8%) | 14 (87.5%) | 14 (87.5%) | 13 (81.3%) | 9 (56.3%) | 7 (43.8%) |
| Phenylketonuria | 16 (100.0%) | 15 (93.8%) | 15 (93.8%) | 14 (87.5%) | 8 (50.0%) | 8 (50.0%) |
| Galactosemia | 16 (100.0%) | 15 (93.8%) | 15 (93.8%) | 14 (87.5%) | 9 (56.3%) | 9 (56.3%) |
| Niemann-Pick Disease | 14 (87.5%) | 13 (81.3%) | 13 (81.3%) | 12 (75.0%) | 9 (56.3%) | 8 (50.0%) |
| VLCAD Deficiency | 15 (93.8%) | 14 (87.5%) | 14 (87.5%) | 13 (81.3%) | 9 (56.3%) | 9 (56.3%) |
| Homocystinuria | 16 (100.0%) | 15 (93.8%) | 15 (93.8%) | 14 (87.5%) | 9 (56.3%) | 9 (56.3%) |
| Tyrosinemia Type 1 | 16 (100.0%) | 15 (93.8%) | 15 (93.8%) | 14 (87.5%) | 9 (56.3%) | 9 (56.3%) |
| Pompe Disease | 16 (100.0%) | 15 (93.8%) | 15 (93.8%) | 14 (87.5%) | 9 (56.3%) | 8 (50.0%) |
| Familial Dysautonomia | 13 (81.3%) | 13 (81.3%) | 12 (75.0%) | 12 (75.0%) | 8 (50.0%) | 8 (50.0%) |
| Biotinidase Deficiency | 15 (93.8%) | 15 (93.8%) | 13 (81.3%) | 13 (81.3%) | 8 (50.0%) | 9 (56.3%) |
| Isovaleric Acidemia | 14 (87.5%) | 14 (87.5%) | 13 (81.3%) | 13 (81.3%) | 8 (50.0%) | 9 (56.3%) |
| LCHAD Deficiency | 13 (81.3%) | 13 (81.3%) | 12 (75.0%) | 12 (75.0%) | 8 (50.0%) | 8 (50.0%) |
| Glutaric Acidemia Type 1 | 14 (87.5%) | 14 (87.5%) | 13 (81.3%) | 13 (81.3%) | 8 (50.0%) | 8 (50.0%) |
| Citrullinemia Type 1 | 15 (93.8%) | 15 (93.8%) | 14 (87.5%) | 14 (87.5%) | 8 (50.0%) | 8 (50.0%) |
| Wilson Disease | 16 (100.0%) | 15 (93.8%) | 15 (93.8%) | 14 (87.5%) | 10 (62.5%) | 9 (56.3%) |
| CPT II Deficiency | 15 (93.8%) | 15 (93.8%) | 13 (81.3%) | 13 (81.3%) | 8 (50.0%) | 9 (56.3%) |
| Mult. Acyl-CoA Dehydrogenase Def. | 14 (87.5%) | 14 (87.5%) | 13 (81.3%) | 13 (81.3%) | 8 (50.0%) | 8 (50.0%) |
| SCAD Deficiency | 13 (81.3%) | 13 (81.3%) | 12 (75.0%) | 12 (75.0%) | 6 (37.5%) | 6 (37.5%) |
| GSD Type 1a | 16 (100.0%) | 15 (93.8%) | 15 (93.8%) | 14 (87.5%) | 8 (50.0%) | 8 (50.0%) |
| Metachromatic Leukodystrophy | 13 (81.3%) | 13 (81.3%) | 12 (75.0%) | 12 (75.0%) | 8 (50.0%) | 7 (43.8%) |
| MPS Type 1 | 14 (87.5%) | 14 (87.5%) | 13 (81.3%) | 13 (81.3%) | 8 (50.0%) | 7 (43.8%) |
| Methylmalonic Acidemia | 14 (87.5%) | 14 (87.5%) | 13 (81.3%) | 13 (81.3%) | 8 (50.0%) | 8 (50.0%) |
| Maple Syrup Urine Disease | 15 (93.8%) | 15 (93.8%) | 14 (87.5%) | 14 (87.5%) | 8 (50.0%) | 8 (50.0%) |
| Fabry Disease | 16 (100.0%) | 14 (87.5%) | 15 (93.8%) | 13 (81.3%) | 10 (62.5%) | 8 (50.0%) |
| OTC Deficiency | 15 (93.8%) | 15 (93.8%) | 14 (87.5%) | 14 (87.5%) | 8 (50.0%) | 8 (50.0%) |
| Neurological/Neuromuscular | ||||||
| LIS1 Lissencephaly | 10 (62.5%) | 10 (62.5%) | 10 (62.5%) | 8 (50.0%) | 5 (31.3%) | 5 (31.3%) |
| CMT Type 1A | 13 (81.3%) | 11 (68.8%) | N/A | N/A | N/A | N/A |
| CMT Type 1B | 13 (81.3%) | 11 (68.8%) | 10 (62.5%) | 9 (56.3%) | 5 (31.3%) | 3 (18.8%) |
| HNLPP | 12 (75.0%) | 8 (50.0%) | 8 (50.0%) | 7 (43.8%) | 5 (31.3%) | 3 (18.8%) |
| Early-Onset Primary Dystonia | 11 (68.8%) | 10 (62.5%) | 8 (50.0%) | 7 (43.8%) | 5 (31.3%) | 5 (31.3%) |
| Epileptic Encephalopathy Infantile 2 | 9 (56.3%) | 9 (56.3%) | 8 (50.0%) | 7 (43.8%) | 5 (31.3%) | 5 (31.3%) |
| Spinal Muscular Atrophy | 13 (81.3%) | 13 (81.3%) | 10 (62.5%) | 9 (56.3%) | 7 (43.8%) | 5 (31.3%) |
| Canavan Disease | 11 (68.8%) | 13 (81.3%) | 10 (62.5%) | 10 (62.5%) | 6 (37.5%) | 5 (31.3%) |
| Dystrophinopathies | 14 (87.5%) | 13 (81.3%) | 12 (75.0%) | 11 (68.8%) | 6 (37.5%) | 5 (31.3%) |
| CMT Type X1 | 12 (75.0%) | 11 (68.8%) | 10 (62.5%) | 9 (56.3%) | 5 (31.3%) | 3 (18.8%) |
| ARX-Related | 11 (68.8%) | 10 (62.5%) | N/A | N/A | N/A | N/A |
| Huntington Disease | 10 (62.5%) | 5 (31.3%) | N/A | N/A | N/A | N/A |
| Dentatorubral-Pallidoluysian Atrophy | 12 (75.0%) | 11 (68.8%) | N/A | N/A | N/A | N/A |
| Spinal and Bulbar Muscular Atrophy | 13 (81.3%) | 10 (62.5%) | N/A | N/A | N/A | N/A |
| SCA Types 1, 3, 6, 7 | 13 (81.3%) | 11 (68.8%) | N/A | N/A | N/A | N/A |
| Myotonic Dystrophy Type 1 | 14 (87.5%) | 13 (81.3%) | N/A | N/A | N/A | N/A |
| Friedreich Ataxia | 12 (75.0%) | 13 (81.3%) | N/A | N/A | N/A | N/A |
| Mitochondrial | ||||||
| Leber Hereditary Optic Neuropathy | 12 (75.0%) | 11 (68.8%) | 10 (62.5%) | 9 (56.3%) | 6 (37.5%) | 5 (31.3%) |
| MELAS | 12 (75.0%) | 12 (75.0%) | 9 (56.3%) | 9 (56.3%) | 4 (25.0%) | 4 (25.0%) |
| MERRF | 12 (75.0%) | 12 (75.0%) | 9 (56.3%) | 9 (56.3%) | 4 (25.0%) | 4 (25.0%) |
| Leigh Syndrome and NARP | 12 (75.0%) | 12 (75.0%) | 10 (62.5%) | 10 (62.5%) | 5 (31.3%) | 5 (31.3%) |
| Developmental | ||||||
| Noonan Syndrome | 12 (75.0%) | 12 (75.0%) | 11 (68.8%) | 11 (68.8%) | 5 (31.3%) | 5 (31.3%) |
| Beckwith-Wiedemann | 14 (87.5%) | 15 (93.8%) | 12 (75.0%) | 14 (87.5%) | 6 (37.5%) | 6 (37.5%) |
| Sotos Syndrome | 12 (75.0%) | 13 (81.3%) | 12 (75.0%) | 11 (68.8%) | 6 (37.5%) | 6 (37.5%) |
| CHARGE | 12 (75.0%) | 13 (81.3%) | 12 (75.0%) | 11 (68.8%) | 6 (37.5%) | 6 (37.5%) |
| MECP2-Related | 10 (62.5%) | 11 (68.8%) | 10 (62.5%) | 11 (68.8%) | 6 (37.5%) | 6 (37.5%) |
| FMR1-Related | 13 (81.3%) | 13 (81.3%) | N/A | N/A | N/A | N/A |
| Inherited Predisposition (non-cancer) | ||||||
| Factor V Leiden Thrombophilia | 12 (75.0%) | 9 (56.3%) | N/A | N/A | N/A | N/A |
| Prothrombin-Related Thrombophilia | 11 (68.8%) | 9 (56.3%) | N/A | N/A | N/A | N/A |
| Familial Hypercholesterolemia | 16 (100.0%) | 14 (87.5%) | 13 (81.3%) | 11 (68.8%) | 7 (43.8%) | 6 (37.5%) |
| Hereditary Hemochromatosis | 14 (87.5%) | 10 (62.5%) | N/A | N/A | N/A | N/A |
| MTHFR | 9 (56.3%) | 8 (50.0%) | N/A | N/A | N/A | N/A |
| Alpha1-Antitrypsin Deficiency | 15 (93.8%) | 13 (81.3%) | N/A | N/A | N/A | N/A |
| Gilbert Syndrome | 11 (68.8%) | 9 (56.3%) | 10 (62.5%) | 8 (50.0%) | 5 (31.3%) | 4 (25.0%) |
| Apolipoprotein E | 8 (50.0%) | 4 (25.0%) | N/A | N/A | N/A | N/A |
| Other | ||||||
| Achondroplasia | 12 (75.0%) | 12 (75.0%) | N/A | N/A | N/A | N/A |
| Hypochondroplasia | 12 (75.0%) | 13 (81.3%) | N/A | N/A | N/A | N/A |
| Marfan Syndrome | 15 (93.8%) | 15 (93.8%) | 13 (81.3%) | 13 (81.3%) | 7 (43.8%) | 7 (43.8%) |
| Dilated Cardiomyopathy | 14 (87.5%) | 13 (81.3%) | 12 (75.0%) | 12 (75.0%) | 7 (43.8%) | 7 (43.8%) |
| Romano-Ward (Long QT) | 16 (100.0%) | 16 (100.0%) | 14 (87.5%) | 14 (87.5%) | 7 (43.8%) | 7 (43.8%) |
| DFNA 3 Nonsyndromic Hearing Loss | 12 (75.0%) | 10 (62.5%) | 9 (56.3%) | 7 (43.8%) | 7 (43.8%) | 7 (43.8%) |
| DFNB 1 Nonsyndromic Hearing Loss | 13 (81.3%) | 11 (68.8%) | 10 (62.5%) | 8 (50.0%) | 8 (50.0%) | 7 (43.8%) |
| CFTR-Related Disorders | 15 (93.8%) | 14 (87.5%) | 13 (81.3%) | 12 (75.0%) | 8 (50.0%) | 7 (43.8%) |
| Familial Mediterranean Fever | 15 (93.8%) | 14 (87.5%) | 11 (68.8%) | 10 (62.5%) | 8 (50.0%) | 7 (43.8%) |
| Beta-Thalassemia | 14 (87.5%) | 13 (81.3%) | 12 (75.0%) | 11 (68.8%) | 7 (43.8%) | 6 (37.5%) |
| Hemoglobin SS | 14 (87.5%) | 13 (81.3%) | N/A | N/A | N/A | N/A |
| Bloom Syndrome | 14 (87.5%) | 15 (93.8%) | 13 (81.3%) | 13 (81.3%) | 7 (43.8%) | 6 (37.5%) |
| Fanconi Anemia (FANCC-Related) | 15 (93.8%) | 15 (93.8%) | 12 (75.0%) | 12 (75.0%) | 7 (43.8%) | 5 (31.3%) |
| Kallmann Syndrome 1 | 12 (75.0%) | 12 (75.0%) | 10 (62.5%) | 10 (62.5%) | 7 (43.8%) | 7 (43.8%) |
| Ichthyosis, X-linked | 13 (81.3%) | 13 (81.3%) | 11 (68.8%) | 11 (68.8%) | 7 (43.8%) | 7 (43.8%) |
| Could reveal unexpected chromosomal sex | ||||||
| 46,XX Testicular DSD | 12 (75.0%) | 12 (75.0%) | N/A | N/A | N/A | N/A |
| Congenital Adrenal Hyperplasia | 14 (87.5%) | 15 (93.8%) | 11 (68.8%) | 12 (75.0%) | 7 (43.8%) | 7 (43.8%) |
| Androgen Insensitivity Syndrome | 13 (81.3%) | 13 (81.3%) | 11 (68.8%) | 11 (68.8%) | 7 (43.8%) | 8 (50.0%) |
| 46,XY DSD and 46,XY CGD | 11 (68.8%) | 12 (75.0%) | N/A | N/A | N/A | N/A |
| Other chromosomal or deletion syndromes | ||||||
| Turner Syndrome | 12 (75.0%) | 13 (81.3%) | N/A | N/A | N/A | N/A |
| Turner mosaic | 12 (75.0%) | 13 (81.3%) | N/A | N/A | N/A | N/A |
| Kleinefelter Syndrome | 12 (75.0%) | 13 (81.3%) | N/A | N/A | N/A | N/A |
| 47,XYY Syndrome | 12 (75.0%) | 12 (75.0%) | N/A | N/A | N/A | N/A |
| Y Chromosome Deletion | 12 (75.0%) | 8 (50.0%) | N/A | N/A | N/A | N/A |
| 22q11.2 Deletion | 13 (81.3%) | 14 (87.5%) | N/A | N/A | N/A | N/A |
| Wolf-Hirschhorn | 12 (75.0%) | 13 (81.3%) | N/A | N/A | N/A | N/A |
The 16 participating specialists were clinical geneticists and/or molecular laboratory directors, most of whom have been involved in early uses of genome-scale data, but there was no attempt to be representative or to include all relevant sub-specialists. Each was asked to assume that they were serving as a consultant to a laboratory that performs clinical WGS, and to decide which variants discovered as incidental findings should be returned in a report to the ordering physician. Specialists were asked to assume that family history was not available, that the patient had no previously recognized clinical features consistent with the disease variant under consideration, that the patient’s gender was known, and that it was known whether the patient was an adult or a minor child (under 18 years), but not the exact age. Specialists were asked to assume that the sequencing was perfectly accurate and could detect translocations and repeat expansions with perfect accuracy, even though this degree of accuracy is not available through current WES/WGS technologies. Details of the consent process, patient preferences regarding results disclosure, and details about how disclosures of results might be handled by the referring physician were not specified. An additional assumption was that family members of the patient being tested would not have been sequenced or genetically tested for the variant under consideration, thus decisions were made on the basis of the genome findings alone without contextualization by patient medical history or family history.
Specialists provided separate responses as to whether incidental findings should be returned to the physicians of adults and of minor children, and where applicable, in each of three categories of variants: a known pathogenic mutation, a truncating variant presumed pathogenic (where other truncating variants were known to be pathogenic), and a missense variant predicted to be deleterious by in silico analysis. For recessive conditions or genes, specialists were asked to specify whether they would return a variant even in a carrier state, or only if the patient were found to be biallelic (for autosomal recessive) or hemizygous (for Xlinked), or not at all. For repeat expansion disorders, specialists indicated whether they would return a finding of a premutation or full mutation, only a full mutation, or neither. For each possible scenario per disease (adult/child, pathogenic/truncating/missense), specialists were also asked if they had difficulty deciding whether to report the variant. Each specialist could decline to respond due to lack of familiarity with the condition or gene. All of the choices described above were made through pull-down, forced-choice menus. For the purposes of this analysis, conditions or genes were counted if the specialist recommended an affirmative response in the dominant disorders or genes, or in any option of the recessive, X-linked or expansion disorders or genes.
The specialists did not communicate with each other about their decisions. All 16 specialists are listed as the co-authors of this paper. One additional co-author (SK) is a genetic counselor who prepared the lists and assisted with data analyses and manuscript preparation, and a second additional co-author (AM) is a contributor in ethics and legal issues who participated in data analyses and manuscript preparation.
RESULTS
Of the 16 specialists who selected conditions, 13 were MDs with or without an additional degree, and 3 were non-MD PhDs; 9 were primarily clinical geneticists, 3 were primarily molecular laboratorians and 4 were active both clinically and in the molecular laboratory.
The conditions and genes proposed and the number and percentage of specialists who would recommend return of incidental findings based upon these is shown in Table 1 (specialist order does not correlate with order of co-authors). For each of the 99 conditions and genes, an average of 13.5 (SD 1.9, range 8–16) specialists suggested incidental genetic findings be returned about adults and 12.8 (SD 2.3, range 4–16) about children if there was a known pathogenic mutation. For each of 72 conditions and genes (excluding trinucleotide repeat expansion, chromosomal, and deletion conditions, and conditions caused by only a specific known mutation), an average of 12.6 specialists (SD 2.3, range 8–16) and 11.9 specialists (SD 2.2, range 7–15) suggested that incidental findings of truncating variants be returned about adults and children, respectively. And an average of 7.4 specialists (SD 1.5, range 4–10) and 6.6 specialists (SD 1.7, range 3–9) suggested that incidental findings of a missense variant predicted to be pathogenic by in silico analysis be returned about adults and children, respectively. The concordance in favor of disclosure was 100% for 21 conditions or genes in the Adult/Known Pathogenic Mutation category, was 80% or higher for 64 conditions or genes, and was at least 50% for all conditions or genes.
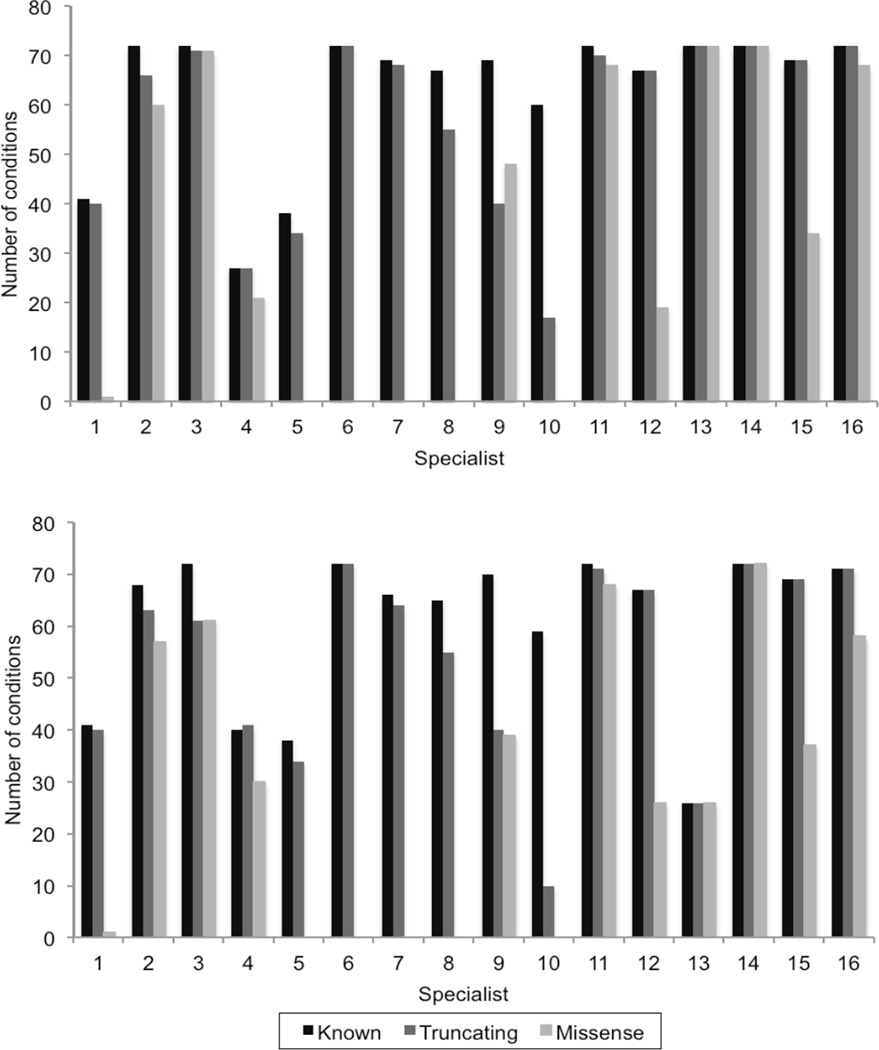
Concordance was higher for returning incidental information about conditions that had potential for medical intervention (such as cancer predisposition syndromes) than for those without (such as developmental or neurodegenerative disorders). Concordance was also higher when returning incidental findings about adults rather than about children, and for returning known pathogenic mutations and presumed pathogenic truncating variants rather than missense variants predicted to be pathogenic. The total number of genes or conditions selected by each specialist varied along these axes (Table 2 and Figure 1) whereas the discomfort that specialists felt in making the decision varied inversely along the same axes (Table 3).
Table 2.
Number (%) of Conditions Selected for Incidental Return By Each Specialist
| Specialist | Known mutation (99 conditions) | Truncating variant (72 conditions) | Missense variant (72 conditions) | |||
|---|---|---|---|---|---|---|
| Adult | Child | Adult | Child | Adult | Child | |
| 1 | 43 (43.4%) | 43 (43.4%) | 40 (55.6%) | 40 (55.6%) | 1 (1.4%) | 1 (1.4%) |
| 2 | 97 (98.0%) | 90 (90.9%) | 66 (91.7%) | 63 (87.5%) | 60 (83.3%) | 57 (79.2%) |
| 3 | 99 (100.0%) | 99 (100.0%) | 71 (98.6%) | 61 (84.7%) | 71 (98.6%) | 61 (84.7%) |
| 4 | 30 (30.3%) | 46 (46.5%) | 27 (37.5%) | 41 (56.9%) | 21 (29.2%) | 30 (41.7%) |
| 5 | 39 (39.3%) | 39 (39.3%) | 34 (47.2%) | 34 (47.2%) | 0 (0.0%) | 0 (0.0%) |
| 6 | 97 (98.0%) | 97 (98.0%) | 72 (100.0%) | 72 (100.0%) | 0 (0.0%) | 0 (0.0%) |
| 7 | 96 (97.0%) | 92 (92.9%) | 68 (94.4%) | 64 (88.9%) | 0 (0.0%) | 0 (0.0%) |
| 8 | 88 (88.9%) | 80 (80.8%) | 55 (76.4%) | 55 (76.4%) | 0 (0.0%) | 0 (0.0%) |
| 9 | 95 (96.0%) | 94 (94.9%) | 40 (55.6%) | 40 (55.6%) | 48 (66.7%) | 39 (54.2%) |
| 10 | 80 (80.8%) | 76 (76.8%) | 17 (23.6%) | 10 (13.9%) | 0 (0.0%) | 0 (0.0%) |
| 11 | 88 (88.9%) | 88 (88.9%) | 70 (97.2%) | 71 (98.6%) | 68 (94.4%) | 68 (94.4%) |
| 12 | 94 (94.9%) | 93 (93.9%) | 67 (93.1%) | 67 (93.1%) | 19 (26.4%) | 26 (36.1%) |
| 13 | 99 (100.0%) | 38 (38.4%) | 72 (100.0%) | 26 (36.1%) | 72 (100.0%) | 26 (36.1%) |
| 14 | 98 (99.0%) | 98 (99.0%) | 72 (100.0%) | 72 (100.0%) | 72 (100.0%) | 72 (100.0%) |
| 15 | 95 (96.0%) | 94 (94.9%) | 69 (95.8%) | 69 (95.8%) | 34 (47.2%) | 37 (51.4%) |
| 16 | 98 (99.0%) | 97 (98.0%) | 72 (100.0%) | 71 (98.6%) | 68 (94.4%) | 58 (80.6%) |
| Mean | 83.5 (84.3%) | 79.0 (79.8%) | 57.0 (79.2%) | 53.5 (74.3%) | 33.4 (46.4%) | 29.7 (41.3%) |
| SD | 23.6 | 23.2 | 18.8 | 19.2 | 31.3 | 27.3 |
| Range | 30–99 | 38–99 | 17–72 | 10–72 | 0–72 | 0–72 |
Figure 1.
Number of conditions/genes out of a total of 72 (excluding repeat expansion, chromosomal, and deletion conditions, and conditions caused by a single known mutation) that each of 16 specialists chose for incidental return in an adult patient (1A) and in a child (1B), by category of variant
Table 3.
Number (%) of Difficult Decisions Cited by each Specialist
| Specialist | Known mutation (99 conditions) | Truncating variant (72 conditions) | Missense variant (72 conditions) | |||
|---|---|---|---|---|---|---|
| Adult | Child | Adult | Child | Adult | Child | |
| 1 | 6 (6.1%) | 6 (6.1%) | 3 (4.2%) | 3 (4.2%) | 1 (1.4%) | 1 (1.4%) |
| 2 | 8 (8.1%) | 28 (28.3%) | 2 (2.8%) | 18 (25.0%) | 46 (63.9%) | 54 (75%) |
| 3 | 27 (27.3%) | 32 (32.3%) | 14 (19.4%) | 18 (25.0%) | 14 (19.4%) | 18 (25.0%) |
| 4 | 25 (25.3%) | 27 (27.3%) | 17 (23.6%) | 20 (27.8%) | 21 (29.2%) | 40 (55.6%) |
| 5 | 15 (15.2%) | 15 (15.2%) | 13 (18.1%) | 12 (16.7%) | 0 (0.0%) | 0 (0.0%) |
| 6 | 1 (1.0%) | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) |
| 7 | 2 (2.0%) | 3 (3.0%) | 0 (0.0%) | 0 (0.0%) | 72 (100.0%) | 72 (100.0%) |
| 8 | 24 (24.2%) | 53 (53.5%) | 13 (18.1%) | 38 (52.8%) | 8 (11.1%) | 3 (4.2%) |
| 9 | 4 (4.0%) | 18 (18.2%) | 8 (11.1%) | 0 (0.0%) | 3 (4.2%) | 14 (19.4%) |
| 10 | 36 (36.4%) | 7 (7.1%) | 50 (69.4%) | 49 (68.1%) | 63 (87.5%) | 65 (90.3%) |
| 11 | 1 (1.0%) | 32 (32.3%) | 4 (5.6%) | 13 (18.1%) | 67 (93.1%) | 65 (90.3%) |
| 12 | 4 (4.0%) | 1 (1.0%) | 3 (4.2%) | 1 (1.4%) | 1 (1.4%) | 2 (2.8%) |
| 13 | 0 (0.0%) | 11 (11.1%) | 0 (0.0%) | 11 (15.3%) | 0 (0.0%) | 11 (15.3%) |
| 14 | 1 (1.0%) | 4 (4.0%) | 1 (1.4%) | 3 (4.2%) | 1 (1.4%) | 3 (4.2%) |
| 15 | 6 (6.1%) | 12 (12.1%) | 14 (19.4%) | 18 (25.0%) | 70 (97.2%) | 44 (61.1%) |
| 16 | 12 (12.1%) | 65 (65.7%) | 8 (11.1%) | 48 (66.7%) | 20 (27.8%) | 48 (66.7%) |
| Mean | 10.8 (10.9%) | 19.6 (19.8%) | 9.4 (13.1%) | 15.8 (21.9%) | 24.2 (33.6%) | 27.5 (38.2%) |
| SD | 11.3 | 18.9 | 12.3 | 16.3 | 28.8 | 27.0 |
| Range | 0–36 | 0–65 | 0–50 | 0–49 | 0–72 | 0–72 |
DISCUSSION
This report presents an exploratory description of concordance and discordance around specific conditions and genes with respect to incidental findings in the context of clinical WGS/WES. There were 21 conditions or genes in which all 16 specialists agreed that known pathogenic mutations should be disclosed if found incidentally in adults, and 4 conditions in which all 16 specialists agreed that known pathogenic mutations should be disclosed if found incidentally in minor children (Table 4). There was considerable concordance among all 99 conditions or genes for known pathogenic mutations with a majority of specialists selecting nearly every condition or gene for return as an incidental finding.
Table 4.
Genes or conditions selected by all specialists to be returned incidentally in adults. Those marked with an asterisk were selected by all specialists to be returned incidentally in children.
| Hereditary Breast and Ovarian Cancer |
| Li-Fraumeni Syndrome |
| Lynch Syndrome |
| APC-Associated Polyposis |
| MUTYH Polyposis |
| Von Hippel-Lindau* |
| MEN 1 |
| MEN 2 |
| PTEN Hamartoma Tumor Syndrome* |
| Retinoblastoma* |
| Gaucher Disease |
| Phenylketonuria |
| Galactosemia |
| Homocystinuria |
| Tyrosinemia Type 1 |
| Pompe Disease |
| Wilson Disease |
| GSD Type 1a |
| Fabry Disease |
| Familial Hypercholesterolemia |
| Romano-Ward (Long QT)* |
Substantial discordance was observed across two major domains. First, some specialists were more reluctant than others to disclose incidental findings. For example, even within the category of adult/known pathogenic mutations, one specialist reported that he or she would recommend disclosure of only 30.3% of the identified genes or conditions, whereas two others thought 100% should be disclosed. Second, there was discordance in judgments about the relative value of different criteria when making decisions about disclosures. For some, whether the result pertained to a child or an adult seemed to be the most relevant factor driving the decision to disclose, while for others, the established pathogenicity of the variant was the most important factor. These criteria also influenced whether contributors found a particular decision to be difficult. Additional research is needed to better understand the source of this discordance; for example, whether it reflects primarily differences in clinical or ethical judgment or, in the case of the potential pathogenicity of variants, whether specialists were fully aware of the limited accuracy of in silico prediction tools for missense variants.
This paper has a number of limitations. The specialists were not representative and were of varying degrees of expertise. Each specialist responded independently and without interaction that would characterize formal consensus building. Specialists were asked to assume that no information was available about family history or knowledge of signs or symptoms in relatives of the proband, which is somewhat artificial.
The degree of discordance observed suggests that whether considering incidental findings in research subjects or incidental findings in clinical WES/WGS, it may be difficult to reach consensus on a specific list of variants that meet a threshold for disclosure, as called for in various consensus statements on incidental findings. However, in an actual consensus group, experts on a subset of disorders would educate the rest of the group, and likely increase concordance. Plans that attempt to factor in subject or patient preference (which were excluded in this exercise) could make consensus easier or more difficult. In this exercise, the choice was whether to provide such information in a report to the ordering physician, not to the patient. This may have provided comfort to the specialists who were wavering because the ordering physician would have the ability to seek preferences from the patient about how much information to share, and the ability to contextualize the report by integrating personal medical history, family history, symptoms and signs, or by referring the patient. The variability in responses surrounding issues for which consensus has already been achieved and published (such as the unreliability of in silico prediction of variant significance) also highlights the need for greater education, even among specialists, as well as the need for extensively curated clinical grade databases.
In summary, there is considerable concordance and considerable discordance in choices about return of incidental genomic information among specialists queried for this report. The variability in responses suggests that a relatively small panel of specialists is unlikely to embody all of the necessary expertise to achieve consensus about the vast number of genetic variants that will be identified by WES/WGS, especially if additional variants were considered that are less well characterized than those we selected. Modifications to a typical consensus approach may need to be considered, such as collaboration among multiple expert panels, with each panel being composed of specialists in a more narrow focus of expertise. It appears that even genetic specialists are a heterogeneous group and may not be yet fully prepared to deal with the implementation of new genetic technologies.
Table 5.
| a: Autosomal Recessive Conditions, Known Mutation— Number of Specialists Selecting Each Incidental Genetic Finding for Return if Biallelic or a Carrier, or Only if Biallelic | ||||
|---|---|---|---|---|
| Condition/Gene | Adult | Child | ||
| Biallelic or Carrier |
Biallelic only | Biallelic or Carrier |
Biallelic only | |
| MUTYH Polyposis | 11 | 5 | 6 | 7 |
| MCAD Deficiency | 12 | 3 | 7 | 8 |
| Gaucher Disease | 13 | 3 | 8 | 7 |
| Tay-Sachs Disease | 13 | 2 | 8 | 6 |
| Phenylketonuria | 13 | 3 | 8 | 7 |
| Galactosemia | 13 | 3 | 8 | 7 |
| Niemann-Pick Disease | 13 | 1 | 8 | 5 |
| VLCAD Deficiency | 13 | 2 | 8 | 6 |
| Homocystinuria | 13 | 3 | 8 | 7 |
| Tyrosinemia Type 1 | 13 | 3 | 8 | 7 |
| Pompe Disease | 13 | 3 | 8 | 7 |
| Familial Dysautonomia | 13 | 0 | 8 | 5 |
| Biotinidase Deficiency | 13 | 2 | 8 | 7 |
| Isovaleric Acidemia | 13 | 1 | 9 | 5 |
| LCHAD Deficiency | 12 | 1 | 8 | 5 |
| Glutaric Acidemia Type 1 | 13 | 1 | 8 | 6 |
| Citrullinemia Type 1 | 13 | 2 | 8 | 7 |
| Wilson Disease | 12 | 4 | 8 | 7 |
| CPT II Deficiency | 13 | 2 | 8 | 7 |
| Mult. Acyl-CoA Dehydrogenase Def. | 12 | 2 | 8 | 6 |
| SCAD Deficiency | 10 | 3 | 6 | 7 |
| GSD Type 1a | 13 | 3 | 8 | 7 |
| Metachromatic Leukodystrophy | 13 | 0 | 8 | 5 |
| MPS Type 1 | 12 | 2 | 8 | 6 |
| Methylmalonic Acidemia | 13 | 1 | 8 | 6 |
| Maple Syrup Urine Disease | 13 | 2 | 8 | 7 |
| Spinal Muscular Atrophy | 13 | 0 | 8 | 5 |
| Canavan Disease | 11 | 0 | 7 | 6 |
| Hereditary Hemochromatosis | 10 | 4 | 7 | 3 |
| MTHFR | 7 | 2 | 5 | 3 |
| Alpha1-Antitrypsin Deficiency | 12 | 3 | 9 | 4 |
| Gilbert Syndrome | 9 | 2 | 6 | 3 |
| CFTR-Related Disorders | 13 | 2 | 8 | 6 |
| DFNB 1 Nonsyndromic Hearing Loss | 12 | 1 | 8 | 3 |
| Familial Mediterranean Fever | 11 | 4 | 8 | 6 |
| Beta-Thalassemia | 13 | 1 | 8 | 5 |
| Hemoglobin SS | 13 | 1 | 7 | 6 |
| Bloom Syndrome | 12 | 2 | 8 | 7 |
| Fanconi Anemia (FANCC-Related) | 13 | 2 | 8 | 7 |
| Congenital Adrenal Hyperplasia | 12 | 2 | 8 | 7 |
| b: Autosomal Recessive Conditions, Truncating Variant Presumed Pathogenic— Number of Specialists Selecting Each Incidental Genetic Finding for Return if Biallelic or a Carrier, or Only if Biallelic | ||||
|---|---|---|---|---|
| Condition/Gene | Adult | Child | ||
| Biallelic or Carrier |
Biallelic only | Biallelic or Carrier |
Biallelic only | |
| MUTYH Polyposis | 10 | 5 | 7 | 6 |
| MCAD Deficiency | 11 | 3 | 7 | 7 |
| Gaucher Disease | 12 | 3 | 8 | 6 |
| Tay-Sachs Disease | 12 | 2 | 8 | 5 |
| Phenylketonuria | 12 | 3 | 8 | 6 |
| Galactosemia | 12 | 3 | 8 | 6 |
| Niemann-Pick Disease | 12 | 1 | 8 | 4 |
| VLCAD Deficiency | 12 | 2 | 8 | 5 |
| Homocystinuria | 12 | 3 | 8 | 6 |
| Tyrosinemia Type 1 | 12 | 3 | 8 | 6 |
| Pompe Disease | 12 | 3 | 8 | 6 |
| Familial Dysautonomia | 12 | 0 | 8 | 4 |
| Biotinidase Deficiency | 12 | 1 | 8 | 5 |
| Isovaleric Acidemia | 12 | 1 | 9 | 4 |
| LCHAD Deficiency | 10 | 2 | 8 | 4 |
| Glutaric Acidemia Type 1 | 12 | 1 | 8 | 5 |
| Citrullinemia Type 1 | 12 | 2 | 8 | 6 |
| Wilson Disease | 10 | 5 | 8 | 6 |
| CPT II Deficiency | 11 | 2 | 8 | 5 |
| Mult. Acyl-CoA Dehydrogenase Def. | 11 | 2 | 8 | 5 |
| SCAD Deficiency | 9 | 3 | 6 | 6 |
| GSD Type 1a | 12 | 3 | 8 | 6 |
| Metachromatic Leukodystrophy | 12 | 0 | 8 | 4 |
| MPS Type 1 | 11 | 2 | 8 | 5 |
| Methylmalonic Acidemia | 12 | 1 | 8 | 5 |
| Maple Syrup Urine Disease | 12 | 2 | 8 | 6 |
| Spinal Muscular Atrophy | 10 | 0 | 5 | 4 |
| Canavan Disease | 10 | 0 | 4 | 6 |
| Gilbert Syndrome | 7 | 3 | 5 | 3 |
| CFTR-Related Disorders | 11 | 2 | 8 | 4 |
| DFNB 1 Nonsyndromic Hearing Loss | 9 | 1 | 7 | 1 |
| Familial Mediterranean Fever | 8 | 3 | 7 | 3 |
| Beta-Thalassemia | 11 | 1 | 8 | 3 |
| Bloom Syndrome | 11 | 2 | 9 | 4 |
| Fanconi Anemia (FANCC-Related) | 10 | 2 | 7 | 5 |
| Congenital Adrenal Hyperplasia | 9 | 2 | 7 | 5 |
| c: Autosomal Recessive Conditions, Missense Variant Predicted Pathogenic— Number of Specialists Selecting Each Incidental Genetic Finding for Return if Biallelic or a Carrier, or Only if Biallelic | ||||
|---|---|---|---|---|
| Condition/Gene | Adult | Child | ||
| Biallelic or Carrier |
Biallelic only | Biallelic or Carrier |
Biallelic only | |
| MUTYH Polyposis | 5 | 4 | 3 | 2 |
| MCAD Deficiency | 9 | 0 | 4 | 4 |
| Gaucher Disease | 9 | 1 | 4 | 5 |
| Tay-Sachs Disease | 8 | 1 | 3 | 4 |
| Phenylketonuria | 8 | 0 | 4 | 4 |
| Galactosemia | 8 | 1 | 4 | 5 |
| Niemann-Pick Disease | 8 | 1 | 3 | 5 |
| VLCAD Deficiency | 8 | 1 | 5 | 4 |
| Homocystinuria | 8 | 1 | 4 | 5 |
| Tyrosinemia Type 1 | 8 | 1 | 4 | 5 |
| Pompe Disease | 8 | 1 | 3 | 5 |
| Familial Dysautonomia | 8 | 0 | 3 | 5 |
| Biotinidase Deficiency | 8 | 0 | 4 | 5 |
| Isovaleric Acidemia | 8 | 0 | 4 | 5 |
| LCHAD Deficiency | 8 | 0 | 3 | 5 |
| Glutaric Acidemia Type 1 | 8 | 0 | 3 | 5 |
| Citrullinemia Type 1 | 8 | 0 | 3 | 5 |
| Wilson Disease | 8 | 2 | 5 | 4 |
| CPT II Deficiency | 8 | 0 | 4 | 5 |
| Mult. Acyl-CoA Dehydrogenase Def. | 8 | 0 | 4 | 4 |
| SCAD Deficiency | 6 | 0 | 3 | 3 |
| GSD Type 1a | 8 | 0 | 5 | 3 |
| Metachromatic Leukodystrophy | 8 | 0 | 3 | 4 |
| MPS Type 1 | 8 | 0 | 3 | 4 |
| Methylmalonic Acidemia | 8 | 0 | 4 | 4 |
| Maple Syrup Urine Disease | 8 | 0 | 4 | 4 |
| Spinal Muscular Atrophy | 7 | 0 | 4 | 1 |
| Canavan Disease | 6 | 0 | 2 | 3 |
| Gilbert Syndrome | 4 | 1 | 3 | 1 |
| CFTR-Related Disorders | 6 | 2 | 5 | 2 |
| DFNB 1 Nonsyndromic Hearing Loss | 6 | 2 | 5 | 2 |
| Familial Mediterranean Fever | 6 | 2 | 5 | 2 |
| Beta-Thalassemia | 7 | 0 | 5 | 1 |
| Bloom Syndrome | 6 | 1 | 5 | 1 |
| Fanconi Anemia (FANCC-Related) | 5 | 2 | 4 | 1 |
| Congenital Adrenal Hyperplasia | 6 | 1 | 5 | 2 |
Table 6.
| a: X-linked Conditions, Known Mutation— Number of Specialists Selecting Each Incidental Genetic Finding for Return if Hemizygous or a Carrier, or Only if Hemizygous | ||||
|---|---|---|---|---|
| Condition/Gene | Adult | Child | ||
| Hemizygous or Carrier |
Hemizygous only |
Hemizygous or Carrier |
Hemizygous only |
|
| Fabry Disease | 13 | 3 | 9 | 5 |
| OTC Deficiency | 14 | 1 | 11 | 4 |
| Dystrophinopathies | 14 | 0 | 9 | 4 |
| CMT Type X1 | 12 | 0 | 8 | 3 |
| ARX-Related | 11 | 0 | 7 | 3 |
| MECP2-Related | 10 | 0 | 11 | 0 |
| Kallmann Syndrome 1 | 12 | 0 | 8 | 4 |
| Ichthyosis, X-Linked | 13 | 0 | 8 | 5 |
| Androgen Insensitivity Syndrome | 12 | 1 | 8 | 5 |
| b: X-linked Conditions, Truncating Variant Presumed Pathogenic— Number of Specialists Selecting Each Incidental Genetic Finding for Return if Hemizygous or a Carrier, or Only if Hemizygous | ||||
|---|---|---|---|---|
| Condition/Gene | Adult | Child | ||
| Hemizygous or Carrier |
Hemizygous only |
Hemizygous or Carrier |
Hemizygous only |
|
| Fabry Disease | 12 | 3 | 9 | 4 |
| OTC Deficiency | 13 | 1 | 11 | 3 |
| Dystrophinopathies | 12 | 0 | 7 | 4 |
| CMT Type X1 | 10 | 0 | 7 | 2 |
| MECP2-Related | 10 | 0 | 10 | 1 |
| Kallmann Syndrome 1 | 10 | 0 | 8 | 2 |
| Ichthyosis, X-Linked | 11 | 0 | 8 | 3 |
| Androgen Insensitivity Syndrome | 10 | 1 | 7 | 4 |
| c: X-linked Conditions, Missense Variant Predicted Pathogenic— Number of Specialists Selecting Each Incidental Genetic Finding for Return if Hemizygous or a Carrier, or Only if Hemizygous | ||||
|---|---|---|---|---|
| Condition/Gene | Adult | Child | ||
| Hemizygous or Carrier |
Hemizygous only |
Hemizygous or Carrier |
Hemizygous only |
|
| Fabry Disease | 9 | 1 | 6 | 2 |
| OTC Deficiency | 8 | 0 | 6 | 2 |
| Dystrophinopathies | 6 | 0 | 5 | 0 |
| CMT Type X1 | 4 | 1 | 3 | 0 |
| MECP2-Related | 6 | 0 | 6 | 0 |
| Kallmann Syndrome 1 | 6 | 1 | 5 | 2 |
| Ichthyosis, X-Linked | 6 | 1 | 5 | 2 |
| Androgen Insensitivity Syndrome | 6 | 1 | 5 | 3 |
Table 7.
Repeat Expansion Conditions— Number of Specialists Selecting Each Incidental Genetic Finding for Return if a Premutation or Full Mutation, or only if a Full Mutation
| Condition/Gene | Adult | Child | ||
|---|---|---|---|---|
| Premutation or Full mutation |
Full mutation only |
Premutation or Full mutation |
Full mutation only |
|
| Huntington Disease | 10 | 0 | 4 | 1 |
| Spinocerebellar Ataxia Types 1, 3, 6, 7 | 13 | 0 | 7 | 4 |
| Myotonic Dystrophy Type 1 | 13 | 1 | 8 | 5 |
| Friedreich Ataxia | 12 | 0 | 9 | 4 |
| Spinal and Bulbar Muscular Atrophy | 13 | 0 | 7 | 3 |
| FMR1-Related Disorders | 13 | 0 | 10 | 3 |
Acknowledgments
Funding for this work provided by NIH grants: HG02213, AG027841, HG005491, HG005092, HG006615, HG003178, HG006500, CA138836, HG006485. Dr. Berry is supported by the Manton Center for Orphan Disease Research. Dr. Biesecker is supported by intramural funding from the National Human Genome Research Institute. Dr. McGuire is supported by the BCM Clinical and Translational Research Program and the Baylor Annual Fund.
REFERENCES
- 1.Guttmacher A, McGuire A, Ponder B, Stefansson K. Personalized genomic information: Preparing for the future of genetic medicine. Nat Rev Genet. 2010 Feb;11(2):161–165. doi: 10.1038/nrg2735. [DOI] [PubMed] [Google Scholar]
- 2.Feero W, Guttmacher A, Collins F. Genomic medicine - An updated primer. N Engl J Med. 2010;362(21):2001–2011. doi: 10.1056/NEJMra0907175. [DOI] [PubMed] [Google Scholar]
- 3.Ginsburg G, Willard H. Genomic and personalized medicine: Foundations and applications. Transl Res. 2009 Dec;154(6):277–287. doi: 10.1016/j.trsl.2009.09.005. [DOI] [PubMed] [Google Scholar]
- 4.Korf B. Genetics and genomics education: The next generation. Genetics In Medicine. 2011 Mar;13(3):201–202. doi: 10.1097/GIM.0b013e31820986cd. 2011. [DOI] [PubMed] [Google Scholar]
- 5.Khoury M. Public health genomics: The end of the beginning. Genetics In Medicine. 2011 2011 Mar;13(3):206–209. doi: 10.1097/GIM.0b013e31821024ca. [DOI] [PubMed] [Google Scholar]
- 6.Green E, Guyer M. Charting a course for genomic medicine from base pairs to bedside. Nature. 2011 Feb 10;470(7333):204–213. doi: 10.1038/nature09764. [DOI] [PubMed] [Google Scholar]
- 7.Mayer AN, Dimmock DP, Arca MJ, et al. A timely arrival for genomic medicine. Genet Med. 2011 Mar;13(3):195–196. doi: 10.1097/GIM.0b013e3182095089. [DOI] [PubMed] [Google Scholar]
- 8.Lupski J, Reid J, Gonzaga-Jauregui C, et al. Whole-genome sequencing in a patient with Charcot-Marie-Tooth Neuropathy. N Engl J Med. 2010 Mar 10;362:1181–1191. doi: 10.1056/NEJMoa0908094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Worthey EA, Mayer AN, Syverson GD, et al. Making a definitive diagnosis: Successful clinical application of whole exome sequencing in a child with intractable inflammatory bowel disease. Genet Med. 2011 Mar;13(3):255–262. doi: 10.1097/GIM.0b013e3182088158. [DOI] [PubMed] [Google Scholar]
- 10.Link DC, Schuettpelz LG, Shen D, et al. Identification of a novel TP53 cancer susceptibility mutation through whole-genome sequencing of a patient with therapy-related AML. JAMA. 2011 Apr 20;305(15):1568–1576. doi: 10.1001/jama.2011.473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bainbridge MN, Wiszniewski W, Murdock DR, et al. Whole-genome sequencing for optimized patient management. Sci Transl Med. 2011 Jun 15;3(87):87re83. doi: 10.1126/scitranslmed.3002243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Berg J, Evans J, Leigh M, et al. Next generation massively parallel sequencing of targeted exomes to identify genetic mutations in primary ciliary dyskinesia: Implications for application to clinical testing. Genetics in Medicine. 2011 Mar;13(3):218–229. doi: 10.1097/GIM.0b013e318203cff2. 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. [Accessed November 10, 2011]; http://www.bcm.edu/geneticlabs/index.cfm?PMID=21319.
- 14. [Accessed November 10, 2011]; http://www.genomeweb.com/sequencing/knome-adds-clia-certified-seqwright-sequencing-provider-personal-genome-services.
- 15. [Accessed November 10, 2011]; http://www.genomeweb.com/sequencing/partners-healthcare-centers-lmm-introduce-clinical-whole-genome-sequencing-inter.
- 16.Kohane I, Masys D, Altman R. The incidentalome: A threat to genomic medicine. JAMA. 2006 Jul 12;296(2):212–215. doi: 10.1001/jama.296.2.212. [DOI] [PubMed] [Google Scholar]
- 17.Berg JS, Khoury MJ, Evans JP. Deploying whole genome sequencing in clinical practice and public health: Meeting the challenge one bin at a time. Genetics in Medicine. 2011 Jun;13(6):499–504. doi: 10.1097/GIM.0b013e318220aaba. [DOI] [PubMed] [Google Scholar]
- 18.GeneTests. Medical Genetics Information Resource. 2011 http://www.genetests.org.