Summary
Here, we exploit the hair follicle to define the point at which stem cells become irreversibly committed along a differentiation lineage. Employing histone and nucleotide double-pulse-chase and lineage tracing, we show that the early SC descendents en route to becoming transit-amplifying cells retain stemness and slow-cycling properties and home back to the bulge niche when hair growth stops. These become the primary SCs for the next hair cycle, while initial bulge SCs become reserves for injury. Proliferating descendents further en route irreversibly lose their stemness, although they retain many SC markers and survive, unlike their transit-amplifying progeny. Remarkably, these progeny also home back to the bulge. Combining purification and gene expression analysis with differential ablation and functional experiments, we define critical functions for these non-SC niche residents, and unveil the intriguing concept that an irreversibly committed cell in an SC lineage can become an essential contributor to the niche microenvironment.
INTRODUCTION
Adult stem cells (SCs) govern tissue homeostasis and wound repair. They reside in a specific niche, defined as the microenviroment that hosts and maintains SCs (Spradling et al., 2008). Most SCs are infrequently cycling, a feature thought to preserve their stemness, namely their ability to self-renew and remain undifferentiated over the animal’s lifetime. During normal homeostasis, they often exit from their niches and progress to become transit-amplifying (TA) cells, undergoing a series of rapid divisions before committing to terminal differentiation (Fuchs, 2009; Morrison and Kimble, 2006).
Determining the point in a lineage hierarchy where SCs lose long-term self-renewing capacity and become irreversibly committed represents a fundamental and challenging question in SC biology. Transitioning from a slow-cycling to more rapidly-cycling state is not indicative, as hematopoietic stem cells (HSCs) and hair follicle (HF) SCs can reversibly switch from dormancy to cycling during normal homeostasis and wound repair (Blanpain et al., 2004; Foudi et al., 2009; Nowak et al., 2008; Taylor et al., 2000; Waghmare et al., 2008; Wilson et al., 2008). Merely exiting their niche is also not a reliable measure, as some HSCs circulate, trafficking between their bone marrow niche and extramedullary tissues (Cao et al., 2004). Even embarking along a differentiation pathway may not be an unequivocal indicator of loss of stemness; studies in Drosophila and mouse testis show that germline SC niche vacancies can be filled by early spermatogonial cells that dedifferentiate when returned to the niche (Brawley and Matunis, 2004; Kai and Spradling, 2004; Nakagawa et al., 2008).
The murine HF offers an excellent system for monitoring an SC lineage and exploring plasticity of SC progenies. During homeostasis, the lower HF cycles through bouts of active hair growth (anagen), destruction (catagen), and rest (telogen) (Lavker et al., 2003; Paus and Cotsarelis, 1999). When the new HF emerges, it grows next to the old hair, which persists into the next cycle. This creates a protrusion or “bulge,” first described >100 years ago (Unna, 1876). In 1990, nucleotide pulse-chase experiments revealed the existence of slow-cycling, label retaining cells (LRCs) in the bulge (Cotsarelis et al., 1990). A decade later, these cells were isolated, characterized and shown to self-renew long-term and contribute to HF lineages and wound-repair (Blanpain et al., 2004; Claudinot et al., 2005; Ito et al., 2005; Morris et al., 2004; Tumbar et al., 2004; Zhang et al., 2009). These findings established the bulge as a bona fide HF-SC niche.
Hair growth is fueled by bulge SCs, which are activated at the start of anagen by the dermal papilla (DP), a cluster of underlying mesenchymal cells. Upon activation, SCs exit the bulge and proliferate downward, creating a long linear trail of cells, the outer root sheath (ORS) (Ito et al., 2005; Zhang et al., 2009). In mature HFs, the ORS extends from bulge to matrix. Enveloping the DP at the HF base, matrix cells cycle rapidly but transiently before differentiating upward to generate the hair and its channel (Figures 1A, S1A).
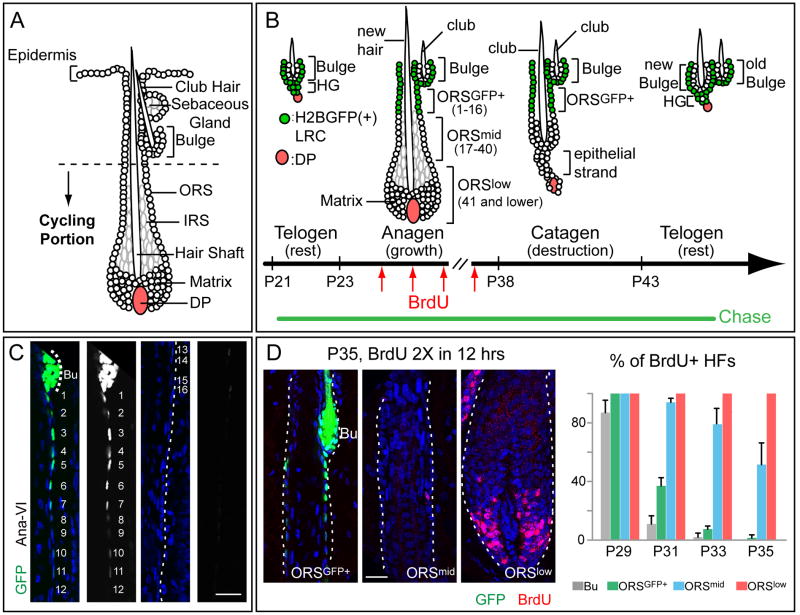
Figure 1. Dynamics of slow and fast cycling cells throughout the hair cycle.
(A) Cycling portions of a mature HF (B-D) Tet-Off H2BGFP mice were chased from P21 to the days/stages noted (P35–P37 corresponds to AnaVI). Before harvesting skin, some mice were given 2X6 hr pulses of BrdU. Schematic depicts H2BGFP cells (green) that will be label-retaining (LRCs) when the chase is stopped at each stage. Cell #s are counting down from the bulge base (=0). In (C), the right panels are duplicates of GFP monochromes of the left panels. Scale bars, 30μm. Bu, bulge. DAPI in blue. Note that S-phase cells in AnaVI are mainly in ORSlow and matrix. %HFs with BrdU+ cells in bulge or in different ORS segments were quantified from P29–P35. For each stage, n=2 mice and ≥23 HFs/mouse were counted. Data are mean ± SD.
Catagen illuminates an unambiguous distinction between long-lived HF-SCs, and short-lived matrix progeny which undergo massive apoptosis. The remaining epithelial strand retracts, drawing the DP upward. Current evidence suggests that at the catagen/telogen transition, a few bulge SCs migrate to meet the DP, generating the hair germ (HG) (Ito et al., 2004; Zhang et al., 2009). Bearing closer resemblance to bulge than matrix, HG cells are activated prior to bulge at the start of anagen (Greco et al., 2009). Prior to activation, HF-SCs undergo an extended rest period which can last for months.
While extensive studies have been performed on bulge SCs and TA-matrix cells, the properties and fates of ORS cells are less clear. Although these cells do not express high levels of CD34, a marker of their SC bulge predecessors, they display many HF-SC markers not found in matrix, including Lgr5, Sox9, Lhx2 and TCF3 (Fuchs, 2009). To date, functional studies on ORS cells have been limited to cultures of microdissected rat whisker HFs, where long-term clones capable of engraftment were obtained not only from bulge and ORS but also matrix (Claudinot et al., 2005; Oshima et al., 2001; Rochat et al., 1994). Whether ORS cells in vivo possess stemness like their predecessors or are committed like their TA progeny remains unknown.
The unique regenerative aspects of HF homeostasis and the transient, but spatially and temporally well-defined ORS, offers an unparalleled opportunity to examine the transition between a SC and a TA cell. Where does the transition from slow-cycling to rapidly-cycling occur along the ORS trail? What happens to ORS cells during catagen? Do they undergo apoptosis like TA cells or do they survive like SCs? If they survive, do they home back to the bulge, and if they do so, do they function as SCs?
Answering these questions could provide fundamental insights into defining not only the importance of bulge to SC survival but also the point where an SC loses its stemness and embarks upon a terminal differentiation pathway. If all ORS cells either die or differentiate during catagen, this would imply that HF-SCs lose stemness once they leave the bulge. If on the other hand, some ORS cells survive and return back to the bulge as functional SCs, this would imply that at least at some point along the progression from bulge to TA compartments, an SC that has left the bulge still possesses intrinsic features of stemness.
In this report, we address these key questions in the unperturbed in vivo confines of the normal hair cycle, as well as in response to injury. In so doing, we unearth surprising and dynamic features of the HF-SC niche and its progeny. First, we show that during anagen, the upper ORS remains slow-cycling, like their SC predecessors, while lower ORS cells cycle more rapidly. Moreover, slow-cycling ORS cells survive catagen and contribute mightily to both HG and a new bulge: in fact, they become the main source of SCs used during the next hair cycle. Lacking a DP, the initial bulge ceases to play a major role in homeostasis but can respond to injury. Finally and perhaps most surprisingly, some actively cycling lower ORS cells not only survive catagen, but also home back to the bulge. Like their upper ORS predecessors, these cells retain many HF-SC markers. However, they irreversibly lose their ability to proliferate in normal homeostasis or upon wounding. Instead, they function decisively in hair anchorage during the resting phase and in providing the quiescent signaling cues that control the hair cycle. Our findings define a point along the ORS where cells lose stemness and become irreversibly fated to differentiate or die, and illuminate how downstream SC progeny can provide negative feedback to the niche and restrict SC self-renewal and tissue formation.
Results
Bulge Descendents in Anagen ORS Can Be Subdivided into Slow-Cycling And Faster Cycling
To dissect the properties of bulge SC descendents along the ORS, we first addressed whether these cells remain slow-cycling like bulge or become faster cycling like matrix. We used bi-transgenic mice expressing Doxycycline (Doxy)-repressible histone H2BGFP controlled by a keratin 5 (K5) promoter. With this Tet-Off system, skin epithelium is uniformly labeled until Doxy exposure, when new H2BGFP synthesis is tightly repressed and cells deplete existing GFP by 2X/division (Tumbar et al., 2004).
Doxy chase was begun at postnatal day P21, just before the start of the 1st hair cycle (Figure 1B). At the last stage of anagen (AnaVI, based on Muller-Rover et al., 2001; P35–P37 in these mice), the bulge but not matrix contained bright H2BGFP LRCs (Figures 1C,D). Interestingly, an LRC trail extended from the bulge base along the ORS. To quantify, we standardized image acquisition for GFP intensities by setting the detector so that brightest bulge cells were maximal but not saturated in a 12 bit (=4096 shades) image. Under this condition, bulge was composed almost exclusively of cells with GFP intensities >256 (GFPbright). Within the first 16 positions counting down along the ORS trail in a planar section, >85% of cells were GFP+ (intensity >16) with the majority of GFPbright cells in #1–12. By contrast, few ORS cells in sites 16–30 were GFP+, and below #30, GFP was not detected (Figures 1C, S1B).
To further probe proliferation heterogeneities during anagen, we performed a series of short BrdU pulse experiments in Tet-Off H2BGFP mice. When labelings were conducted at intervals between P27 and P29, i.e. after HFs displayed an emerging inner root sheath (IRS) and typical anagen shape, BrdU+ cells were detected in bulge, ORS and matrix. By P31 however, most bulge cells had ceased proliferation. Although occasional BrdU+ cells were still seen in ORS positions 1–16, proliferation was mostly beneath this zone in ORS and matrix. By P33, most BrdU+ ORS cells were below #30, and from P35–P37, BrdU+ cells were largely restricted to matrix, with few labeled ORS cells <#40 (Figures 1D, S1C). The transition between ORS and matrix was at ~#75–85.
To summarize: ORSGFP+(#1–16) behaved similarly to bulge, displaying slow-cycling characteristics and returning to quiescence ~2d after bulge proliferation ceased; ORSmid (#17–40) remained proliferative 2d after ORSGFP+; and ORSlow cells (#40–80) and matrix cycled continuously.
SCs That Exit the Bulge During the Growth Phase And Remain Slow-Cycling (ORSGFP+) Are Spared From Apoptosis During the Destructive Phase
Since ORSGFP+ cells divide only a few times during anagen, we next wondered what happens to these cells during the destructive phase. Throughout catagen, the ORSGFP+ trail showed no signs of migration or expansion (Figure 2A). Three possible lineage behaviors could explain this result: 1) GFP+ ORS cells are static in catagen; 2) the influx of bulge cells to ORSGFP+ is compensated by an efflux of ORSGFP+ cells, proliferating and moving towards matrix; 3) ORSGFP+ cells undergo apoptosis and bulge cells migrate downward to replace them.
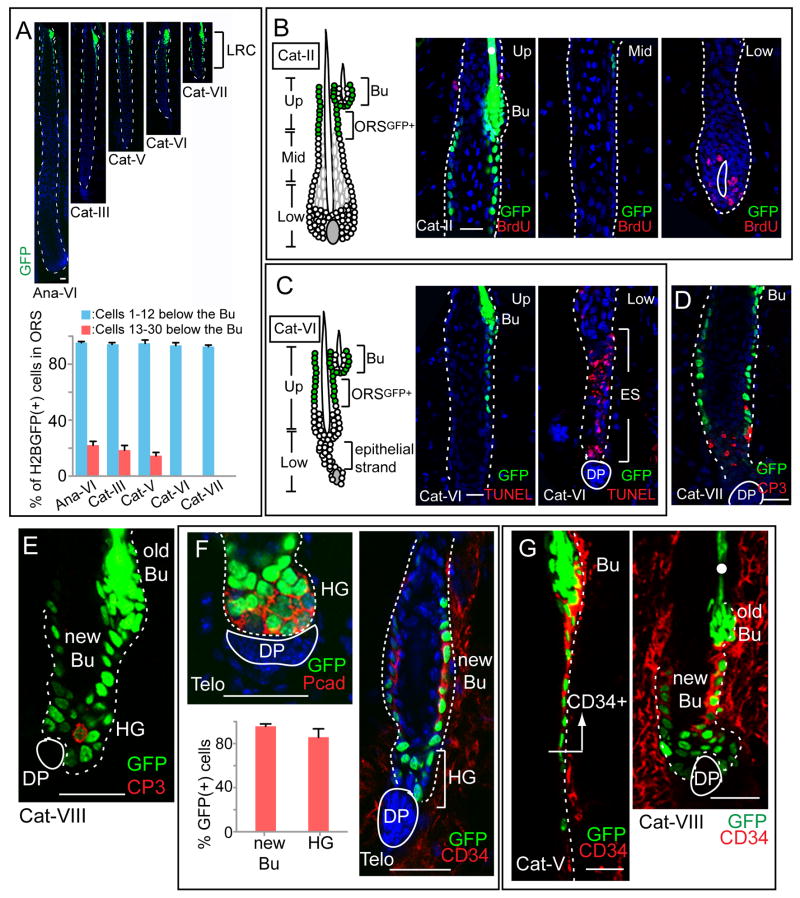
Figure 2. Label-retaining HF-SC descendents in the ORS are spared from apoptosis during catagen and form a new bulge and HG at telogen.
Tet-Off H2BGFP mice were chased from P21 to the HF stages indicated. Prior to analysis, some mice were given a 4hr BrdU pulse. (A) H2BGFP LRCs maintain the same distribution from anagen VI to catagen’s end. (B) BrdU labeling in early catagen (Cat-II) shows proliferation only in matrix and ORSlow and not ORSGFP+ or ORSmid. (C-E) TUNEL and CP3 immunolabeling in catagen VI–VIII reveals cell death in the retracting epithelial strand (ES) but rarely in LRCs. (F) Fate of H2BGFP LRCs at catagen to 2nd telogen transition. Like the old bulge, the emerging new bulge and HG contain LRCs. Quantifications: n=3 mice; 42 total HFs analyzed for bulge; 87 for HG. (G) HF-SC bulge marker CD34 is upregulated in upper ORSGFP+ beginning at catagen V. The shape of the future new bulge and HG can already be distinguished at Cat-VIII. HFs are outlined by dotted white line. White dots mark autofluorescent hair shafts. Scale bars, 30μm. Data are mean ± SD.
To distinguish between these models, we monitored apoptotic and proliferative events during this period. Early catagen was marked by dramatic reduction in matrix proliferation (Figure 2B). Cells within bulge and ORS were mostly quiescent during catagen. These data rule out model 2. Soon thereafter, cell death was detected by DNA fragmentation (TUNEL) and activated caspase 3 (CP3). Apoptosis began within matrix and expanded upward into the retracting epithelial strand (Figures 2C). However, even towards catagen’s end, only a few ORSGFP+ and bulge LRCs scored positive for TUNEL or CP3, and ultrastructural analysis suggested that these were likely healthy cells that had engulfed apoptotic debris (Figures 2D, S2A-D). These data rule out model 3. Together, these differences revealed a striking molecular boundary between the TUNEL/CP3-positive epithelial strand, and the apparently unscathed LRCs in bulge and ORS. These data are consistent with model 1.
ORSGFP+ Cells Form a New Bulge and Hair Germ at the End of Catagen
Since most ORSGFP+ cells survive catagen, we next addressed where they go. Near the end of catagen, the LRC trail was still located below the initial bulge. When HFs were followed into telogen and DP adopted its characteristic position directly below the HG, it became apparent that the ORSGFP+ trail had adopted a form resembling the HG and a new bulge (Figure 2E).
Imaging and quantifications coupled with FACS analysis confirmed that ~98% of CD34+ cells within this new bulge and ~80–90% of those in HG were GFP+ (Figure 2F, S3C). Since in the Tet-Off H2BGFP system, a GFP(-) cell from an earlier time cannot be the source for a GFP+ cell at a later point, this leaves only ORSGFP+ and initial bulge as possible sources for this structure.
Since from catagen to telogen, the initial bulge showed no major change in size or cell number (Figure S2E), our collective evidence favored ORSGFP+ as the major source of both this new bulge and HG. These results were surprising however, since anagen ORSGFP+ cells did not show strong immunostaining for either bulge marker CD34 or HG marker P-cadherin. After first confirming that new bulge and HG express these markers (Figure 2F), we traced the temporal roots of these differences. Towards catagen’s end, the first ~8 cells in the ORSGFP+ zone acquired strong CD34 immunostaining, while the lower part of ORSGFP+ remained CD34(-) but became P-cadherin-bright (Figures 2G; data not shown).
To further monitor these events, we performed a lineage tracing experiment with Lgr5-CreER-IRES-EGFP/Rosa-stop-LacZ-stop mice (Barker et al., 2007). At full anagen, Lgr5 was expressed by both bulge and ORS; however, since ORS is >15X larger than bulge, we could preferentially mark ORS cells by giving a low dose of tamoxifen (Figures 3A, S3A,B). Indeed when examined by thick sections (90μm) at anagen’s end, >70% of HFs had LacZ+ ORS cells, while only ~4% had LacZ+ bulge cells. When traced into telogen, >20% HFs had LacZ+ cells in new bulge or HG, while HFs with LacZ+ cells in the old bulge remained at ~5%. These data add to the evidence that ORSGFP+ is the main source for both new bulge and HG.
Figure 3. ORSGFP+ LRCs form the HG and CD34+ SCs of the new bulge.
Schematics summarize experiments and results. (A) Lineage tracing with Lgr5CreER/Rosa-LacZ to monitor ORS cell fate. Tamoxifen was given in full-anagen. 3d later (AnaVI), most HFs had LacZ+ cells in ORS (both up and low), but not bulge or matrix. When chased to telogen, LacZ+ cells showed up mainly in new bulge (both CD34+ and K6+ layers). n=2 mice, 176 HFs. Quantifications were on 90μm sections; images shown are 20μm sections (hematoxylin counterstain). (B) H2BGFP/BrdU double-label, double-pulse-chase experiment to monitor fate of upper ORSGFP cells. Tet-Off H2BGFP mice were chased from P21 and BrdU pulses during mid-anagen (P30–32) preferentially labeled cells in the upper ORSGFP trail. When chased to the anagen/catagen transition (P38), many HFs had BrdU+ LRCs in their ORSGFP+ (arrowhead); BrdU+ LRCs were rare elsewhere (n=3 mice, 42 HFs). When chased to telogen (P43), BrdU+/GFP+ cells (arrowhead) were mainly in the new bulge or HG (n=3 mice, 58 HFs). Scale bars, 30μm. new Bu and K6+Bu denote CD34+ and K6+ layers of new bulge. (C) Relative epifluorescence intensities of cells from different LRC populations of Tet-Off H2BGFP mice chased from the 1st telogen through one hair cycle. Raw data sets are plotted to right of each box-and-whisker diagram: median, 25th and 75th percentiles are denoted by notch, bottom and top boxes; 5th and 95th percentile are whiskers; minimum and maximum measurements are x’s. Asterisks indicate significant differences between data sets (**: P<0.01). Data are mean ± SD.
Based upon our proliferation analyses of ORS during anagen, we next began with pulse-chased Tet-Off H2BGFP mice and superimposed a series of BrdU pulse-chases that allowed us to differentiate old bulge and ORSGFP+ and selectively label different ORS segments. Since proliferation within the initial bulge occurs during early anagen (~P25–P29), we preferentially labeled ORS by administering BrdU at P30–P32 (Figure 3B). Moreover, since cells below ORSGFP+ cycle frequently, a 6d chase at anagen’s end and into catagen (P38) restricted most BrdU+ cells to the ORSGFP+ zone. When chased further into telogen, these GFP/BrdU double+ cells were found in new bulge and HG, thereby demonstrating that the ORSGFP+ of the prior cycle contributes substantially to the new bulge and HG.
Finally, we used epifluorescence intensity analyses to estimate the average #divisions that GFP+ LRCs within each compartment undergo during a hair cycle. Since cell divisions are silenced during catagen and telogen, GFP intensities within each compartment are retained, and thus, their intensities in telogen serve as a guide to their origin nearing anagen’s end. Analyses of GFP+ cells chased from the start of the first postnatal anagen to telogen revealed that cells which maintained their residence in the initial bulge divided the least (~2X) during the hair cycle: their epifluorescence corresponded to the old bulge (Figure 3C). By contrast, cells that resided in the ORS and acquired CD34 during catagen corresponded in epifluorescence to new bulge, and on average divided 1–2X more than the old bulge. LRCs that resided in the ORS, but remained CD34(−), displayed epifluorescence corresponding to HG and reflective of ~1–2X more divisions than new bulge. Quantifications in control mice showed that without Doxy, HG, new and old bulges displayed uniform levels of the highest H2BGFP signal, underscoring tight Tet-Off regulation and validating the efficacy of the data (Figure S3C).
Together, these results establish that cells in ORSGFP+ are the major source of HG and new bulge. Furthermore, they unveil a hitherto unrecognized relation between HF-SCs that exit the bulge during the growth phase, and the new bulge and HG that form during the destructive phase.
Some Mid-Zone ORS Cells Contribute to the HG
The ability of cells within the ORSGFP+ to survive catagen led us to wonder whether more rapidly cycling bulge progeny located in the middle or lower ORS might also be spared and possibly be the source of the ~10–20% of HG cells with very low GFP. To test this hypothesis, we delayed our BrdU pulse to P34–P36, a time when most bulge and ORSGFP+ cells had ceased proliferation. While ORSmid cells were proliferative at this time, they became quiescent soon thereafter, as evidenced by their label-retention after a 4d chase. These BrdU+ LRCs were uniformly low for GFP and could be found in the HG following a chase into telogen (Figure S3D). Thus while most of the HG was comprised of ORSGFP+ cells, the mid-zone was the source of HG cells undergoing the most divisions in the prior hair cycle. This cut-off was further defined by delaying the BrdU pulse so that cells below the mid-zone were preferentially labeled: under these conditions, no BrdU+ cells were detected in the HG (see below).
Together, these experiments delineate the ORSGFP+ zone as the point at which an HF-SC can exit the initial bulge and still be recycled to the CD34+ new bulge. The point at which HF-SCs can be recycled to the HG extends to the mid-zone. Moreover, these data reveal a strong inverse correlation between the number of divisions progeny have undergone and whether they are recycled to bulge vs HG.
A Potential Conundrum: The Return of Some Fast Cycling Bulge Descendents Below the ORSmid Zone to the Niche
During our double-label pulse-chase studies, we noticed that some chased BrdU+ cells started to show up in the new bulge when BrdU was pulsed at P34–36. Their number increased dramatically when the BrdU pulse was delayed to P36–38 (end of anagen) and then chased through catagen, suggestive that these cells came from a position below the ORS mid-zone (Figure 4A). Closer inspection revealed that in contrast to ORSGFP+ cells, these BrdU+ cells were restricted to the innermost bulge. Immunolabeling and FACS analysis showed that the cells were CD34(−) and hence distinct from suprabasal CD34+ bulge cells described previously (Blanpain et al., 2004). Instead, these cells were positive for keratin 6 (K6) (Figures 4A, S4A-C).
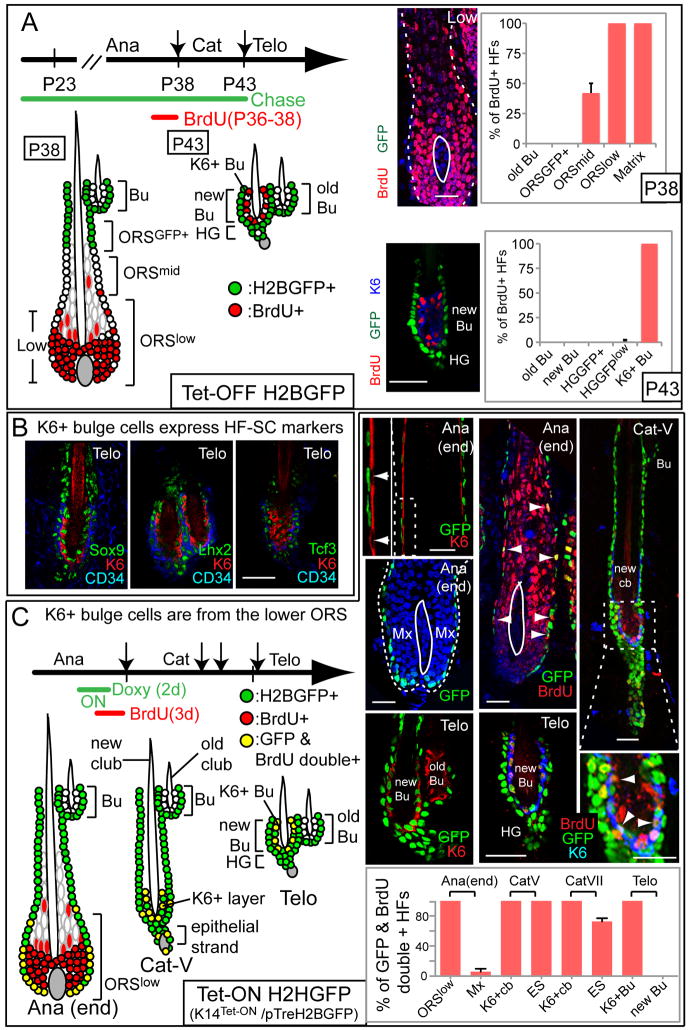
Figure 4. K6+ bulge cells express HF-SC markers and are derived from actively cycling lower ORS during catagen.
(A) H2BGFP/BrdU double-label, double pulse-chase scheme to monitor fate of cells below the mid-zone ORS. BrdU was administered to Tet-Off H2BGFP mice at anagen’s end (P36–38) and analyzed directly, or after chasing to telogen. At P38, many lower ORS and matrix cells are BrdU+ (n=2 mice, 38 HFs). When chased to P43, BrdU+ LRCs are restricted to the K6+ layer of the new bulge (n=4 mice, 129 HFs). (B) K6+ bulge cells express key transcription factors characteristic of bulge SCs and their ORS progeny. (C) Lineage tracing with K14Tet-On/H2BGFP coupled with BrdU pulse-chase. Doxy was administered to turn on H2BGFP in the entire ORS, but not K6+ companion layer or matrix, in late anagen. From catagen V through telogen, H2BGFP is detected in K6+ cells at the tip of the newly formed club hair. This layer retracts with the club hair during catagen and winds up in the new bulge by telogen. Note that K6+ old bulge cells are not GFP+. If BrdU is given at anagen’s end, BrdU+/H2BGFP+ cells are found in ORSlow. At catagen, double+ cells are in K6+ layer, which by telogen has moved inside new bulge. Scale bars, 30μm. K6+ club=K6+ layer enclosing new club hair. new Bu and K6+Bu denote CD34+ and K6+ layers of new bulge (n=2 mice; >23 HFs per stage). Data are mean ± SD.
K6 is also known to mark the terminally differentiated companion layer, which is derived from matrix during anagen and sandwiched between ORS and IRS (Figure S1A). The K6+ bulge cells differed from companion layer by their expression of key HF-SC transcription factors (Figure 4B). Ultrastructurally, K6+ bulge cells also more closely resembled CD34+ bulge SCs than companion layer cells (Figure S4D-F). However, they differed from bulge SCs in making unusual adhesive contacts with the club hair and extensive desmosomal contacts with their neighbors. Thus, the cells fated to enter the innermost layer of the new bulge at the end of catagen were distinct from any of the known HF residents.
The K6+ Bulge Layer is Derived from Actively Cycling Cells in the Lower ORS
A priori, the K6+ bulge layer could be a bona fide companion layer, derived from matrix but displaying distinct molecular features at different hair cycle stages. Alternatively, it could be derived from actively cycling lower ORS cells, which face a barrier to lineage progression when matrix apoptoses (catagen IV). The latter possibility was intriguing given that the ORS expresses many HF-SC markers that matrix and anagen companion layers do not (Rendl et al., 2005) (Figure S5A).
To identify the source of K6+ bulge cells, we first revisited our Lgr5-CreER/Rosa-LacZ lineage tracings, which labeled cells in both upper and lower ORS, but not matrix when induced near the end of anagen (Figure 3A). In telogen, LacZ+ cells were present in both CD34+ and K6+ bulge layers (Figures 3A, S5B). Similar results were obtained when lineage tracings were conducted with K14creER/Rosa26-LacZ mice (Figure S5C). Together with the BrdU pulse results, these data suggest that the K6+ bulge layer comes from lower ORS and not matrix.
To further demonstrate the lower ORS→matrix cell step that occurs during anagen is bypassed in catagen, we used K14-Tet-On/H2BGFP mice, which upon Doxy, turn on H2BGFP in ORS (Nguyen et al., 2006). When Doxy was given at mid anagen and HFs were examined at anagen’s end, H2BGFP was nicely expressed in ORS (including its CD34+ bulge layer) but not matrix or companion layer (Figure 4C). Since H2BGFP is stable and ORS cells don’t divide during catagen, the fate of label could be monitored. Through catagen IV, bright H2BGFP persisted only in ORS. At catagen V after the matrix apoptosed, GFP+ cells appeared in the K6+ layer at the tip of the retracting club hair (shown).
These cells survived catagen, remained K6+ and GFP+, expressed HF-SC/ORS markers and wound up in the telogen bulge (Figures 4C, S5F,G). Moreover, since when chased into telogen, K6+ cells of new bulge were GFP+ while those of old bulge were GFP(−), the GFP-labeling reflected lineage fate mapping and not ectopic K14 promoter activity (shown). We also confirmed this by inducing H2BGFP in telogen and verifying absence of GFP+ cells in the K6+ bulge layer (Figures 4C, S5D). These results exclude both matrix and anagen companion layer as possible precursors of the K6+ bulge. Moreover, the catagen-imposed bypassing of the ORS→matrix transition explains the dramatically different biology between cells of the K6+ bulge (ORS-derived) and companion layer (matrix-derived).
To specifically follow the fate and movement of ORSlow cells, we combined a BrdU pulse-chase scheme (pulse at the end of anagen) with this Tet-On H2BGFP strategy. ORSlow was the only population with BrdU/H2BGFP double+ cells in every HF. As expected, starting from catagen V, double+ cells appeared in the K6+ layer around the newly formed club hair, and retracted upward with it as the new bulge formed. During catagen, a few double+ cells also appeared in the epithelial strand and in the external layer flanking newly formed K6+ cells (Figure 4C, S5G). In sharp contrast to these layers, however, no TUNEL+ or Cp3+ cells were detected in the ORS-derived K6+ layer (Figure S5E, 0/352 HF examined). Thus, although some ORSlow cells survived catagen and wound up in new bulge, they took up residence in the K6+CD34(−) layer.
K6+ Bulge Cells Do Not Participate in Normal Homeostasis
We next wondered whether niche environment might restore HF-SC function to lower ORS-derived bulge cells, enabling them to utilize the remaining proliferative capacity they possessed prior to catagen. The importance of testing this possibility was heightened when we discovered that the K6+ layer displayed not only HF-SC markers Sox9, Lhx2 and TCF3, still expressed by lower ORS, but also nuclear NFATc1, previously found only in bulge SCs (Horsley et al., 2008) (Figure S6A).
We first addressed whether K6+ bulge cells proliferate in the next hair cycle. To capture proliferation if it existed, we pulsed with BrdU throughout the 2d window of early anagen when HF-SCs are most active. Under conditions where >60% of CD34+ bulge cells incorporated BrdU, no proliferation was detected in K6+ bulge cells of >190 HFs examined (Figure 5A).
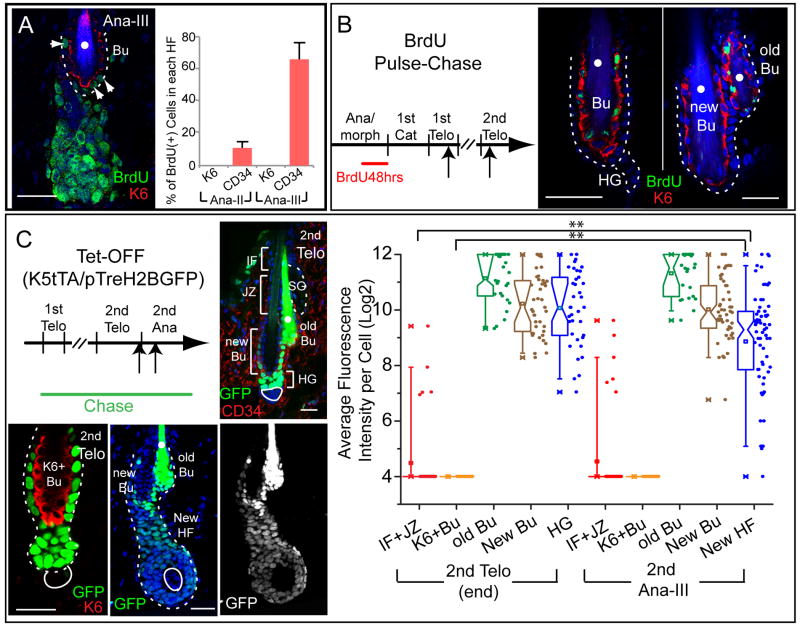
Figure 5. CD34+ bulge SCs and HG are the only cells used in normal HF homeostasis.
(A) Representative example and quantifications of HFs in early anagen (AnaII-AnaIII) with 2d BrdU pulse. Note BrdU in some CD34+ HF-SCs (arrowheads) but not inner K6+ bulge cells. White dots denote autofluorescent club hair (n=2 mice, 61 HFs for AnaII; n=3 mice, 133 HFs for AnaIII). (B) A 2d BrdU pulse given at anagen’s end is followed through 2 hair cycles. While not proliferative, K6+ bulge cells retain label from late anagen predecessors. K6+/BrdU+ cells persist through the next hair cycle and become the K6 layer of old bulge. (C) Lineage tracing using Tet-Off H2BGFP mice shows that the new HF comes from bulge LRCs. Chase was begun at 1st telogen; analysis began at end of 2nd telogen when H2BGFP selectively label old, new bulge and HG. Developing HFs are composed solely of H2BGFP+ cells. IF, infundibulum; JZ, junctional zone; SG, sebaceous gland. Scale bars, 30μm. Box-and-whisker plots measure H2BGFP epifluorescence intensity within different HF populations. Note paucity of GFP intensity in IF, JZ and K6+ bulge, revealing their lack of contribution to (GFP+) newly forming HF. **: P<0.01. Data are mean ± SD.
Although the K6+ bulge layer was quiescent, it was possible that some of its cells might migrate to the CD34+ layer or ORS and contribute to the hair cycle. We ruled this out by administering BrdU at the end of anagen and then chasing into telogen to selectively label K6+ bulge cells (Figure 5B). When chased throughout the following hair cycle, no migration of BrdU labeled cells was observed. In the next telogen, BrdU label remained within the K6+ layer of what became the old bulge (Figures 5B,S6B).
Our data infer that even though highly proliferative cells in anagen can wind up in the bulge, slow-cycling CD34+ cells are the source of new HFs. To further substantiate this, we took Tet-Off H2BGFP mice that were chased from P21 to the 2nd telogen, so that H2BGFP labeled CD34+ old and new bulge and CD34(−) HG, but not K6+ bulge cells or HF cells above the bulge (Figure 5C). When the 2nd adult anagen began, the emerging follicle was composed of GFP+ cells. Even though proliferation rapidly diluted out label, the fluorescence intensity was still higher than all other HF compartments except CD34+ bulge and HG. Thus while prior lineage tracings have shown contributions of CD34+ bulge and HG cells to the new HF (Morris et al., 2004; Zhang et al., 2009), these new data further suggest that cells in the K6+ bulge layer, junctional zone, sebaceous gland and infundibulum do not contribute to normal HF homeostasis. Based upon these data, CD34+ slow-cycling SCs appear to be the sole source of SCs used for HF homeostasis.
CD34+ HF-SCs in Both Old and New Bulge Participate in Wound Repair, While K6+ Bulge Cells Do Not Respond to Either Wounding or CD34+ Bulge Cell Ablation
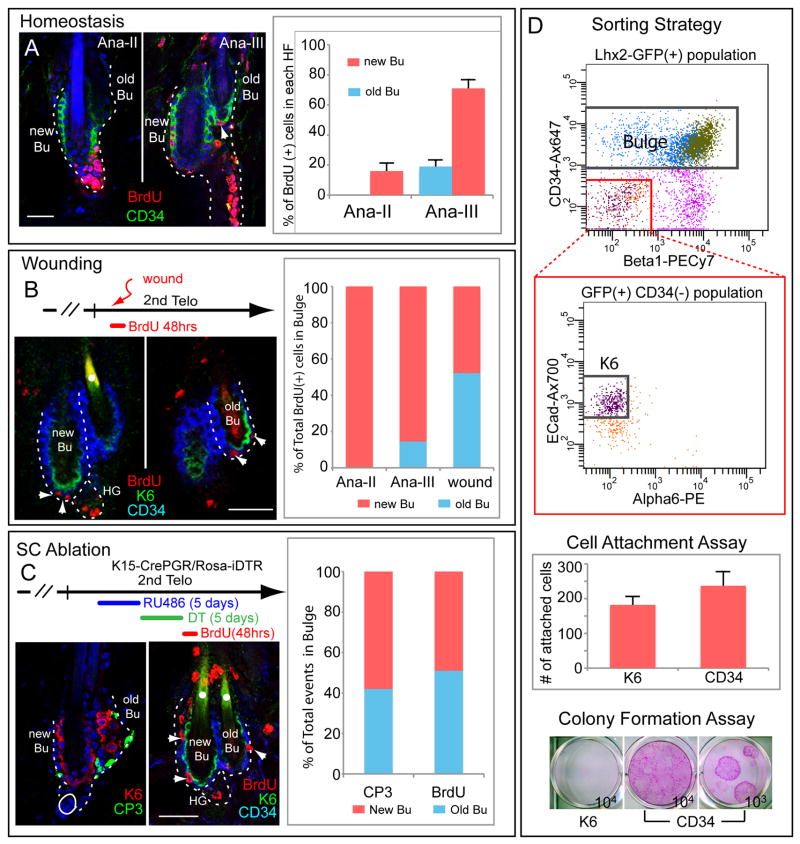
The new HF always emerged from new and not old bulge, leading us to wonder whether the old bulge functions in normal homeostasis. Careful inspection revealed an occasional proliferative cell at the base of the old bulge, but only when BrdU was pulsed at anagen III, i.e. the height of new bulge activity (Figure 6A). However when quantified, this constituted <15% of combined bulge activity, indicating that the CD34+ new bulge is primarily responsible for fueling new HF downgrowth.
Figure 6. CD34+ but not K6+ bulge cells are activated in wound repair and upon HF-SC ablation.
(A) Homeostasis. Mice given a 2d pulse with BrdU at start of 2nd anagen were monitored to AnaIII. Arrowhead marks BrdU+ cell in old bulge, which is >3X less active than new bulge (n=3 mice, 62 HFs). (B) Wounding. Punch biopsies were from 2nd telogen. BrdU given over 2d shows that old and new bulges participate comparably in injury responses (n=3 mice, 31 HFs). (C) SC ablation. K15-CrePGR was used to express DTR in CD34+ bulge during 2nd telogen. After DT and BrdU, mice were analyzed (n=3 mice, 64 HFs). Note that when bulge SCs are ablated, remaining CD34+ cells from old and new bulges proliferate, while K6+ bulge cells are refractory. Scale bars, 30μm. (D) K6+ and CD34+ bulge cells were isolated by FACS from telogen HFs of Lhx2-EGFP mice and subjected to culture experiments, performed in triplicate. 1000 cells of each population were assayed for attachment 16hr post-plating. K6+ and CD34+ cells were also plated and a/ 3 wks, colonies were fixed and stained w/1% Rhodamine B. Data are mean ± SD.
CD34+ bulge cells are known to contribute to wound repair (Blanpain et al., 2004; Ito et al., 2005; Morris et al., 2004). Two additional questions now surfaced: 1) Since the old bulge still contains CD34+ HF-SCs, does it contribute to wound-repair and if so, does it do so equivalently with the new bulge? 2) While refractile to normal homeostasis, do K6+ bulge cells participate in wound repair?
To address these questions, we introduced punch wounds during the extended 2nd telogen, administered BrdU, and then examined the skin 2d later (Figure 6B). BrdU+ cells were found in the (CD34+) outer layer of new bulge and its associated HG. Proliferating CD34+ cells were also detected in old bulge. Quantifications revealed similar contributions from the bulges, but only from CD34+ and not K6+ cells.
Next, we tested responses under more extreme conditions: HF-SC ablation (Figure 6C). K15-CrePGR mice were used to induce expression of Diphtheria Toxin Receptors (DTRs) (Buch et al., 2005) in CD34+ bulge cells. RU486 was given to activate CrePGR, followed by DT to ablate CD34+ bulge cells. Activated CP3 was prevalent in CD34+ but not K6+ bulge layers, verifying selective targeting. Death was equivalent for both new and old bulges, and a 2d BrdU pulse prior to analyses further revealed proliferation in CD34+ bulge cells that had not been ablated (Figure 6C; data not shown). By contrast, K6+ bulge cells did not respond even when the HF-SC reservoir was depleted.
K6+ Bulge Cells Fail to Grow In Vitro
To address K6+ bulge cell potential, we devised a strategy to isolate them by FACS and then carried out colony formation assays (Figure 6D). An Lhx2-GFP transgene behaved similarly to endogenous Lhx2, displaying GFP in both K6+ and CD34+ bulge layers of telogen HFs (Figure S6C). K6+ and CD34+ cells in the GFP+ pool were then further fractionated by differential CD34, E-cadherin and integrin levels: K6+ bulge cells were gated as GFP+CD34−α6−β1−Ecadhigh, while CD34+ cells were sorted as GFP+ and CD34+. Post-sort marker analysis revealed >94% purity, and consistent with immunofluorescence, RT-PCR showed that K6+ and CD34+ cells expressed similar levels of Sox9, Lhx2, Tcf3 and Nfatc1 (Figure S6C,D). In vitro, even though both populations attached to their substratum, only CD34+ and not K6+ bulge cells formed colonies (Figure 6D). Collectively, these data suggest that lower ORS cells that home back to the bulge have irreversibly lost their proliferative and regenerative potential.
K6+ Bulge Cells Function to Retain the Hair Coat and Maintain HF-SC Quiescence
If K6+ bulge cells do not function as HF-SCs, what are their functions in the bulge niche? The first clues came from hair plucking studies (Figure S7A). When telogen hairs were plucked, the entire K6 layer came with the club hair, demonstrating a tight association. By contrast, most CD34+ cells remained in the bulge.
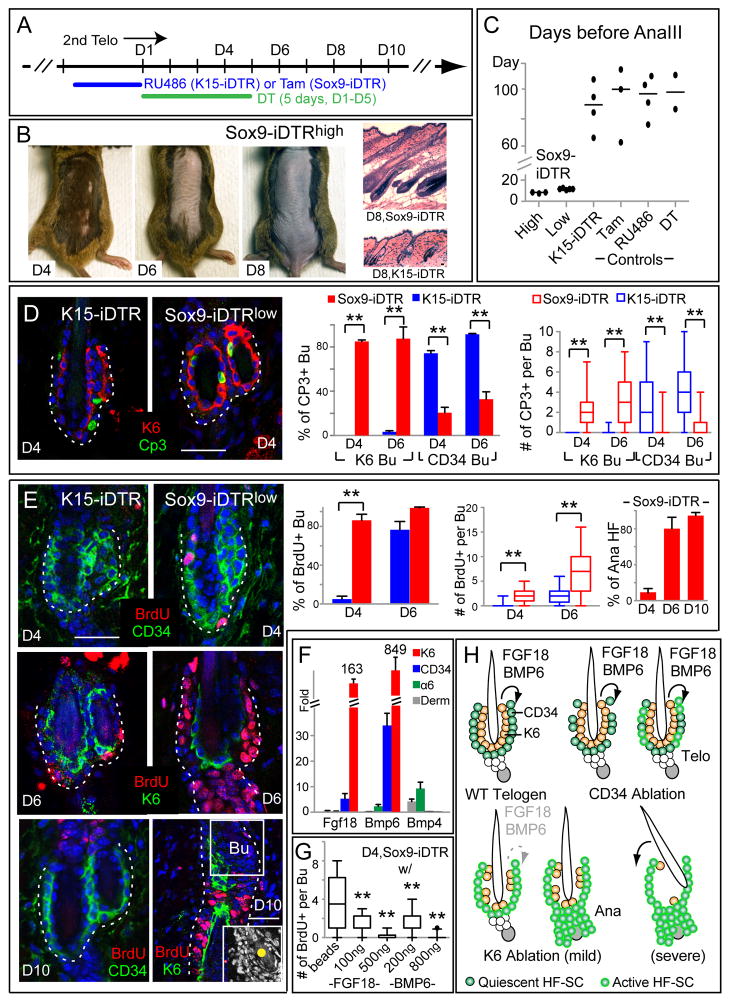
To test whether the K6+ layer functions to anchor the club hair to the bulge, we devised a strategy to preferentially ablate K6+ vs CD34+ bulge cells (Figures 7A). Sox9-CreER (Soeda et al., 2010) is expressed in both K6+ and CD34+ bulge layers. However, in Sox9-CreER/Rosa-iDTR mice, when hairs were first trimmed and tamoxifen was applied topically, drug entered the hair pores and preferentially induced DTR in K6+ cells. By contrast, RU486 induced DTR in CD34+ bulge cells of K15-CrePGR/Rosa-iDTR mice (Figure 7D and data not shown). Following DTR induction, mice were then treated with DT. Within 4d, Sox9-iDTR mice began to show hair loss and by D6 the coat was gone (Figure 7B; 5 of 5 mice tested). During this time, K15-iDTR mice kept their hair coat even when slightly more bulge cells were ablated (Figure S7B).
Figure 7. K6+ bulge cells anchor the club hair and serve as a signaling center for the HF-SC niche.
(A) Scheme to differentially express DTR in K6+ and CD34+ bulge cells and selectively DT-ablate each layer during extended 2nd telogen. D1=d1 of DT injection. Sox9-iDTRhigh was treated with Tamoxifen 4d while Sox9-iDTRlow was treated 2d (see text). (B) In contrast to K15-iDTR mice, Sox9-iDTR mice lose their hair coat at D6 (anchoring function) and skin turns black at D8, reflective of precocious entry into anagen and confirmed by histology. (C) Quantifications of anagen entry (signaling function). Graph shows #d before HFs entered anagen III for various genotypes/treatments. Horizontal bars denote median. (D, E) DT-treated K15- and Sox9-iDTR mice were given a 1d BrdU pulse at times indicated prior to immunodetection of apoptotic and proliferation markers on skin sections. In D10 Sox9-iDTR image, K6 marks the anagen companion layer. Scale bars, 30μm. Graphs quantify %bulges positive for CP3, BrdU and anagen. Box-and-whisker plots indicate #events per HF/bulge: mid-line, median; box, 25th to 75th percentiles; whiskers, minimum and maximum. (F) Real-time PCR on FACS-isolated populations. Values are normalized to total skin (epidermis+dermis) mRNAs. Bmp4 is higher in dermis (derm) and α6+ cells as previously reported (Plikus et al., 2008). Note high Fgf18 and Bmp6 expression in K6+ bulge cells. (G) Sox9-iDTR mice were treated 3d w/DT and growth factors and then 1d w/BrdU before analysis of proliferation (>24 HFs/experiment in duplicate). (H) Model summarizes different outcomes of ablating K6+ vs CD34+ bulge cells in telogen. When CD34+ cells are ablated, remaining HF-SCs become activated briefly, but then return to quiescence. When K6+ cells are ablated, quiescent signals (FGF18, BMP6) from K6+ bulge are lost, greatly reducing the threshold for anagen-activation. The K6+ bulge layer also functions to anchor the club hair. **: P<0.01. Data are mean ± SD.
Interestingly, 2d after hair loss, skins of Sox9-iDTR mice became dark (Figure 7B). This indicator of anagen entry (Muller-Rover et al., 2001) was substantiated by histological analyses (shown). It occurred ≥50d earlier than K15-iDTR or control mice (Figure 7C). By mechanisms unknown, hair plucking also triggers anagen reentry. Interestingly, Sox9-iDTR mice were >4d quicker to enter anagen compared to hair plucking, but K6+ bulge cells were also lost by plucking (Figure S7A). These collective findings led us to consider the possibility that it is loss of K6+ bulge that triggers this precocious anagen, and to assess whether K6+ bulge cells actively participate in maintaining telogen.
To focus specifically on the effects of K6+ cell loss rather than hair loss or mechanical trauma, we identified conditions where sufficient K6+ bulge cells survived in our Sox9-iDTR mice so that club hairs were retained longer. We then adjusted RU486 so that total bulge cell death events were similar between Sox9- and K15-iDTR mice (Figure S7B). Under these conditions, Cp3+ cells were detected in CD34+ (K15-iDTR) and K6+ (Sox9-iDTR) layers beginning at D2, peaking ~D4–D6 and waning by D10 (Figure 7D, data not shown).
In K15-iDTR mice, apoptosis was accompanied by brief proliferation from surviving CD34+ bulge cells, as detected by sequential BrdU pulses over this interval. This appeared to be a repair response, as HFs thereafter returned to quiescence and anagen was not induced (Figure 7E, S7C). By contrast, despite club hair retention (Figure 7E inset), Sox9-iDTR HFs not only displayed more robust CD34+ proliferation, but soon afterwards they entered anagen. Throughout these treatments, K6+ cells showed no signs of proliferation. These effects were not observed with RU486, tamoxifen or DT alone (Figure 7C; data not shown), confirming that K6+ bulge cells contribute markedly in maintaining the resting state of the hair cycle. Intriguingly, the effects of K6+ cell loss were transient: following anagen entry, CD34+ bulge cells gradually returned to quiescence (data not shown).
To explore what potential signals from the K6+ bulge might influence CD34+ cell behavior, we focused on Fgf18 and Bmp6. These genes were previously shown to be upregulated in CD34+ telogen bulge cells in vivo, and their encoded factors inhibit HF-SC cycling in vitro (Blanpain et al., 2004; Greco et al., 2009). Remarkably, purified K6+ bulge cells displayed enormous enrichment of these genes compared to other epidermal or dermal cells that might impact on bulge microenvironment. Moreover, fgf18 and bmp6 expression levels were >25X in K6+ vs CD34+ bulge cells (Figure 7F).
To test whether these factors account for the ability of K6+ bulge cells to maintain CD34+ bulge quiescence, we ablated the K6+ layer in Sox9-iDTR mice and injected these growth factors intradermally using fluorescent beads as a guiding reference. In contrast to buffer and beads alone, each of these factors potently suppressed CD34+ bulge activation upon ablation the K6+ layer (Figures 7G, S7D).
DISCUSSION
Recycling SCs Caught in Transit From Their Niche to The TA Compartment
In this study, we examined the fate of HF-SCs that departed from their niche but had not yet reached the TA pool when the destructive phase began. In contrast to the prevailing view, we discovered that many ORS cells in transit between bulge and matrix survive the massive apoptosis that follows anagen. By using H2B-GFP as a sensitive and stable label for lineage tracing, we not only tracked the fate of CD34+ HF-SCs but also distinguished descendents on the basis of cell divisions that occur after departing the niche. By combining this powerful approach with classical lineage tracing and nucleotide pulse-chase, we discovered that descendents closest to the bulge and which undergo the fewest cell divisions are recycled and contribute to the long-term SC pool that fuels the subsequent hair cycle. Ones a little further en route become activated SCs of the HG (Greco et al., 2009). Fast-cycling ORS cells nearer to matrix return to the bulge and function, but not as bona fide SCs.
Recycled SCs Make a New Niche
It was hitherto unrecognized that HF-SCs that depart their niche in one cycle become CD34+ bulge SCs for the next cycle. Our findings further imply that HF-SCs with fewer divisions are set aside in reserve within the old bulge, while those undergoing more divisions are recycled into a new bulge for homeostasis. In this regard, it is intriguing that in hematopoiesis, the most quiescent HSCs participate in injury repair while less quiescent ones are used in homeostasis (Foudi et al., 2009; Wilson et al., 2008). It will be interesting in the future to probe deeper into the impact of cell divisions on HF-SC stemness, and to explore how melanoblast and other SCs in the bulge migrate and function in this dynamic niche environment.
The Origin of the HG
Two major theories propose how the HG originates: (1) The lateral disc hypothesis posits that the HG comes from Shh(+) cells within matrix (Panteleyev et al., 2001); and (2) the bulge migration hypothesis suggests that HG cells migrate from the bulge at the catagen/telogen transition (Ito et al., 2004; Zhang et al., 2009). Both models are attractive, and while Shh(+) matrix cells are not the source of HF-SCs or HG (Greco et al., 2009), we show that cells outside the bulge clearly survive the destructive phase, as initially speculated (Panteleyev et al., 2001). Moreover, our study extends the similarities between HG and bulge (Greco et al., 2009; Ito et al., 2004; Zhang et al., 2009) even though HG appears to be derived from ORS-SCs which exited the bulge during the prior growth phase.
Our findings agree with prior studies emphasizing close proximity to DP stimuli in explaining why HG is activated so quickly at the start of a new cycle (Greco et al., 2009). However, our new data suggest that this enhanced sensitivity may also rely upon the intrinsic feature of having traveled further along the lineage and divided 1–2X more than new bulge cells. Moreover, since the ORS mid-zone is at the nexus along the lineage at which long-term self-renewing potential begins to wane, this could explain why HG cells do not sustain stemness in vitro (Greco et al., 2009).
Unexpected Origins and Properties of the K6+ Bulge: Insights Into Hair Cycle Control
Our findings suggest that when catagen is initiated and matrix undergoes apoptosis, proliferative cells in the lower ORS are short-circuited and return to the bulge. This finding helps to resolve the paradox as to why there are fast-cycling cells from anagen that return to the bulge (Jaks et al., 2008). Since these cells do express many HF-SC markers, it is tempting to conclude that they are SCs coming home to roost. However, our data clearly show that once catagen sets in, the proliferative potential of these cells ceases and cannot be reactivated for either homeostasis or wound-repair.
The most interesting facet of these surviving lower ORS cells is that despite their failure to regain proliferative potential, they perform essential non-SC functions in the bulge. We posit that their special ability to sense the bulge niche and thereby halt upward protrusion of hair from the skin surface could originate from retention of HF-SC markers when normal lineage progression is bypassed. Additional environmental cues (Figure S7E) along the lineage may endow them with differentiation features that enable their anchorage to the hair shaft and protect the animal against cyclical alopecia.
Perhaps even more remarkable is that lower ORS-derived bulge cells act as a signaling center in the niche. FGF18 and BMP6 were previously identified as autocrine factors able to maintain bulge SC quiescence (Blanpain et al., 2004; Greco et al., 2009). Our findings confirm these data but also point to a paracrine loop, that likely transmits a burst of these key factors to bulge SCs at the catagen/telogen transition when the K6+ layer forms. By imposing potent quiescence signals to the niche, the K6 bulge layer counterbalances the activating role provided by the DP at this time. In this way, the K6 bulge layer establishes the need for prolonged crosstalk between HF-SCs and DP to generate the blocking factors that enable HF-SCs to overcome this threshold (Greco et al., 2009). Moreover at anagen, the DP moves away from the niche towards the matrix but the K6 layer remains with the bulge, thereby launching a regulatory switch between bulge SCs and TA matrix. We posit that the smaller autocrine FGF18/BMP6 signals may further provide a means for HF-SCs along the upper ORS to retain their slow-cycling status until the new bulge is formed. Our studies now pave the way to test such hypotheses in the future.
Lessons Learned About Stem Cell Biology
By taking advantage of the distinct cycling properties of the HF lineage, we designed unambiguous lineage tracing experiments to determine the sources and fates of HF-SCs and their descendents throughout homeostasis. By superimposing a temporal series of pulse-chase experiments, we adopted the principles of a movie to monitor HF-SC movement during the hair cycle. This strategy could be useful for many other SCs that display proliferative differences with their progeny.
While the system has its advantages, the timing of labeling can determine whether the slow-cycling SCs retain the label (e.g. Cotsarelis et al., 1990; Morris and Potten, 1999) or their committed rapidly cycling progeny do (e.g. Jaks et al., 2008). Our studies also reveal an example of SC progeny that have become irreversibly committed, and yet which still express SC markers and home back to their SC niche. These revelations may help to resolve some controversies which have arisen from label-retaining and conventional lineage tracing experiments in this and other SC systems.
Several other concepts derived from our study could have broad implications for SC biology: First, our data show that a downstream lineage of SCs can be a critical component of the niche microenvironment and regulate the rate of SC divisions. However, our data also illustrate the power of intrinsic factors on stemness. As exemplified by the K6+ bulge layer and in contrast to the Drosophila germline (Brawley and Matunis, 2004; Kai and Spradling, 2004), once cells lose these features, regaining stemness is not assured by returning to an unperturbed niche, nor by depleting the niche of its SCs. Thus, although the niche can markedly affect SC behavior, it may not be sufficient to reset the stemness clock once SCs have passed the point of no return in a lineage. Future studies will be valuable in further defining the distinctions and the parallels among different SCs and their niches.
Experimental Procedures
Mice and Labeling Experiments
Lhx2-EGFP was generated by the GENSAT project (Heintz, 2004). See Supplementary Materials for all other strains used. Tet-Off and Tet-On were activated by continuously feeding mice with Doxy (2mg/kg) starting at P21 or times specified. CreER was activated by intraperitoneal injection (150 μg/g tamoxifen in corn oil) or topical application (10mg/ml in ethanol) as specified. CrePGR activation was by topical application of RU486 (1% in ethanol). For 5-Bromo-2′-deoxyuridine (BrdU) pulse-chase experiments, mice were injected intraperitoneally (50 μg/g) (Sigma-Aldrich) and chased for times specified. To test for rare proliferations, injections were supplemented with dietary BrdU water (0.8mg/ml).
Hair Cycle Timing
Subdivisions of the hair cycle into 6 anagen and 8 catagen stages were based on (Muller-Rover et al., 2001). Since hair cycles vary among strains and sexes, stages instead of mouse ages were usually evaluated. For K5-tTA/pTRE-H2BGFPK5-tTA (Tet-Off) mice, both are provided. Typically 3–4 mice of matched sex were analyzed.
Wounding and Ablation
0.6cm punch biopsy wounds were created on backs of anesthetized mice. For ablation of bulge cells, K15-CrePGR or Sox9-CreER X Rosa-iDTR mice were first treated topically to induce Cre, and then injected intraperitoneally (i.p.) with Diphtheria toxin (200ng DT/injection, Sigma) 1X/d for 5d.
Quantifications of H2BGFP Intensities
Different stages of Tet-Off H2BGFP HFs were co-stained for CD34 and imaged by LSM510 laser-scanning confocal (same laser input and gain). Fluorescence intensities were measured using MetaMorph 7 (Universal Imaging Corp.). Cell division numbers were estimated based on H2BGFP epifluorescence intensity (brightest cells assigned 0 divisions). To reflect cell proliferation histories, native H2BGFP intensities were measured. The exception is Figure 5C where signals were enhanced with GFP antibody and laser input and gain were increased as necessary to detect weakly fluorescent cells. Individual data points were plotted and statistic analyses (Student’s t test) were performed using OriginLab 7.5 or Prism 5 software. Box-and-whisker plots are used to describe the entire population without assumptions on statistical distribution (Schober et al., 2007).
Supplementary Material
Acknowledgments
We are grateful to colleagues who generously donated mice, especially B.de Crombrugghe (UT MD Anderson) and H. Akiyama (Kyoto University) for sharing Sox9-CreER mice prior to publication; S. Mazel, C. Bare and RU’s FCRC for FACS sorting; A. North (Bioimaging Resource Center) for advice in image acquisition; Comparative Biology Center (ALAAC-accredited) for health care to our mice; and members of the Fuchs’ lab, in particular: T. Chen, D. Devenport E. Ezhkova and B. Keyes for comments on the manuscript; M. Schober for advice on image analyses and quantifications; N. Stokes and D. Oristian for assistance in mouse research. Y-C. H. is a Starr Stem Cell Scholars Postdoctoral Fellow. This work was supported by grants from NIH (R01AR050452), Starr Foundation and NYSTEM (N09G074) to E.F., who is an HHMI Investigator. RU FCRC is supported by the Empire State Stem Cell fund through NYSDOH Contract #C023046.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Literature Cited
- Barker N, van Es JH, Kuipers J, Kujala P, van den Born M, Cozijnsen M, Haegebarth A, Korving J, Begthel H, Peters PJ, Clevers H. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature. 2007;449:1003–1007. doi: 10.1038/nature06196. [DOI] [PubMed] [Google Scholar]
- Blanpain C, Lowry WE, Geoghegan A, Polak L, Fuchs E. Self-renewal, multipotency, and the existence of two cell populations within an epithelial stem cell niche. Cell. 2004;118:635–648. doi: 10.1016/j.cell.2004.08.012. [DOI] [PubMed] [Google Scholar]
- Brawley C, Matunis E. Regeneration of male germline stem cells by spermatogonial dedifferentiation in vivo. Science. 2004;304:1331–1334. doi: 10.1126/science.1097676. [DOI] [PubMed] [Google Scholar]
- Buch T, Heppner FL, Tertilt C, Heinen TJ, Kremer M, Wunderlich FT, Jung S, Waisman A. A Cre-inducible diphtheria toxin receptor mediates cell lineage ablation after toxin administration. Nat Methods. 2005;2:419–426. doi: 10.1038/nmeth762. [DOI] [PubMed] [Google Scholar]
- Cao YA, Wagers AJ, Beilhack A, Dusich J, Bachmann MH, Negrin RS, Weissman IL, Contag CH. Shifting foci of hematopoiesis during reconstitution from single stem cells. Proc Natl Acad Sci U S A. 2004;101:221–226. doi: 10.1073/pnas.2637010100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claudinot S, Nicolas M, Oshima H, Rochat A, Barrandon Y. Long-term renewal of hair follicles from clonogenic multipotent stem cells. Proc Natl Acad Sci U S A. 2005;102:14677–14682. doi: 10.1073/pnas.0507250102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotsarelis G, Sun TT, Lavker RM. Label-retaining cells reside in the bulge area of pilosebaceous unit: implications for follicular stem cells, hair cycle, and skin carcinogenesis. Cell. 1990;61:1329–1337. doi: 10.1016/0092-8674(90)90696-c. [DOI] [PubMed] [Google Scholar]
- Foudi A, Hochedlinger K, Van Buren D, Schindler JW, Jaenisch R, Carey V, Hock H. Analysis of histone 2B-GFP retention reveals slowly cycling hematopoietic stem cells. Nat Biotechnol. 2009;27:84–90. doi: 10.1038/nbt.1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs E. The Tortoise and the Hair: Slow-Cycling Cells in the Stem Cell Race. Cell. 2009;137:811–819. doi: 10.1016/j.cell.2009.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greco V, Chen T, Rendl M, Schober M, Pasolli HA, Stokes N, Dela Cruz-Racelis J, Fuchs E. A two-step mechanism for stem cell activation during hair regeneration. Cell Stem Cell. 2009;4:155–169. doi: 10.1016/j.stem.2008.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintz N. Gene expression nervous system atlas (GENSAT) Nat Neurosci. 2004;7:483. doi: 10.1038/nn0504-483. [DOI] [PubMed] [Google Scholar]
- Horsley V, Aliprantis AO, Polak L, Glimcher LH, Fuchs E. NFATc1 balances quiescence and proliferation of skin stem cells. Cell. 2008;132:299–310. doi: 10.1016/j.cell.2007.11.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito M, Kizawa K, Hamada K, Cotsarelis G. Hair follicle stem cells in the lower bulge form the secondary germ, a biochemically distinct but functionally equivalent progenitor cell population, at the termination of catagen. Differentiation. 2004;72:548–557. doi: 10.1111/j.1432-0436.2004.07209008.x. [DOI] [PubMed] [Google Scholar]
- Ito M, Liu Y, Yang Z, Nguyen J, Liang F, Morris RJ, Cotsarelis G. Stem cells in the hair follicle bulge contribute to wound repair but not to homeostasis of the epidermis. Nat Med. 2005;11:1351–1354. doi: 10.1038/nm1328. [DOI] [PubMed] [Google Scholar]
- Jaks V, Barker N, Kasper M, van Es JH, Snippert HJ, Clevers H, Toftgard R. Lgr5 marks cycling, yet long-lived, hair follicle stem cells. Nat Genet. 2008;40:1291–1299. doi: 10.1038/ng.239. [DOI] [PubMed] [Google Scholar]
- Kai T, Spradling A. Differentiating germ cells can revert into functional stem cells in Drosophila melanogaster ovaries. Nature. 2004;428:564–569. doi: 10.1038/nature02436. [DOI] [PubMed] [Google Scholar]
- Lavker RM, Sun TT, Oshima H, Barrandon Y, Akiyama M, Ferraris C, Chevalier G, Favier B, Jahoda CA, Dhouailly D, et al. Hair follicle stem cells. J Investig Dermatol Symp Proc. 2003;8:28–38. doi: 10.1046/j.1523-1747.2003.12169.x. [DOI] [PubMed] [Google Scholar]
- Morris RJ, Liu Y, Marles L, Yang Z, Trempus C, Li S, Lin JS, Sawicki JA, Cotsarelis G. Capturing and profiling adult hair follicle stem cells. Nat Biotechnol. 2004;22:411–417. doi: 10.1038/nbt950. [DOI] [PubMed] [Google Scholar]
- Morris RJ, Potten CS. Highly persistent label-retaining cells in the hair follicles of mice and their fate following induction of anagen. J Invest Dermatol. 1999;112:470–475. doi: 10.1046/j.1523-1747.1999.00537.x. [DOI] [PubMed] [Google Scholar]
- Morrison SJ, Kimble J. Asymmetric and symmetric stem-cell divisions in development and cancer. Nature. 2006;441:1068–1074. doi: 10.1038/nature04956. [DOI] [PubMed] [Google Scholar]
- Muller-Rover S, Handjiski B, van der Veen C, Eichmuller S, Foitzik K, McKay IA, Stenn KS, Paus R. A comprehensive guide for the accurate classification of murine hair follicles in distinct hair cycle stages. J Invest Dermatol. 2001;117:3–15. doi: 10.1046/j.0022-202x.2001.01377.x. [DOI] [PubMed] [Google Scholar]
- Nakagawa M, Koyanagi M, Tanabe K, Takahashi K, Ichisaka T, Aoi T, Okita K, Mochiduki Y, Takizawa N, Yamanaka S. Generation of induced pluripotent stem cells without Myc from mouse and human fibroblasts. Nat Biotechnol. 2008;26:101–106. doi: 10.1038/nbt1374. [DOI] [PubMed] [Google Scholar]
- Nguyen H, Rendl M, Fuchs E. Tcf3 governs stem cell features and represses cell fate determination in skin. Cell. 2006;127:171–183. doi: 10.1016/j.cell.2006.07.036. [DOI] [PubMed] [Google Scholar]
- Nowak JA, Polak L, Pasolli HA, Fuchs E. Hair follicle stem cells are specified and function in early skin morphogenesis. Cell Stem Cell. 2008;3:33–43. doi: 10.1016/j.stem.2008.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oshima H, Rochat A, Kedzia C, Kobayashi K, Barrandon Y. Morphogenesis and renewal of hair follicles from adult multipotent stem cells. Cell. 2001;104:233–245. doi: 10.1016/s0092-8674(01)00208-2. [DOI] [PubMed] [Google Scholar]
- Panteleyev AA, Jahoda CA, Christiano AM. Hair follicle predetermination. J Cell Sci. 2001;114:3419–3431. doi: 10.1242/jcs.114.19.3419. [DOI] [PubMed] [Google Scholar]
- Paus R, Cotsarelis G. The biology of hair follicles. N Engl J Med. 1999;341:491–497. doi: 10.1056/NEJM199908123410706. [DOI] [PubMed] [Google Scholar]
- Plikus MV, Mayer JA, de la Cruz D, Baker RE, Maini PK, Maxson R, Chuong CM. Cyclic dermal BMP signalling regulates stem cell activation during hair regeneration. Nature. 2008;451:340–344. doi: 10.1038/nature06457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rendl M, Lewis L, Fuchs E. Molecular dissection of mesenchymal-epithelial interactions in the hair follicle. PLoS Biol. 2005;3:1910–1924. doi: 10.1371/journal.pbio.0030331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rochat A, Kobayashi K, Barrandon Y. Location of stem cells of human hair follicles by clonal analysis. Cell. 1994;76:1063–1073. doi: 10.1016/0092-8674(94)90383-2. [DOI] [PubMed] [Google Scholar]
- Schober M, Raghavan S, Nikolova M, Polak L, Pasolli HA, Beggs HE, Reichardt LF, Fuchs E. Focal adhesion kinase modulates tension signaling to control actin and focal adhesion dynamics. J Cell Biol. 2007;176:667–680. doi: 10.1083/jcb.200608010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snippert HJ, Haegebarth A, Kasper M, Jaks V, van Es JH, Barker N, van de Wetering M, van den Born M, Begthel H, Vries RG, et al. Lgr6 marks stem cells in the hair follicle that generate all cell lineages of the skin. Science. 2010;327:1385–1389. doi: 10.1126/science.1184733. [DOI] [PubMed] [Google Scholar]
- Soeda T, Deng JM, de Crombrugghe B, Behringer RR, Nakamura T, Akiyama H. Sox9-expressing precursors are the cellular origin of the cruciate ligament of the knee joint and the limb tendons. Genesis. 2010 doi: 10.1002/dvg.20667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spradling AC, Nystul T, Lighthouse D, Morris L, Fox D, Cox R, Tootle T, Frederick R, Skora A. Stem cells and their niches: integrated units that maintain Drosophila tissues. Cold Spring Harb Symp Quant Biol. 2008;73:49–57. doi: 10.1101/sqb.2008.73.023. [DOI] [PubMed] [Google Scholar]
- Taylor G, Lehrer MS, Jensen PJ, Sun TT, Lavker RM. Involvement of follicular stem cells in forming not only the follicle but also the epidermis. Cell. 2000;102:451–461. doi: 10.1016/s0092-8674(00)00050-7. [DOI] [PubMed] [Google Scholar]
- Tumbar T, Guasch G, Greco V, Blanpain C, Lowry WE, Rendl M, Fuchs E. Defining the epithelial stem cell niche in skin. Science. 2004;303:359–363. doi: 10.1126/science.1092436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unna PG. Beitrage zur Histologie und Entwicklungsgeschichte der menschlichen Oberhaut und ihrer Anhangsgebilde. Arch Mikroskop Anat Entwicklungsmech. 1876;12:665–741. [Google Scholar]
- Waghmare SK, Bansal R, Lee J, Zhang YV, McDermitt DJ, Tumbar T. Quantitative proliferation dynamics and random chromosome segregation of hair follicle stem cells. Embo J. 2008;27:1309–1320. doi: 10.1038/emboj.2008.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson A, Laurenti E, Oser G, van der Wath RC, Blanco-Bose W, Jaworski M, Offner S, Dunant CF, Eshkind L, Bockamp E, et al. Hematopoietic stem cells reversibly switch from dormancy to self-renewal during homeostasis and repair. Cell. 2008;135:1118–1129. doi: 10.1016/j.cell.2008.10.048. [DOI] [PubMed] [Google Scholar]
- Zhang YV, Cheong J, Ciapurin N, McDermitt DJ, Tumbar T. Distinct Self-Renewal and Differentiation Phases in the Niche of Infrequently Dividing Hair Follicle Stem Cells. Cell Stem Cell. 2009;5:267–278. doi: 10.1016/j.stem.2009.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.