Abstract
In skin, multipotent stem cells generate the keratinocytes of the epidermis, sebaceous gland, and hair follicles. In this paper, we show that Tcf3 and Lef1 control these differentiation lineages. In contrast to Lef1, which requires Wnt signaling and stabilized β-catenin to express the hair-specific keratin genes and control hair differentiation, Tcf3 can act independently of its β-catenin interacting domain to suppress features of epidermal terminal differentiation, in which Tcf3 is normally shut off, and promote features of the follicle outer root sheath (ORS) and multipotent stem cells (bulge), the compartments which naturally express Tcf3. These aspects of Tcf3's action are dependent on its DNA binding and Groucho repressor-binding domains. In the absence of its β-catenin interacting domain, Lef1's behavior (ΔNLef1) seems to be markedly distinct from that of ΔNTcf3. ΔNLef1 does not suppress epidermal differentiation and promote ORS/bulge differentiation, but rather suppresses hair differentiation and gives rise to sebocyte differentiation. Taken together, these findings provide powerful evidence that the status of Tcf3/Lef complexes has a key role in controlling cell fate lineages in multipotent skin stem cells.
Keywords: β-Catenin, groucho, CtBP, Wnt/wingless, hair follicle
The canonical Wnt/Wingless signaling pathway directs cell fate in a variety of cell types and has an integral role in organogenesis. At the core of the pathway is the stability of β-catenin, a protein that serves a dual role in intercellular junction formation and in transcriptional regulation. Wnt signaling inhibits the amino-terminal phosphorylation and subsequent degradation of β-catenin, allowing cytoplasmic pools of β-catenin to enter the nucleus and bind to members of the Lef1/Tcf family of HMG domain-containing DNA-binding proteins. β-catenin's activation domain and Lef1/Tcf's DNA-binding domain combine to form a bipartite transcriptional activator of target genes (for review, see Willert and Nusse 1998; Korswagen and Clevers 1999; Waltzer and Bienz 1999).
Although Lef1/Tcf proteins often function as transactivators, these proteins can also interact with, and behave as, transcriptional repressors. CtBP represses a Wnt/Wingless response and selectively interacts with the carboxy-terminal domain of Tcf proteins, missing in Lef1 (Brannon et al. 1999). In contrast, the Groucho family of repressor proteins associate with the amino-terminal domain of all known Tcf/Lef family members (Cavallo et al. 1998; Roose et al. 1998; Brantjes et al. 2001). Genetically, a Drosophila dTcf–Groucho interaction antagonizes Wnt/Wingless signaling (Cavallo et al. 1998), and expression of Groucho proteins in Xenopus embryos can block axis formation and activation of β-catenin target genes, such as siamois and xnr3 (Roose et al. 1998). In zebrafish, embryonic Tcf3 appears to function as a transcriptional repressor, as an amino-terminally truncated Tcf3 (missing its β-catenin binding domain) complements the headless phenotype caused by loss-of-function of wild-type Tcf3 (Kim et al. 2000). Therefore, in the absence of a Wnt/Wingless signal, the association of repressor proteins with Tcfs can in some instances repress the function of genes regulated by Tcf/Lef-binding sites.
Most studies in mammals have emphasized the transactivating powers of Tcf/Lef proteins. Mutations that lead to constitutive stabilization of β-catenin are associated with a stunning variety of tumors, including those of the hair follicle (Chan et al. 1999; Roose and Clevers 1999). Even in situations such as this, however, repressor actions of Tcf complexes have been described. Therefore, for example, in a human colon cancer line, even though the levels of stabilized β-catenin are high and lead to Tcf4–β-catenin transactivation of proliferation-associated genes, the cells also produce a truncated form of Tcf1 that is unable to bind β-catenin, and in turn causes the attenuation of Tcf4-mediated target gene transcription (Roose et al. 1999). This repressor feedback mechanism explains why Tcf1−/− mice develop intestinal and colon neoplasms that have superactivated Tcf4–β-catenin complexes (Roose et al. 1999). Naturally occurring, alternatively spliced forms of Lef1 transcripts have also been described (Hovanes et al. 2000, 2001). This raises the possibility that such forms might act as repressors in normal developmental processes, affording an additional mechanism by which Tcf/Lef proteins might mediate repression of genes regulated by Tcf/Lef-binding sites.
The skin expresses several members of the Tcf/Lef family as well as a number of Wnts (van Genderen et al. 1994; Zhou et al. 1995; Millar et al. 1999; St. Jacques et al. 1998; DasGupta and Fuchs 1999). The skin epithelium is derived from multipotent stem cells, which in the adult, reside in a region of the outer root sheath (ORS) of the follicle known as the bulge (Taylor et al. 2000; Oshima et al. 2001). These stem cells display differentiation properties similar to those of the ORS. The stem cells, however, change these properties as they differentiate and give rise to the epidermis, sebaceous gland, and the rest of the hair follicle (Fig. 1A).
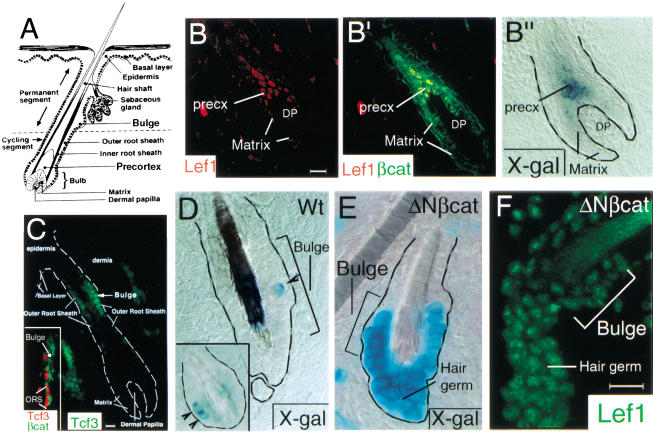
Figure 1.
Expression of Tcf3, Lef1, nuclear β-catenin, and TOPGAL activity in the hair follicle. (A) Structure of the hair follicle. (B) The bulb region of an anagen hair follicle. Double immunofluorescent staining of Lef1 (red in B, B‘) and β-catenin (green in B‘) showed that β-catenin is present at cell borders in all cells of the hair follicle, but nuclear β-catenin localization is apparent only in the precortex cells, which contain nuclear Lef1. (B“) X-gal staining of TOPGAL transgenic mice showed that activation of this reporter construct was associated with the colocalization of nuclear β-catenin and Lef1. (C) Immunofluorescent staining of Tcf3 with a mouse monoclonal antibody (green; large panel) showing prominent staining in the bulge with staining extending to the ORS. (C, inset) Double immunofluorescent localization of Tcf3 with a guinea pig polyclonal antibody (red) and β-catenin (green) showed Tcf3 present in bulge and ORS cell nuclei and β-catenin present primarily at cell borders. (D–F) The bulge region of the hair follicle during the hair cycle. (D) X-gal staining of a TOPGAL single transgenic hair follicle showed the activation of the reporter in a small number of bulge cells (arrowheads). (E) In TOPGAL/K14–ΔNβcat double transgenic hair follicles, most bulge cells of the hair follicle displayed TOPGAL activity. (F) Immunofluorescent staining of Lef1 in skin sections from K14–ΔNβcat mice. Note that Lef1 protein was up-regulated in the bulge region and secondary hair germ of K14–ΔNβcat skin. (precx) Precortex; (DP) dermal papilla.
Stem cells of the skin express Tcf3 (Fig. 1C; DasGupta and Fuchs 1999). In the majority of these cells, β-catenin appears restricted to cell borders (Fig. 1C, inset). The cells that exit the bulge and migrate upward to populate the epidermis and sebaceous gland lose Tcf3 expression. At the start of the hair cycle, as the mesenchymal dermal papilla cells come into contact with the bulge, a downgrowth from the bulge (called the secondary hair germ) represents the first signs of the regrowth of the “cycling segment” of the hair follicle (Fig. 1A). Cells that stream from the bulge partition into those that maintain Tcf3 expression, which then go on to form the ORS of the follicle, and those that switch from Tcf3 to Lef1, which then go on to form the transiently amplifying matrix cells that produce the inner root sheath (IRS) and the hair (see Fig. 1A; DasGupta and Fuchs 1999). Concomitant with the differentiation of matrix cells to hair cells, β-catenin and Lef1 concentrate in the nucleus (Fig. 1B). In transgenic mice, these events coincide with the induction of the Lef1/Tcf reporter gene (TOPGAL: Lef1/Tcf optimal promoter-β-galactosidase), suggesting that the hair-specific lineage may be controlled by Wnt signaling and the transactivation of Lef1-regulated genes (Fig. 1B“; DasGupta and Fuchs 1999).
The functional significance of Lef1 and β-catenin to hair follicle morphogenesis is supported by the fact that Lef1−/− mice display a paucity of hair follicles (van Genderen et al. 1994). Moreover, when the keratin 14 (K14) promoter, active in the epidermis, ORS and bulge, was used to express Lef1 in transgenic mice, hair germ-like invaginations were detected in the epidermis (Zhou et al. 1995). When a constitutively stable form of β-catenin (ΔN87βcat) was expressed under the control of the same K14 promoter, hair follicle morphogenesis and hair differentiation occurred in cells derived from the epidermis, ORS, and bulge (Gat et al. 1998). Lef1 expression coincided with hair differentiation in these mice (Gat et al. 1998). Taken together, these findings suggest that Lef1 and β-catenin are transactivators necessary for manifestation of the hair cell fate.
Given the importance of Lef1 and β-catenin in hair differentiation, we investigated what the role of Tcf3 might be in this regard. In examining the Tcf3 positive bulge compartment and ORS at different stages of the hair cycle, TOPGAL activity appeared silent. At the start of the hair cycle, however, one or two bulge cells of most follicles displayed signs of TOPGAL activity (examples shown in Fig. 1D). In mice expressing high levels of stabilized β-catenin (K14–ΔN87βcat; Gat et al. 1998) in the bulge, many more stem cells activated TOPGAL specifically at the start of the cycle (Fig. 1E). Intriguingly, this induction occurred concomitantly with the appearance of Lef1 in these stem cells and in the matrix cells of the hair germ that formed (Fig. 1F). These findings suggest that the signal that initiates each hair cycle may have an impact directly on changes in Tcf3/Lef1 gene expression and also on the activity of genes that are regulated by Tcf/Lef proteins.
The largely distinctive patterns of TOPGAL and Tcf3 activity also suggested that Lef/Tcf target genes may often be repressed or inactive when Tcf3 is expressed. To explore further the underlying significance of Tcf3 expression in the stem cells and the potential importance of changes in Tcf/Lef-mediated gene expression to other programs of differentiation in keratinocytes, we examined the behavior of Tcf3 and Lef1 on Lef1/Tcf-regulated genes in keratinocytes in vitro and on stem cell differentiation in transgenic mice. We also explored the behavior of Tcf3 and Lef1 when their β-catenin-binding domains are missing, thereby crippling their ability to transactivate genes in response to Wnt signaling. Our data suggest that Tcf3 and ΔNTcf3 function as repressors, Lef1 functions as an activator, and ΔNLef1 acts to interfere with Lef/Tcf activator function. These behaviors manifest themselves in remarkable alterations in the differentiation pathways taken by the stem cells when various Lef1/Tcf3 transgenes are expressed.
Results
In cultured primary keratinocytes, Lef1 and Tcf3 exhibit different activities that cannot be explained by their ability to associate with and localize β-catenin to the nucleus
To express Tcf3 and Lef1 in skin epithelial cells, we engineered the vectors shown in Figure 2A. The K14 promoter/enhancer is active in primary cultured keratinocytes, in multipotent embryonic skin epithelium, in the stem cell compartment (bulge) of adult skin and its ORS and epidermal progeny (Vasioukhin et al. 1999). Because in vivo and in vitro, the skin epithelia are derived from keratinocytes in which the K14 promoter/enhancer is active, it was optimal for assessing the consequences to stem cell differentiation when Tcf3 and/or Lef1 expression are altered. For comparative purposes, we engineered mammalian Lef1/Tcf3 cDNAs to either encode or not a myc epitope tag at the amino terminus of the protein. Immunoblot analyses with an anti-myc antibody revealed roughly comparable levels of expression of all transgene products (data not shown). Both in vitro (these data) and in vivo (see below), the addition of the epitope tag did not appear to affect the activity of the proteins or their stability.
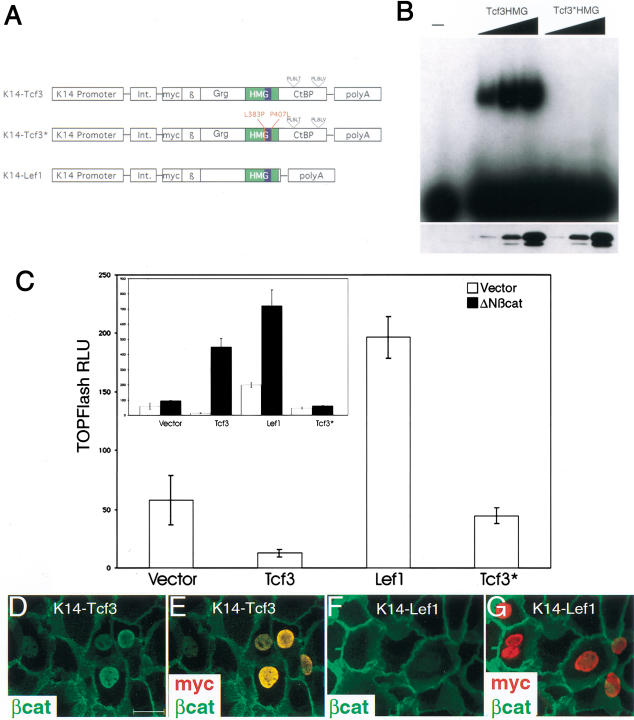
Figure 2.
In keratinocytes, Tcf3 and Lef1 have different effects on target gene activation and on nuclear localization of β-catenin. (A) Schematic diagrams of constructs illustrating key protein domains of Tcf3, Tcf3*, and Lef1. The α-helical DNA-interacting segment is denoted in blue and the mutated amino acids in Tcf3* are indicated in red. The two CtBP-binding motifs in the carboxyl terminus of Tcf3 and Tcf3* (PLSLT and PLSLV) are inidicated. (Int.) Intron; (myc) myc-epitope tag; (β) β-catenin-binding domain; (Grg) Grg-binding domain; (CtBP) CtBP-binding region. (B, top) EMSA using recombinant 6×HisTcf3 HMG and 6×HisTcf3* HMG proteins. Recombinant Tcf3 HMG retarded the mobility of the 32P-labeled TCRα oligo in a concentration dependent manner, whereas Tcf3* HMG did not bind the DNA. (B, bottom) Western blot using anti-pentaHis monoclonal antibody to detect the Tcf3 HMG and Tcf3* HMG at the concentrations used for the EMSA. (C) Primary human keratinocytes were transfected with TOPFlash reporter, the indicated Lef1/Tcf3, and either empty vector (open bars) or K14–ΔNβcat (solid bars, inset). Values represent the average of duplicate transfections +/− standard deviation, and this graph is a representative from three separate experiments. Tcf3 repressed TOPFlash activation, whereas Lef1 activated it. Tcf3* had no major effect on TOPflash activity. (D–G) Keratinocytes grown on coverslips were transfected with either K14–Tcf3 (D,E) or K14–Lef1 (F,G), given high Ca++ to induce junction formation, and processed for immunofluorescence to detect endogenous β-catenin (green) and transfected Lef1 or Tcf3 (anti-myc; red). Note endogenous β-catenin was restricted to the junctions in nontransfected and Lef1 transfected cells; note also that in Tcf transfected cells, β-catenin also displayed nuclear localization.
To verify that any inhibitory effects of Tcf3 would be attributable to its ability to bind DNA, we used a mutant form of Tcf3 (designated Tcf3*) harboring L383P and P407I mutations in the HMG DNA-binding domain (Fig. 2A). Flanking an α-helix required for high-affinity DNA binding, these key residues are thought to stabilize the contacts made with the minor groove of the DNA (Love et al. 1995). We confirmed this by using an electrophoretic mobility-shift assay (EMSA) performed with lysates from bacteria expressing either wild-typeTcf3 HMG or Tcf3* HMG (amino acids 331–427) (Fig. 2B). The Tcf3* mutation did not affect protein stability or interaction with β-catenin, as judged by SDS-PAGE of the expressed proteins (Fig. 2B, bottom), and like Tcf3, Tcf3* elicited nuclear localization of an epitope-tagged version of one of the Groucho proteins (Grg5) when coexpressed in keratinocytes (data not shown; see mouse data below).
Keratinocytes were cotransfected with the TOPFlash (Tcf Optimal Promoter + luciferase) reporter plasmid and K14–Lef1, K14–Tcf3*, and K14–Tcf3. Under normal conditions, that is, in the absence of exogenous β-catenin, Tcf3 repressed whereas Lef1 stimulated the basal level of TOPFlash activity in primary human keratinocytes (Fig. 2C). When cotransfected with K14–ΔN87βcat to deliver high levels of stable β-catenin, however, both Lef1 and Tcf3 stimulated TOPFlash activity (Fig. 2C, inset). The repressor and activator activity of mouse Tcf3 was dependent on its ability to bind DNA, as judged by the inactivity of Tcf3* in these assays (shown) and as judged by the failure of the proteins to transactivate or repress FOPFlash, containing mutations in the DNA-binding sites (data not shown; see Molenaar et al. 1996; DasGupta and Fuchs 1999). These data with mouse Tcf3 are consistent with those of Molenaar et al. (1996), who showed that high levels of β-catenin could activate Xenopus Tcf3 in cultured B cells.
To determine whether the results reflected an increased efficiency of Lef1 over Tcf3 to bind to and/or localize β-catenin in the nucleus, we processed transfections for immunofluorescence microscopy. Unexpectedly, ∼80% of Tcf3-expressing keratinocytes displayed nuclear anti-β-catenin staining (Fig. 2D,E) under conditions where <20% of Lef1 expressing cells showed discernable nuclear staining (Fig. 2F,G). In cells cotransfected with K14–ΔN87βcat (Gat et al. 1998), nuclear β-catenin was seen in most cells expressing either Lef1 or Tcf3 (data not shown). Therefore, although Lef1 was the more potent transactivator, Tcf3 was more efficient at concentrating β-catenin in the nucleus. To convert Tcf3 from a repressor to a transactivator in keratinocytes, very high concentrations of β-catenin seemed to be required, perhaps to titrate out or compete with some negative regulator in Tcf3 activation. Immunofluorescence staining with a panel antibody showed nuclear localization of the Groucho repressor proteins (data not shown; see in vivo data below).
Tcf3 and Tcf3* both translocate endogenous β-catenin in nuclei of transgenic mouse epidermis, ORS, and bulge, but only Tcf3 affects skin function
Normally, Tcf3 is restricted to the stem cells and the lower ORS, but is down-regulated during upward migration of cells from the bulge to the upper ORS and basal layer of the epidermis (DasGupta and Fuchs 1999). To examine the consequences of maintaining Tcf3 in this compartment, we generated mice expressing the genes shown in Figure 2A. To select mice for study that expressed comparable transgene expression levels, we used the myc-epitope tagged genes after verifying that the K14–Lef1 myc-tagged transgene gave a comparable phenotype to that reported for the untagged K14–Lef1 transgene (for details, see Zhou et al. 1995).
A total of seven K14–Tcf3, two K14–Tcf3*, and two K14–Lef1 transgenic mice were born and determined to express the epitope-tagged transgene products. Figure 3A shows anti-myc immunoblot detections of representative examples for equivalent loadings of skin protein extracts. Of our Tcf3-expressing animals, some expressed higher and some lower levels than the example shown. Over a range of ∼10×, the phenotypes were comparable. The sizes of the bands detected with the anti-myc antibody were as expected, with Tcf3 being considerably larger (∼80 kD) than Lef1 (∼55 kD) (Zhou et al. 1995; Pukrop et al. 2000). In both cultured epithelial cells and transgenic skin, Tcf3* consistently migrated slightly faster than Tcf3. Such variation in migration is relatively common even with slight variations in protein sequences, although differences in posttranslational modifications have not been ruled out.
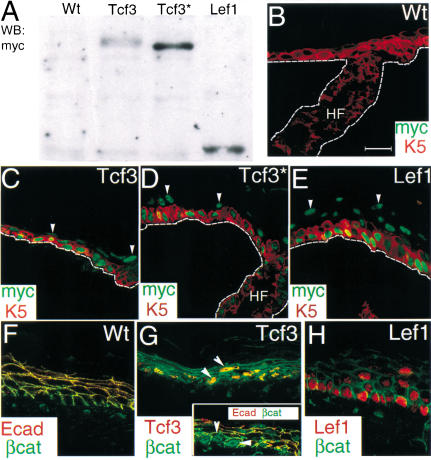
Figure 3.
Expression of K14–Tcf3, K14–Tcf3*, and K14–Lef1 in transgenic mice. (A) Western blot analysis of total skin protein from transgenic mice. Equal levels of protein extracts were resolved by 7% SDS-PAGE gel, and transgene products were detected by anti-myc monoclonal antibody. (B–E) Double immunofluorescent staining of newborn mouse back skin sections. Mouse anti-myc (green) was used to detect transgene expression and rabbit anti-K5 (red) used to highlight the basal layer of the epidermis and the ORS/bulge. Note that transgenic protein was stable even in suprabasal layers (arrowheads). The broken lines demarcate dermal–epidermal boundaries. (F–H) Localization of β-catenin in newborn back skin sections. (F) β-catenin (green) was restricted to the cell borders where it colocalized with E-cadherin (red) in wild-type epidermis. (G) Double immunofluorescence using anti-β-catenin (green) and anti-Tcf3 (red) antibodies shows colocalization of Tcf3 and β-catenin in epidermal nuclei (arrowheads) of K14–Tcf3 transgenic mice. (G, inset) Nuclear localization of β-catenin (green; arrowheads) was distinct from cell borders identified by E-cadherin staining (red). (H) Double immunofluorescent staining using anti-β-catenin (green) and anti-Lef1 (red) antibodies shows that transgenic expression of Lef1 did not appear to cause nuclear localization of β-catenin in K14–Lef1 mice.
Anti-myc antibodies did not detect the endogenous c-myc protein present in skin (Fig. 3B). Anti-myc did detect transgene expression, however, which in epidermis was targeted to the basal layer but persisted in some suprabasal layers, suggesting that the proteins were relatively stable (Fig. 3C–E). In all cases, the transgene product concentrated in the nucleus, consistent with the nuclear localization sequences located in these HMG proteins (Prieve et al. 1996). This was also true for Tcf3*, suggesting that nuclear localization was not dependent on the ability of the protein to bind DNA.
Consistent with our in vitro observations, endogenous β-catenin was detected in the nuclei of Tcf3- but not Lef1-expressing epidermis (Fig 3F–H). This behavior occurred irrespective of the ability of Tcf3 to bind DNA (data not shown). Nuclear export of β-catenin is regulated, at least in part, by the binding of APC (Henderson 2000; Rosin-Arbesfeld et al. 2000). Because the association of β-catenin with APC and Lef1/Tcf proteins is mutually exclusive, it seems likely that this increase in nuclear β-catenin reflects an increase in Tcf3–β-catenin complexes. We did not pursue this, however, because we later show that this localization is not relevant to the Tcf3-mediated changes in differentiation program that we observe.
In ORS (Tcf3+) and bulge (Tcf3+), K14–Tcf3 expression has no apparent affect, but in epidermis (Tcf3−), it suppresses barrier function and terminal differentiation
Transgenic mice expressing Tcf3 failed to thrive and they either died or were sacrificed within 48 h after birth. These features were not observed in the nontransgenic littermates, nor were they characteristics of K14–Lef1 or K14–Tcf3* transgenic mice, all of which remained healthy and fertile throughout life.
To explore the nature of the neonatal lethality, we investigated the integrity of the skin and oral epithelia, where the K14 promoter is active. Epidermis functions as a barrier to maintain body hydration and exclude harmful external agents. The barrier can be measured by the ability of skin epidermis to exclude penetration of dyes, such as 5-bromo-4-chloro-3-indoyl-β-D-galactopyranoside (Xgal) (Hardman et al. 1998; Segre et al. 1999). In wild-type littermates, the barrier initiated at embryonic day 16 (E16), and was complete by E18.5 (Fig. 4A,B; see also Hardman et al. 1998). All the K14–Tcf3 transgenic embryos examined failed to acquire barrier function by E18.5 (Fig. 4C,D). This barrier defect did not appear to be caused by a delay in development, because the Tcf3 embryos were of normal size, had opened eyelids, and exhibited a distinctive Zorro-like mask of blue dye not characteristic of the normal developmental pattern of barrier acquisition. Additionally, K14–Tcf3 embryos were as large or larger than wild-type littermates.
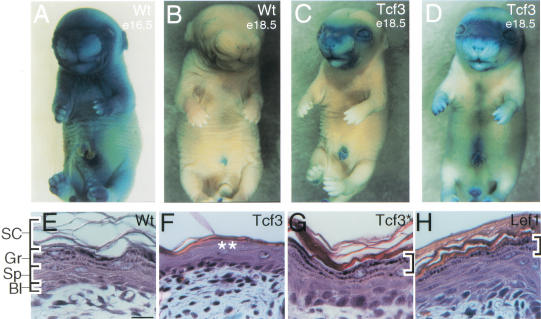
Figure 4.
Transgenic expression of Tcf3 perturbs epidermal barrier formation and late-stage epidermal differentiation. (A–D) Barrier function assays. An endogenous β-galactosidase activity in embryonic skin allows for detection of permeability to small molecules (i.e., Xgal). (A,B) Wild-type embryos acquire barrier function between E16.5–18.5 (Hardman et al. 1998). The dorsal–ventral progression is characteristic of nontransgenic littermates. (C,D) Barrier formation in K14–Tcf3 was incomplete, even at E18.5. Note the Zorro-like mask of blue staining and the opened eyelids. (E–H) Hematoxylin and eosin stained sections of newborn back skins of mice with genotypes indicated in the upper right corners. Basal (Bl) and spinous (Sp) layers of the epidermis appear histologically normal in each sample. The basophilic keratohyalin granules (brackets in E,G,H) are distinctly absent from the K14–Tcf3 epidermis (asterisks in F).
The skin barrier is acquired by the process of epidermal terminal differentiation. On exhaustion of their proliferative potential, basal cells exit the cell cycle, detach from the underlying basement membrane and terminally differentiate as they move toward the skin surface (Fuchs 1997). In transit, they undergo a series of transcriptional changes, which culminate in the production of dead cells. The cells consist of a proteinaceous sac, called the cornified envelope, which encases densely packed keratin filaments cemented by the protein filaggrin. These cellular ghosts are physically sealed through lipids that are secreted in the latter stages of the differentiation process.
The inner layers, that is, the basal (BL) and spinous (SP) layers, of wild-type and K14–Tcf3 epidermis appeared morphologically similar (Fig. 4E,F). In contrast, late-stage terminal differentiation was affected by Tcf3 expression. Wild-type granular layers (Gr) display prominent keratohyalin granules composed of the large precursor protein, profilaggrin. Late in differentiation, profilaggrin is extensively processed to liberate filaggrin, which then associates with and bundles keratin filaments (for review, see Fuchs 1997). In K14–Tcf3 epidermis, keratohyalin granules were either attenuated or completely absent (Fig. 4F, asterisks). Neither K14–Tcf3* nor K14–Lef1 skin displayed this defect (Fig. 4G,H), suggesting that the effect was dependent on both the Lef1/Tcf member involved and its ability to bind DNA. Similar defects were seen in the dorsal tongue epithelium of K14–Tcf3 mice, perhaps explaining why the K14–Tcf3 mice failed to feed.
Because the mice died shortly after birth, we were unable to assess the consequences of Tcf3 transgene expression to the hair cycle. However, no obvious abnormalities were seen in either the morphology or the biochemistry of the early postnatal hair follicles (data not shown). Therefore, rather than generate hair follicle-like structures as seen in K14–Lef1 and K14–ΔN87βcat mice (Zhou et al. 1995; Gat et al. 1998), K14–Tcf3 appeared to elicit defects in late-stage epidermal differentiation. To some extent, the morphological characteristics of the epithelium seen in K14–Tcf3-expressing mice resembled those of the stem cell compartment (bulge) and the lower portion of the ORS of the hair follicle, that is, the cells that normally express Tcf3. Interestingly, although K14–Tcf3 resulted in nuclear localization of β-catenin in both ORS and bulge, no obvious abnormalities were detected in these cells. Therefore, transgenic Tcf3 expression did not appear to cause deleterious effects to the keratinocytes that naturally expressed Tcf3, but it did affect the epidermal keratinocytes, which normally cease to express Tcf3 when they become committed to their epidermal fate.
Biochemical changes in K14–Tcf3 epidermis are consistent with suppression of terminal differentiation and promotion of features characteristic of the bulge/ORS
As expected from the morphology of K14–Tcf3 skin, cell proliferation markers (Ki67) and early markers of differentiation (K5, K14, K1, and involucrin) were normal (data not shown). Late stage markers, however, revealed biochemical signs indicative of partial repression of epidermal differentiation restricted to the K14–Tcf3 transgenic skin. Therefore, the paucity of keratohyalin granules was reflected by a marked reduction in anti-filaggrin staining (Fig. 5A–D), and the major constituent of the cornified envelope, loricrin, also appeared to be atypical (Fig. 5E–H). Cornified envelopes were still made, however, and transglutaminase was expressed in K14–Tcf3 epidermis (data not shown). Taken together, these results suggest that expression of Tcf3, but not Lef1, suppresses some, but not all, aspects of late-stage terminal differentiation in epidermis.
Figure 5.
The appearance of differentiation features characteristic of ORS/bulge in transgenic epidermis expressing Tcf3, but not Lef1 nor Tcf3*. Each panel (A–P) shows double immunofluorescent staining of newborn back skin sections from the mice with the genotype indicated in upper right corner. Mouse anti-myc (green) staining was used to detect transgene expression, and rabbit polyclonal antibodies (red) were used to detect filaggrin, loricrin, K17, or K6 as indicated. (A–D) Compared with the normal punctate pattern of anti-filaggrin staining (red in A,C,D), filaggrin staining was less intense and more diffuse in K14–Tcf3 epidermis (B). (E–H) Compared with the normal pattern of anti-loricrin staining (red in E,G,H) which is localized to cell borders and distinct from keratohyalin granules, the loricrin staining of K14–Tcf3 epidermis was more intense and nearly uniform in suprabasal layers (F). (I–L) Expression of K17 (red) was restricted to the ORS of nontransgenic (I), K14–Tcf3* (K), and K14–Lef1 (L) newborn back skin. In contrast, K17 was expanded to all layers of the epidermis in K14–Tcf3 mice (J). (M–P) K6 (red) was restricted to the inner layer of the ORS in non-transgenic (M), K14–Tcf3* (O), and K14–Lef1 (P) newborn back skin, but it was induced in suprabasal layers of the epidermis in K14–Tcf3 (N) mice.
K5, K14, transglutaminase, and involucrin have been identified as components of the ORS/bulge, as well as epidermis. Therefore, we examined K17, a keratin whose expression is down-regulated in epidermis and elevated in the ORS and bulge just before birth (Fig. 5I) (McGowan and Coulombe 1998). In postnatal K14–Tcf3 epidermis, K17 expression persisted (Fig. 5I–L). Similarly, K6 and its partner K16, which are normally restricted to the inner (suprabasal) layers of the ORS, were also induced (Fig. 5M–P). Analogous to its expression pattern in the ORS and bulge, K6 was seen only in the suprabasal epidermal layers. The induction of K6 and K17 expression in K14–Tcf3 transgenic skin was not pronounced in K14–Tcf3* or K14–Lef1 transgenic skin, suggesting its specificity for Tcf3 expression. Taken together, the morphological and biochemical data suggest that transgenic expression of Tcf3 suppresses features characteristic of skin epithelial cell types lacking Tcf3 (epidermis) and promotes those typical of cell types expressing Tcf3 (ORS and bulge).
In keratinocytes, the repressive effects of Tcf3 are dependent upon its Grg-interaction domain
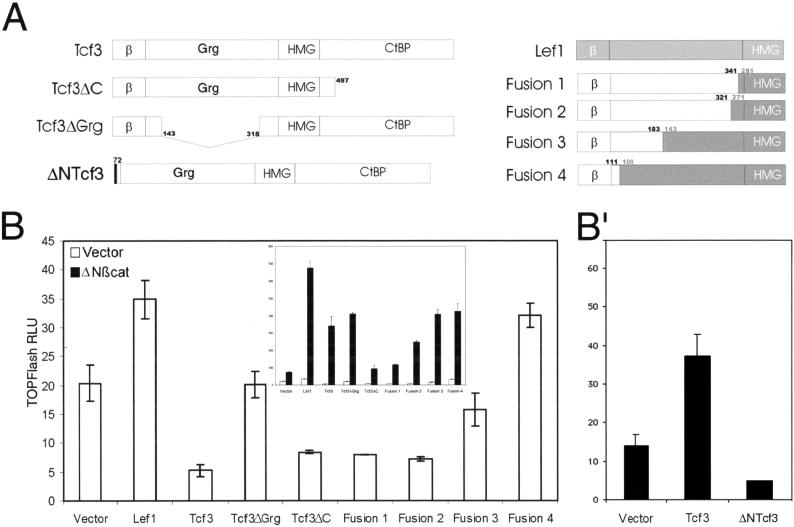
XTcf3 interacts with at least two different repressor proteins, CtBP and members of the Groucho family (Brannon et al. 1999; Brantjes et al. 2001), making their mouse homologs likely candidates to mediate mammalian Tcf3 repression of TOPFlash activity in keratinocytes. To investigate this possibility, we generated and tested Tcf3 mutants harboring deletions in the putative CtBP (Tcf3ΔC), Grg (Tcf3ΔGrg), and β-catenin (ΔNTcf3) interacting domains (Fig. 6). As judged by immunoblot analyses, these and other mutant forms of Tcf3 and Lef1 shown in Figure 6A were comparably expressed in cultured epithelial cells (data not shown). WhereasTcf3ΔC only modestly affected Tcf3-mediated repression of basal TOPFlash activity, Tcf3ΔGrg relieved repression. These findings suggested that of the two, Grg interactions may be more relevant in mediating Tcf3 repression in keratinocytes. Recent studies by Brantjes et al. (2001) have shown that like Drosophila Groucho, mammalian Groucho proteins associate with histone deacetylases and with vertebrate Tcfs, thereby suggesting a mechanism for the repressor activity observed here.
Figure 6.
Tcf3 requires the Grg-binding domain but not the CtBP-binding domains for repressor activity in keratinocytes. (A) Diagram of Tcf3 mutants and Tcf3–Lef1 fusions used in frame B (see below). For fusion proteins, Tcf3 regions are white, and Lef1 regions are gray. Numbers above each fusion correspond to the amino acids of Tcf3 (black numbers) and Lef1 (gray numbers) at the junctions of the fusions. (B) Primary human keratinocytes were transfected and processed as described in Fig. 2. Note that Tcf3-mediated repression of basal TOPFlash activity required the Grg-interaction domain, but not the carboxy-terminal segment. Note also that the ΔNβcat-mediated activation of TOPFlash required the carboxy-terminal segment, but not the Grg-interacting domain. The requirement of the carboxy-terminal segment for ΔNβcat-mediated activation was influenced by residues 183–341 of the Tcf3 protein, as judged by the fact that Fusion 3 activated, whereas Fusion 1 did not. (B‘) The amino-terminal 71 amino acids of Tcf3 were not required for repression, but were required for ΔNβcat-mediated activation. Transfections in B′ were conducted in the presence of ΔNβcat.
To further characterize the region of Tcf3 required for repression and assess the basis for the difference in activity between Lef1 and Tcf3, we engineered and tested a series of Lef1–Tcf3 fusion proteins for their effect on the TOPFlash reporter (Fig. 6). When the majority of Tcf3's amino-terminal segment (321 or 341 amino acids) was fused to the carboxy-terminal segment of Lef1 including the HMG domain (Fig. 6B, Fusions 1 or 2, respectively), these hybrid proteins effectively repressed basal TOPFlash activity in a fashion similar to that of Tcf3ΔC. With only part of the amino-terminal segment spanning the Grg domain (183 amino acids), the Tcf3–Lef1 hybrid lost most of its ability to repress (Fig. 6B, Fusion 3). With only 111 amino acids of Tcf3's unique amino-terminal segment, The Tcf3–Lef1 hybrid protein behaved as a transactivator (Fig. 6B, Fusion 4). Taken together, these findings illuminated a marked difference in the segment between the β-catenin and DNA-binding domains of these proteins.
Additional insights were obtained when the Tcf3 mutants and Tcf3–Lef1 hybrids were coexpressed with the ΔN87βcatenin. Although Tcf3ΔC and Tcf3 had repressed basal TOPFlash activity to a similar degree, Tcf3ΔC did not stimulate TOPFlash like Tcf3 did in the presence of elevated levels of stabilized β-catenin (Fig. 6B, inset). Therefore, in keratinocytes, the carboxy-terminal segment of Tcf3 seemed to be necessary to enable full-length Tcf3 to activate target genes in the presence of high levels of β-catenin. In the absence of the putative Grg-interacting domain, this requirement was alleviated, as Tcf3ΔGrg and the Tcf3–Lef1 fusion proteins lacking this segment of Tcf3 were able to transactivate TOPFlash. Based on these data, it seems that in the presence of high levels of stabilized β-catenin, the carboxy- and amino-terminal segments of Tcf3 interact either indirectly or directly to adopt a conformation that does not interfere with the β-catenin and/or HMG domain transactivating functions.
Finally, we examined the consequences to Tcf3 function on removal of its β-catenin-interacting domain (Fig. 6B“). In the presence of exogenous β-catenin, ΔNTcf3 repressed TOPFlash activity below levels of ΔN87βcatenin alone.
The transdifferentiation seen in the epidermis of K14–Tcf3 mice is dependent on the Groucho interacting domain but not on the CtBP or the β-catenin interacting domains
Our in vitro studies led to the hypothesis that in keratinocytes, Tcf3 acts as a repressor and that this effect is mediated primarily through the putative Grg-interacting domain in the amino-terminal segment of the protein. The induction of K17 and K6 and the nuclear localization of β-catenin on Tcf3 expression in skin, however, left open the possibility that in vivo, Tcf3 might function as a transactivator. To evaluate this possibility, we engineered transgenic mice expressing K14–Tcf3ΔC, K14–Tcf3ΔGrg, and K14–ΔNTcf3 (Fig. 7A).
Figure 7.
Repressor activity of Tcf3 is required for its potent effects in transgenic mice. (A) Schematic diagrams as described in the legend to Fig. 2. (B,C) Hematoxylin and eosin stained sections of back skin. Note the lack of keratohyalin granules in K14–Tcf3ΔC (asterisks in B) and the normal granular layer in K14–Tcf3ΔGrg (bracket in C). (D–G) Double immunofluorescent detection of transgene protein (green) and either filaggrin (red; D,E) or loricrin (red; F,G). Note that defects in anti-filaggrin and anti-loricrin staining were even more severe in K14–Tcf3ΔC skin than in K14–Tcf3 skin. Note normal staining in K14–Tcf3ΔGrg skin. (H–K) SDS-PAGE and Western blot analysis of equal amounts of total skin protein extracts from 4-day-old nontransgenic, K14–Tcf3ΔC and K14–Tcf3ΔGrg mice. (H) Coomassie-stained gel showing comparable loading of samples. (I) Anti-myc Western blot to illustrate levels of transgene expression. (J) Anti-filaggrin Western blot. Note that mature filaggrin and proteolytically processed pro-filaggrin were decreased in K14–Tcf3ΔC mice but were normal in K14–Tcf3ΔGrg mice. (K) Anti-loricrin Western blot confirming that loricrin levels were diminished in K14–Tcf3ΔC mice but normal in K14–Tcf3ΔGrg mice. (L) Immunohistochemical detection of Grg proteins in nontransgenic epidermis using a panTLE antibody that recognizes all known Grg/TLE proteins. Note that staining was restricted to nuclei and was more intense in suprabasal layers compared with the basal layer. Anti-Grg antibody staining was also seen in the nuclei of cultured keratinocytes. (M) Hematoxylin and eosin stained skin section from K14–ΔNTcf3 mouse. Note the lack of keratohyalin granules.
As predicted from our in vitro studies, the overall phenotype of K14–Tcf3ΔC transgenic mice was similar to that of K14–Tcf3 mice. Two mice survived until postnatal day 3 and one died at postnatal day 4. Tcf3ΔC-expressing epidermis displayed a lack of keratohyalin granules in both newborn (data not shown) and four-day-old mouse samples (Fig. 7B). In contrast, all five independent K14–Tcf3ΔGrg founder animals survived to adulthood, produced F1 offspring, and displayed no apparent defects in viability or fertility, although high expressing lines did show an approximately one day premature opening of eyelids. Tcf3ΔGrg-expressing mice also displayed normal skin morphology (Fig. 7C). Despite these differences between Tcf3ΔC and Tcf3ΔGrg, transgene expression levels were comparable (Fig. 7H,I; Coomassie and anti-myc immunoblot of skin proteins).
The lack of keratohyalin granules in Tcf3ΔC-expressing skin was supported by the reduction in anti-filaggrin and anti-loricrin staining observed in skin sections and by the reduction in these proteins detected by immunoblot analyses with these antibodies (Fig. 7D–G,J,K). In addition, as suggested by the bulge/ORS morphology of K14–Tcf3ΔC epidermis, it displayed K17 and K6 staining in patterns analogous to that noted for K14–Tcf3 transgenic epidermis (data not shown). In contrast, Tcf3ΔGrg-expressing skin displayed normal staining and protein expression patterns. The loss of K14–Tcf3 phenotype associated with mutation of the Grg-interacting domain suggested that Grg proteins may exist in the skin and contribute to the Tcf3-mediated abnormalities in epidermal differentiation. Consistent with this notion was the fact that a panel antibody (pan-TLE) against mammalian Grg proteins labeled the nuclei of wild-type epidermis (Fig. 7L).
As we had observed previously for the Tcf3 and Tcf3*, Tcf3ΔC and Tcf3ΔGrg expression resulted in the nuclear localization of endogenous β-catenin (data not shown). Because both Tcf3* and Tcf3ΔGrg transgenic mice exhibited generally good health, the nuclear colocalization of Tcf3 and β-catenin, per se, did not appear to be sufficient to cause the phenotypic changes associated with the K14–Tcf3 and K14–Tcf3ΔC transgenic mice. To assess whether the β-catenin interaction domain of Tcf3 was necessary for manifesting the K14–Tcf3 phenotype in vivo, we examined K14–ΔNTcf3 transgenic mice. K14–ΔNTcf3 mice displayed open eyelids at birth, were frail, and died or were sacrificed within a day. As shown in Figure 7M, the morphology of the skin of these mice was comparable to that seen in the K14–Tcf3 and K14–Tcf3ΔC animals. Furthermore, K17 and K6 expression were still seen in the K14–ΔNTcf3 skin (data not shown). Taken together, it seems most likely that the activation of genes associated with ORS/bulge differentiation in Tcf3 transgenic epidermis arises indirectly from repression of Tcf3-regulated genes. A further understanding of this process awaits identification of Tcf3 target genes.
In the absence of the β-catenin-interacting domain, transgenic Lef1 blocks hair cell differentiation and induces sebaceous cell differentiation in its place
Given that Tcf3 and ΔNTcf3 behaved similarly in vitro and in vivo, we wondered whether Lef1 might behave similarly to Tcf3 when its ability to associate with β-catenin was compromised. We first examined the difference in behavior of ΔNTcf3 and ΔNLef1 on activation of target genes in vitro. Because Lef1 normally functions in hair follicle morphogenesis, and because the hair keratin genes were shown previously to possess sequences that bind to Lef1 (Zhou et al. 1995), we used the promoter/enhancer for one of these genes (mHK1a) driving luciferase as a reporter. This gene is normally expressed in the precortical cells that exhibit TOPGAL activity and nuclear β-catenin localization (see Fig. 1).
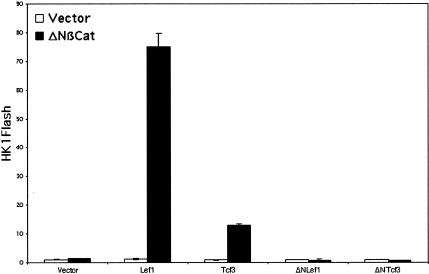
As shown in Figure 8, neither Lef1 nor β-catenin (ΔN87βcat) alone stimulated luciferase activity appreciably when transfected in cultured keratinocytes. Activity levels rose nearly 80×, however, when Lef1 and β-catenin were coexpressed. Tcf3's effect on this promoter was considerably less potent, and in this way, was similar to what we observed with the artificial TOP promoter/enhancer (see Fig. 2). The mouse HK1a promoter/enhancer was not activated by ΔNLef1 or by ΔNTcf3, indicating that most, if not all, of the transactivating potential of Lef1 and Tcf3 are dependent on the β-catenin binding domain at the amino terminus of the protein.
Figure 8.
Activation of the mouse hair keratin 1a promoter by Lef1 and β-catenin. Primary mouse keratinocytes were transfected and processed as described in the legend to Fig. 2, except HK1Flash, containing the mouse hair keratin 1a promoter/enhancer, was used instead of TOPFlash. β-catenin had little effect on the hair keratin promoter unless cotransfected with Lef1. Both ΔNLef1 and ΔNTcf3 failed to activate the promoter when cotransfected with ΔNβcat.
We next engineered K14–ΔNLef1 transgenic mice and compared their phenotype with K14–ΔNTcf3 animals. In striking contrast to the neonatal K14–ΔNTcf3 mice (see Fig. 7M), young K14–ΔNLef1 mice exhibited no obvious epidermal defects or signs of ORS differentiation. These animals survived well into adulthood (>9 mo), and exhibited the characteristic thin epidermis of aging mice. With age, however, the hair coat became increasingly sparse. To be able to understand the basis for the defect in K14–ΔNLef1 animals, we traced the histological signs of abnormalities back to a relatively early age, at approximately the start of the first postnatal hair cycle (3 wk).
On initiation of the normal hair cycle, a stream of new cells emerges from the bulge stem cell compartment of each follicle (Fig. 9A). This stream, referred to as the secondary hair germ, soon progresses to form a mature new hair follicle (Fig. 9B). Biochemically, nuclear β-catenin localization coincides with hair keratin production in the new follicle (Fig. 9C–E; β-catenin and hair keratins in green, ORS keratin in red). This staining pattern is analogous to that which occurs during the anagen phase of the initial hair follicle (Fig. 1).
Figure 9.
K14–ΔNLef1 expression promotes sebocyte cell-fate during the hair cycle. (A–E) Secondary hair germ growth in wild-type littermate. (A) Hematoxylin and Oil Red-O stained section of tail shows a primary hair follicle with sebaceous glands (SG; red stain) and secondary hair germ extending below the club hair. (B) Hematoxylin and eosin stained section shows the formation of the developing new follicle, replete with matrix surrounding the dermal papillae (DP), and a discernable outer root sheath (ORS). (C) Immunofluorescent staining using a β-catenin antibody showing nuclear accumulation of β-catenin (arrowheads) in cells that formed a crescent around the dermal papilla (DP); note that the demarcation between matrix and precortex is not yet complete, but the ORS is already distinct. (D) Similar stage to panel B, revealing more prominent nuclear β-catenin in precortex (precx) and more diffuse staining in matrix (mx). (E) Double immunofluorescent detection of K5 (red) and hair keratin (AE13; green) indicates precortex (precx) formation in the region containing nuclear β-catenin; Hair keratins are extremely stable and can also be seen in the hair shaft, containing dead hair cells (see club hair). (F–I) Abnormal cell fate determination in K14–ΔNLef1 skin. (F) Hematoxylin and eosin stained section from the toe of a K14–ΔNLef1 mouse shows the formation of vacuous structures encased by the epithelial cells of the secondary hair germs. (F, top inset) Hematoxylin and Oil Red-O staining of back skin sections show that the vacuous structures appeared to be filled with sebum, as judged by the fact that they accumulate Oil Red-O stain. (F, bottom inset) Immunofluorescent staining for transgene product (anti-myc) shows that it is present in the crescent of secondary hair germ (HG) cells that abut the dermal papilla (arrowheads). (G) Hematoxylin and Oil Red-O staining of tail follicles shows the presence of oil-producing cells (arrow) at the base of the newly developed follicle, which is replete with an outer root sheath (ORS), but lacks hair structures. Note also the oil-producing cells of the sebaceous glands (SG), similar to those seen in A. (H) Immunofluorescent staining for β-catenin shows that nuclear β-catenin is still found in the crescent of cells surrounding the dermal papilla (DP). At this stage, the secondary hair germ is similar to that seen in the wild type (cf. with C). (I) Anti-K5 and AE13 double staining of developing secondary follicle. Note that the cells surrounding the dermal papilla formed, but they did not give rise to hair keratin-expressing cells, as judged by the absence of AE13 staining. In the place of the hair shaft are differentiated cells (*) that appear to be the ones that stain with Oil Red-O.
Whereas K14–ΔNLef1 follicles initiated their new hair cycle and produced a new hair germ, they soon displayed dramatic alterations in the process. Rather than a normal follicle emerging beneath the bulge, large cysts appeared (Fig. 9F). These cysts stained with oil red O, a typical marker of differentiated cells within the sebaceous gland (upper right inset). Despite their resemblance, the natural sebaceous glands of the follicle always appear above the bulge in normal follicles (Fig. 9A; see also 9G). As shown in Figure 9, F and G, the oil red O-staining cells that were specific to the ΔNLef1-expressing skin were located at the tips of the new hair follicles. In older animals, these cysts became very large, until the point where they visibly affected the skin surface and appeared as bumps.
To further pinpoint the step in the hair cycle that was defective in the younger ΔNLef1 animals, we examined hair follicles as they progressed through the cycle. As judged by immunofluorescence, the crescent of nuclear β-catenin-staining precortical cells still formed (Fig. 9H). The cells that exhibit nuclear β-catenin are those that also concentrate nuclear Lef1 (see Fig. 1), and although the K14 promoter is not active in these cells, they retained ΔNLef1 as judged by anti-myc staining (lower left inset in Fig. 9F). Most notably, despite the presence of nuclear β-catenin and Lef1 in these cells, they did not express the hair-specific keratin genes, as judged by the lack of immunofluorescence seen with a panel hair keratin antibody (Fig. 9, cf. I with E).
Taken together, these data showed that ΔNLef1 blocked the activation of hair-specific gene expression and remarkably induced what appeared to be sebum production. Because ΔNLef1 did not exhibit transactivation potential in our in vitro assays, we suspect that its effect on sebum-producing genes is indirect, resulting from the failure to activate certain key Lef1/β-catenin-regulated proteins that normally prevent sebum gene expression. The principle is remarkably similar to that argued for ΔNTcf3. Whereas ΔNTcf3 promoted ORS/bulge differentiation, however, ΔNLef1 resulted in differentiation more characteristic of sebum-producing cells. In revealing marked new differences in behavior of ΔNTcf3 and ΔNLef1, these data further underscore the importance of these factors in directing different stem cell lineages in skin.
Discussion
Lef1 and Tcf3 are found in distinct epithelial cell compartments of the skin that display contrasting biochemical properties. Lef1 is abundant in the hair-producing progenitors of the follicle, whereas Tcf3 resides in the stem cells of the bulge and in the lower ORS (DasGupta and Fuchs 1999). In this study, we discovered that the two proteins have distinct effects on cell fate when transgenically expressed in the skin epithelial stem cells, and in their ORS and epidermal progeny. Lef1 behaved as a transcriptional activator and promoted hair follicle differentiation in response to Wnt signals that stabilize β-catenin (van Genderen et al. 1994; Zhou et al. 1995; Gat et al. 1998; Millar et al. 1999). Inhibition of the Wnt response blocked hair differentiation and promoted sebaceous cell differentiation, underscoring the importance of Wnt signaling and β-catenin stabilization to the pilosebaceous unit of the skin.
In contrast, Tcf3 inhibited certain features of epidermal differentiation while promoting other differentiation features characteristic of the bulge and ORS. In ORS and bulge that normally express Tcf3, transgenic Tcf3 had no obvious morphological effects, despite yielding nuclear localization of β-catenin in these cells. We found that in striking contrast to Lef1, Tcf3's behavior did not rely on Wnt signaling and β-catenin stabilization: ΔNTcf3 and Tcf3 rendered similar phenotypes in our animals. These data suggest that the status of Tcf3 activity in the epidermis, ORS, and bulge influences these pathways of skin epithelial differentiation and in these cells, Tcf3-regulated genes may not require Wnt signaling. Figure 10 outlines a model based on the results reported in this paper.
Figure 10.
Model suggesting roles of Lef1 and Tcf3 in controlling cell fate in the skin stem cell lineage. Each large circle represents a different cell type arising from the stem cell. The color of sectors in the center of the inner circles represents nuclear localization of Lef1, Tcf3, or β-catenin as indicated by the legend. The outer ring represents the cytoplasm and the status of TOPGAL activity in each cell type (blue: ON). See text for detailed description of events depicted. Note that in the model, Tcf3 is associated with maintaining stem cell and ORS cell fate, and Lef1 and β-catenin promote hair cell fate. Blocking a Wnt signal response (Lef1–β-catenin activation) through ΔNLef1 expression allows induction of sebocyte cell fate during the hair cycle. Note also that it remains unclear whether Tcf3–β-catenin and/or Lef1–β-catenin complexes are responsible for the hair cycle-dependent induction of TOPGAL activity in the bulge cells.
In keratinocytes in vitro, Tcf3 seemed to act as a transcriptional repressor in the presence of the appropriate cofactors. The transdifferentiation effects of Tcf3 in the epidermis in vivo appeared to be dependent on the ability of Tcf3 to bind DNA and to interact with the Groucho family of repressor proteins. They were not dependent on the carboxy-terminal domain of Tcf3, thought to associate with CtBP repressor proteins, and as indicated above, they did not rely on the amino terminus of Tcf3, known to bind β-catenin. Based on these findings, the data support the argument that in vivo as in vitro, Tcf3 acts as a repressor in keratinocytes. A role for Tcf3 as a transcriptional repressor in mammals is consistent with its function in other model organisms and the involvement of Groucho proteins in this action is consistent with the fact that mammalian Groucho proteins can interact with histone deacetylases (Brantjes et al. 2001). Given the parallels between the differentiation seen in the epidermis of K14–Tcf3 transgenic mice and that which occurs in the bulge and ORS of naturally Tcf3-positive cells, it seems likely that both in the wild-type hair follicle and in the K14–Tcf3 transgenic epidermis, Tcf3 functions similarly.
Although our in vitro mutagenesis studies did not support a role for CtBP in Tcf3-mediated repression, they did uncover a role for a potential interaction between proteins that associate with Grg and CtBP domains that may be similar to that seen for the Hairy protein, required for embryonic segmentation in Drosophila (Poortinga et al. 1998; Zhang and Levine 1999). Hairy protein physically interacts with both dCtBP and Groucho proteins, and whereas Groucho mutations enhance defects in hairy mutants, reduced dCtBP activity suppresses defects in hairy mutants (Phippen et al. 2000). In mammalian keratinocytes, the Grg-interaction domain of Tcf3 was required for repression, and the CtBP-interaction domain was required for β-catenin mediated activation of TOPFlash. Interestingly, we also observed the converse, namely that the CtBP-interaction domain was not required for repression, and the Grg-interaction domain was not required for β-catenin-mediated activation. These data suggest a model in which CtBP moderates a competition between β-catenin and Grg proteins to determine whether Tcf3 will activate or repress target genes. Consistent with this model is the recent finding that CtBP can act either as a coactivator or as a corepressor when tethered to DNA, depending on the cell type in which it is expressed (Phippen et al. 2000). Therefore, differences in CtBP levels/activity could explain why in culture the behavior of Tcf3 was markedly influenced by β-catenin levels.
The apparent differences in the way in which Tcf3 and Lef1 interact with various repressor proteins in vitro may explain the in vivo differences observed in the behavior of ΔNTcf3 and ΔNLef1. In this regard, it was notable that whereas ΔNTcf3 suppressed features of epidermal differentiation and promoted aspects of ORS/bulge differentiation, ΔNLef1 showed no obvious effects on epidermis. This result leads us to conclude that either the nontransactivating effects of Lef1 are distinct from those of Tcf3 or they are considerably milder. At present, we have no unequivocal resolution to this issue, but the results could be explained if ΔNTcf3 acts to repress genes in keratinocytes in vivo, whereas ΔNLef1 is only able to block transactivation. The differential behavior between the Groucho-interacting domains of Lef1 and Tcf3 in vitro provides a starting point for future studies needed to assess the extent to which this might be the case.
Our findings also indicated that both in vitro and in vivo, Lef1 uses β-catenin interactions to transactivate the expression of genes encoding hair-specific keratins and presumably some other factors involved in promoting the hair cell fate. In the presence of amino-terminally truncated Lef1, hair keratin gene expression and hair cell differentiation was quantitatively blocked. Unexpectedly, however, ΔNLef1 expression not only blocked hair cell production but resulted in transdifferentiation of these cells into sebum-producing cells. This finding predicts the existence of other key differentiation factors that must be controlled by Lef1 complexes in matrix cells when the steps that are downstream of β-catenin stabilization in the Wnt signal transduction pathway are blocked. At least one of these genes seems to encode a protein that converts the hair-producing cell to a sebum-producing cell. Whereas ΔNLef1-related interference of Tcf3-mediated repression may contribute to the transdifferentiation observed, it also may be relevant that in tissue culture, preadipocytes and myoblasts can both differentiate into adipocytes when Wnt signaling is inhibited (Ross et al. 2000). Adipogenesis shares a number of similar features to sebocyte differentiation, making the parallels quite striking. An additional observation that also may have bearing on our finding is that the Lef1 gene encodes multiple mRNAs, some of which are likely to produce Lef1 proteins similar to the ΔNLef1 that we describe in this paper (Hovanes et al. 2000, 2001). That these forms are expressed in the natural sebaceous gland cells of the skin seems likely and awaits future investigation.
In summary, our overall findings argue that Lef1, ΔNLef1, and Tcf3 all have their greatest impact on controlling genes that orchestrate the differentiation status of keratinocytes. Although some of these genes, for example, the hair-specific keratin genes, seem to be activated directly through Lef1 complexes that are dependent on β-catenin and Wnt signaling, other genes, such as those controlling ORS/bulge and sebocyte differentiation, seem to require nontransactivating functions of Lef1/Tcf3. The dramatically different effects of transgenic Tcf3 and Lef1 (and their various mutant forms) on the cell fate of the stem cells suggests a functional importance for the down-regulation of Tcf3 (epidermis), the maintenance of Tcf3 (lower ORS), and the switch from Tcf3 to Lef1 (in matrix cells) as normal stem cells exit the bulge and migrate to their appropriate destinations in the skin. In the future, as target genes for these protein complexes are identified, the complex regulatory pathways that lead to lineage progression should begin to unfold.
Materials and methods
Plasmid construction
Molecular biology techniques were performed according to conventional protocols (Ausubel et al. 2000). All constructs made with PCR were sequenced to verify their authenticity.
The K14-BG expression cassette (Gat et al. 1998) was used for construction of K14–Tcf3* and its derivatives. The myc-tagged mTcf3* cDNA was purified as a blunted EcoRV–BglII fragment from pCDNA3–myc–mTcf3* (a kind gift of Dr. Hans Clevers, University Hospital, The Netherlands) and ligated into BamHI (blunted)-cut K14-BG cassette. To make K14–Tcf3, a fragment encoding the mTcf3 HMG domain was amplified from newborn mouse epidermal cDNA, which was used to correct the mutated region of mTcf3*. To make K14–Tcf3ΔC, PCR primers were used to place a stop codon after nucleotide 1491 of mTcf3 cDNA, thereby encoding a truncated version of the protein. To make Tcf3ΔGrg, an artificial BssHII site was inserted before nucleotide 957 of mTcf3 cDNA, and a subsequent digestion with BssHII and PmlI and ligation into BssHII–PmlI-digested K14–Tcf3 resulted in a cDNA encoding an in-frame deletion of amino acids 143–318 of mTcf3. To make K14ΔNTcf3, a PCR product placing a SacII site at the 5′ end of a truncated Tcf3 cDNA was cloned into SacII–DraIII digested K14–Tcf3 such that the amino-terminal 71 amino acids of Tcf3 were deleted.
The construction of the K14–Lef1 expression plasmid with a myc-epitope tag at the 5′ end of the human Lef1 cDNA used the K14-BG–hLef1 plasmid described previously (Zhou et al. 1995). PCR primers were used to make a hLef1 fragment with a myc-epitope tag at its 5′ end. The tagged hLef1 region was digested with XhoI and EcoRI and used to replace untagged hLef1 sequence from XhoI–EcoRI (partial)-digested K14-BG–hLef1. The K14–ΔNLef1 construct was made by PCR amplifying the hLef1 sequence encoding amino acid 63 onward, using primers placing a BamHI site and myc-tag at the 5′ end of the truncated hLef1 cDNA.
To construct the mTcf3/hLef1 fusion protein expression plasmids, the K14–Lef1 and K14–Tcf3 plasmids were used as templates. For fusion 1, the 3′ end of mTcf3 was replaced with the 3′ end of hLef1 by replacing a PmlI (base pair 1021 of mTcf3)–HindIII (cuts 3′ of polyA in K14-BG cassette) deletion of K14–Tcf3 with a PmlI (base pair 841 of hLef1)–HindIII fragment from K14–Lef1. For fusion 2 and fusion 3, a PCR gene stitching method was used in which recombinant hLef1–mTcf3 PCR products were digested with BssHII and PmlI, and ligated into BssHII–PmlI-digested K14–Tcf3. For fusion 4, the 5′ end of hLef1 was deleted from K14–Lef1 by digesting with MfeI (cuts 5′ of myc-epitope tag) and XhoI (base pair 324 of hLef1; blunted), and it was replaced with the 5′ end mTcf3 with a MfeI–Asp718 (base pair 353 of mTcf3; blunted) from K14–Tcf3.
To construct bacterial expression plasmids for the HMG domains of Tcf3 and Tcf3* proteins, PCR primers were used to amplify base pairs 991–1281 from either K14–Tcf3 or K14–Tcf3* templates. PCR products were digested with BamHI and XhoI and ligated into BamHI–XhoI-cut pTRIEX (Novagen) such that the 6×His tag of the pTRIEX vector was in-frame with the HMG domain encoding PCR product. The HK1Flash plasmid was constructed by removing the 3.5-kb fragment of the mouse hair keratin 1a fragment from plasmid pmHK113.3.3R (Upjohn) with EcoRI and inserting the blunted fragment into the SmaI site of pGL3 basic (Promega).
Generation and analysis of transgenic mice
Transgenic mice were engineered as described (Vassar et al. 1989). Genomic DNA was isolated from tail or toe samples of mice and analyzed by PCR with primers specific for the human K14 promoter. For histology of mouse skin, samples of dorsal backskin were fixed overnight in freshly prepared 4% paraformaldehyde at 4°C, dehydrated, embedded in paraffin, and sectioned for hematoxylin and eosin staining. Oil Red-O staining was performed on frozen cryosections as described (Carson 1990).
Dye penetration assays
Dye penetration assays were performed as described (Hardman et al. 1998; Segre et al. 1999). Embryos from timed pregnancies were isolated at E16.5–17.5 for experiments with nontransgenic mice. Transgenic embryos at E18.5 were compared with nontransgenic littermates. Tails were removed for genotyping and for determining levels of transgene expression, and embryos were placed in a solution of containing X-Gal at pH 4.5. Embryos were allowed to incubate 5 h to overnight at 37°C, washed in PBS, and post-fixed overnight in 4% PFA at 4°C.
EMSA
Protein was extracted from BL21 codon plus bacteria (Stratagene) transformed with Tcf3HMG or Tcf3* HMG constructs in 1× DNA-binding buffer (25 mM HEPES at pH 7.9, 75 mM KCl, 1 mM EDTA, 0.1% Nonidet P-40, 10% glycerol, 0.5 mM dithiothreitol, 1× protease inhibitors; Boehringer), and levels of expression were determined by Western blot analysis using an anti-pentaHIS antibody (QIAGEN). In standard 20 μL reactions in 1× DNA-binding buffer, protein extracts were incubated with 1μg poly(dI-dC) and 1 μL 32P-labeled TCRalpha oligo (Waterman et al. 1991) for 30 min at 4°C. The protein–DNA complexes were resolved by gel electrophoresis on 5% polyacrylamide gels, and band shifts were visualized on dried gels by autoradiography.
Immunofluorescence and antibodies
For indirect immunofluorescent detection of proteins in mouse backskin, the mouse on mouse kit (MOM kit; Vector Laboratories) was used according the manufacturers' protocol with minor variations. All immunofluorescent staining was performed on either 8–10-μm frozen sections of back skin embedded in OCT or 5 μm paraffin-embedded sections. Frozen sections were fixed in 4% PFA, and antigens were visualized with FITC- or TxR-conjugated secondary antibodies (Jackson Labs). All fluorescent images were taken with a Zeiss LSM 410 system.
Primary antibodies used were mouse anti-myc tag (Invitrogen); rabbit anti-K5; rabbit anti-filaggrin (BAbCO); rabbit anti-loricrin (BAbCO); mouse anti-β-catenin (Sigma); rat anti-E-cadherin (Zymed); rabbit anti-Lef1; guinea pig anti-Tcf3; rabbit anti-K6 (BAbCO); rabbit anti-K17 (McGowan and Coulombe 1998); rabbit anti-TLE2 (Santa Cruz); and rat anti-PanTLE (Stifani et al. 1992).
Cell culture and transient transfections
Primary human keratinocytes were grown in serum-free keratinocyte growth media (Clonetics), and transfections were performed only with cultures of five passages or less. Transfections were performed with the Fugene (Roche) reagent according to the manufacturers' protocol, and 40 h after transfection cells were harvested for luciferase assays (Promega Dual-Luciferase Kit). Firefly luciferase values (TOPFlash) were standardized to Renilla luciferase values (pRL–CMV, or pRL–TK; Promega) to account for differences in transfection efficiency between samples. In addition, FOPflash or FOPGAL (DasGupta and Fuchs 1999) was also cotransfected with expression plasmids to ensure that TOPFlash values were dependent on the Lef1/Tcf-binding sites in the promoter.
Acknowledgments
Special thanks are extended to Dave Padua for his expert assistance with immunostaining of back skin samples and critical reading of the manuscript, to Linda Degenstein and Jie Fan for their work in generating and observing the transgenic mice, and to Horace Rhee for his studies on Groucho-Tcf3 interactions in keratinocytes. For generous gifts of antibodies, we thank Dr. Pierre Coulombe and Dr. Stefano Stifani. We also thank the other members of the Fuchs lab for their genuine scientific curiosity and willingness to share ideas, reagents, and protocols. B.J.M. was supported by research scholar grant no. PF-01-043-01-DDC from the American Cancer Society. E.F. is an investigator of the Howard Hughes Medical Institute. This work was supported in part from a grant from the National Institutes of Health (R01-AR31737).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL [email protected]; FAX (773) 702-0141.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.891401.
References
- Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. Current protocols in molecular biology. New York, NY: John Wiley & Sons; 2000. [Google Scholar]
- Brannon M, Brown JD, Bates R, Kimelman D, Moon RT. XCtBP is a XTcf-3 co-repressor with roles throughout Xenopus development. Development. 1999;126:3159–3170. doi: 10.1242/dev.126.14.3159. [DOI] [PubMed] [Google Scholar]
- Brantjes H, Roose J, van De Wetering M, Clevers H. All Tcf HMG box transcription factors interact with Groucho-related co-repressors. Nucleic Acids Res. 2001;29:1410–1419. doi: 10.1093/nar/29.7.1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carson FL. Histotechnology: A self-instructional text. Chicago, IL: American Society of Clinical Pathologists; 1990. [Google Scholar]
- Cavallo RA, Cox RT, Moline MM, Roose J, Polevoy GA, Clevers H, Peifer M, Bejsovec A. Drosophila Tcf and Groucho interact to repress Wingless signalling activity. Nature. 1998;395:604–608. doi: 10.1038/26982. [DOI] [PubMed] [Google Scholar]
- Chan EF, Gat U, McNiff JM, Fuchs E. A common human skin tumour is caused by activating mutations in β-catenin. Nat Genet. 1999;21:410–413. doi: 10.1038/7747. [DOI] [PubMed] [Google Scholar]
- DasGupta R, Fuchs E. Multiple roles for activated LEF/TCF transcription complexes during hair follicle development and differentiation. Development. 1999;126:4557–4568. doi: 10.1242/dev.126.20.4557. [DOI] [PubMed] [Google Scholar]
- Fuchs E. Beauty is skin deep: The fascinating biology of the epidermis and its appendages. Harvey Lect. 1997;94:47–77. [PubMed] [Google Scholar]
- Gat U, DasGupta R, Degenstein L, Fuchs E. De Novo hair follicle morphogenesis and hair tumors in mice expressing a truncated beta-catenin in skin. Cell. 1998;95:605–614. doi: 10.1016/s0092-8674(00)81631-1. [DOI] [PubMed] [Google Scholar]
- Hardman MJ, Sisi P, Banbury DN, Byrne C. Patterned acquisition of skin barrier function during development. Development. 1998;125:1541–1552. doi: 10.1242/dev.125.8.1541. [DOI] [PubMed] [Google Scholar]
- Henderson BR. Nuclear-cytoplasmic shuttling of APC regulates β-catenin subcellular localization and turnover. Nature Cell Biol. 2000;2:653–660. doi: 10.1038/35023605. [DOI] [PubMed] [Google Scholar]
- Hovanes K, Li TW, Waterman ML. The human LEF-1 gene contains a promoter preferentially active in lymphocytes and encodes multiple isoforms derived from alternative splicing. Nucleic Acids Res. 2000;28:1994–2003. doi: 10.1093/nar/28.9.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hovanes K, Li TW, Munguia JE, Truong T, Milovanovic T, Lawrence Marsh J, Holcombe RF, Waterman ML. β-catenin-sensitive isoforms of lymphoid enhancer factor-1 are selectively expressed in colon cancer. Nat Genet. 2001;28:53–57. doi: 10.1038/ng0501-53. [DOI] [PubMed] [Google Scholar]
- Kim CH, Oda T, Itoh M, Jiang D, Artinger KB, Chandrasekharappa SC, Driever W, Chitnis AB. Repressor activity of Headless/Tcf3 is essential for vertebrate head formation. Nature. 2000;407:913–916. doi: 10.1038/35038097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korswagen HC, Clevers H. Activation and repression of wingless/Wnt target genes by the TCF/LEF-1 family of transcription factors. Cold Spring Harb Symp Quant Biol. 1999;64:141–147. doi: 10.1101/sqb.1999.64.141. [DOI] [PubMed] [Google Scholar]
- Love JJ, Li X, Case DA, Giese K, Grosschedl R, Wright PE. Structural basis for DNA bending by the architectural transcription factor LEF-1. Nature. 1995;376:791–795. doi: 10.1038/376791a0. [DOI] [PubMed] [Google Scholar]
- McGowan KM, Coulombe PA. Onset of keratin 17 expression coincides with the definition of major epithelial lineages during skin development. J Cell Biol. 1998;143:469–486. doi: 10.1083/jcb.143.2.469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar SE, Willert K, Salinas PC, Roelink H, Nusse R, Sussman DJ, Barsh GS. WNT signaling in the control of hair growth and structure. Dev Biol. 1999;207:133–149. doi: 10.1006/dbio.1998.9140. [DOI] [PubMed] [Google Scholar]
- Molenaar M, van de Wetering M, Oosterwegel M, Peterson-Maduro J, Godsave S, Korinek V, Roose J, Destree O, Clevers H. XTcf-3 transcription factor mediates β-catenin-induced axis formation in Xenopus embryos. Cell. 1996;86:391–399. doi: 10.1016/s0092-8674(00)80112-9. [DOI] [PubMed] [Google Scholar]
- Oshima H, Rochat A, Kedai C, Kobayashi K, Barrandon Y. Morphogenesis and renewal of hair follicles from adult stem cells. Cell. 2001;104:233–245. doi: 10.1016/s0092-8674(01)00208-2. [DOI] [PubMed] [Google Scholar]
- Phippen TM, Sweigart AL, Moniwa M, Krumm A, Davie JR, Parkhurst SM. Drosophila C-terminal binding protein functions as a context-dependent transcriptional co-factor and interferes with both mad and groucho transcriptional repression. J Biol Chem. 2000;275:37628–37637. doi: 10.1074/jbc.M004234200. [DOI] [PubMed] [Google Scholar]
- Poortinga G, Watanabe M, Parkhurst SM. Drosophila CtBP: A Hairy-interacting protein required for embryonic segmentation and hairy-mediated transcriptional repression. EMBO J. 1998;17:2067–2078. doi: 10.1093/emboj/17.7.2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prieve MG, Guttridge KL, Munguia JE, Waterman ML. The nuclear localization signal of lymphoid enhancer factor-1 is recognized by two differentially expressed Srp1-nuclear localization sequence receptor proteins. J Biol Chem. 1996;271:7654–7658. doi: 10.1074/jbc.271.13.7654. [DOI] [PubMed] [Google Scholar]
- Pukrop T, Gradl D, Henningfeld KA, Knoechel W, Wedlich D, Kuehl M. Identification of two regulatory elements within the HMG-box transcription factor XTCF-4. J Biol Chem. 2000;276:8968–8978. doi: 10.1074/jbc.M007533200. [DOI] [PubMed] [Google Scholar]
- Roose J, Clevers H. TCF transcription factors: Molecular switches in carcinogenesis. Biochim Biophys Acta. 1999;1424:M23–M37. doi: 10.1016/s0304-419x(99)00026-8. [DOI] [PubMed] [Google Scholar]
- Roose J, Huls G, van Beest M, Moerer P, van der Horn K, Goldschmeding R, Logtenberg T, Clevers H. Synergy between tumor suppressor APC and the β-catenin-Tcf4 target Tcf1. Science. 1999;285:1923–1926. doi: 10.1126/science.285.5435.1923. [DOI] [PubMed] [Google Scholar]
- Roose J, Molenaar M, Peterson J, Hurenkamp J, Brantjes H, Moerer P, van de Wetering M, Destree O, Clevers H. The Xenopus Wnt effector XTcf-3 interacts with Groucho-related transcriptional repressors. Nature. 1998;395:608–612. doi: 10.1038/26989. [DOI] [PubMed] [Google Scholar]
- Rosin-Arbesfeld R, Townsley F, Bienz M. The APC tumour suppressor has a nuclear export function. Nature. 2000;406:1009–1012. doi: 10.1038/35023016. [DOI] [PubMed] [Google Scholar]
- Ross SE, Hemati N, Longo KA, Bennett CN, Lucas PC, Erickson RL, MacDougald OA. Inhibition of adipogenesis by Wnt signaling. Science. 2000;289:950–953. doi: 10.1126/science.289.5481.950. [DOI] [PubMed] [Google Scholar]
- Segre JA, Bauer C, Fuchs E. Klf4 is a transcription factor required for establishing the barrier function of the skin. Nat Genet. 1999;22:356–360. doi: 10.1038/11926. [DOI] [PubMed] [Google Scholar]
- Stifani S, Blaumueller CM, Redhead NJ, Hill RE, Artavanis-Tsakonas S. Human homologs of a Drosophila Enhancer of split gene product define a novel family of nuclear proteins. Nat Genet. 1992;2:119–127. doi: 10.1038/ng1092-119. [DOI] [PubMed] [Google Scholar]
- St-Jacques B, Dassule HR, Karavanova I, Botchkarev VA, Li J, Danielian PS, McMahon JA, Lewis PM, Paus R, McMahon AP. Sonic hedgehog signaling is essential for hair development. Curr Biol. 1998;8:1058–1068. doi: 10.1016/s0960-9822(98)70443-9. [DOI] [PubMed] [Google Scholar]
- Taylor G, Lehrer MS, Jensen PJ, Sun TT, Lavker RM. Involvement of follicular stem cells in forming not only the follicle but also the epidermis. Cell. 2000;102:451–461. doi: 10.1016/s0092-8674(00)00050-7. [DOI] [PubMed] [Google Scholar]
- van Genderen C, Okamura RM, Farinas I, Quo RG, Parslow TG, Bruhn L, Grosschedl R. Development of several organs that require inductive epithelial-mesenchymal interactions is impaired in LEF-1-deficient mice. Genes & Dev. 1994;8:2691–2703. doi: 10.1101/gad.8.22.2691. [DOI] [PubMed] [Google Scholar]
- Vasioukhin V, Degenstein L, Wise B, Fuchs E. The magical touch: Genome targeting in epidermal stem cells induced by tamoxifen application to mouse skin. Proc Natl Acad Sci. 1999;96:8551–8556. doi: 10.1073/pnas.96.15.8551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassar R, Rosenberg M, Ross S, Tyner A, Fuchs E. Tissue-specific and differentiation-specific expression of a human K14 keratin gene in transgenic mice. Proc Natl Acad Sci. 1989;86:1563–1567. doi: 10.1073/pnas.86.5.1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterman ML, Fischer WH, Jones KA. A thymus-specific member of the HMG protein family regulates the human T cell receptor C alpha enhancer. Genes & Dev. 1991;5:656–669. doi: 10.1101/gad.5.4.656. [DOI] [PubMed] [Google Scholar]
- Waltzer L, Bienz M. The control of β-catenin and TCF during embryonic development and cancer. Cancer Metastasis Rev. 1999;18:231–246. doi: 10.1023/a:1006321324190. [DOI] [PubMed] [Google Scholar]
- Willert K, Nusse R. Beta-catenin: A key mediator of Wnt signaling. Curr Opin Genet Dev. 1998;8:95–102. doi: 10.1016/s0959-437x(98)80068-3. [DOI] [PubMed] [Google Scholar]
- Zhang H, Levine M. Groucho and dCtBP mediate separate pathways of transcriptional repression in the Drosophila embryo. Proc Natl Acad Sci. 1999;96:535–540. doi: 10.1073/pnas.96.2.535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P, Byrne C, Jacobs J, Fuchs E. Lymphoid enhancer factor 1 directs hair follicle patterning and epithelial cell fate. Genes & Dev. 1995;9:700–713. doi: 10.1101/gad.9.6.700. [DOI] [PubMed] [Google Scholar]