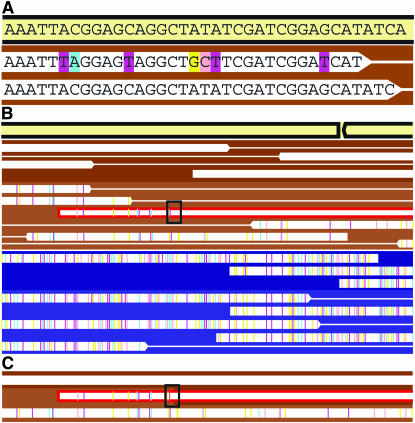
Figure 1.—
Diagram illustrating the representation of single-nucleotide polymorphisms (SNPs) that distinguish individual sequence variants within the community (white boxes signify sequencing reads, lines link mate pairs). Reads are aligned to a reference sequence that is either an isolate genome sequence or a composite sequence derived from a community genomic data set (cream-colored box). (A) Over the region shown one read has 7 SNPs relative to the reference sequence and the other is identical to it. Colored bars highlight the substituted base at each location; cyan, pink, magenta, and yellow represent substitutions of the bases A, C, T, and G, respectively. (B and C) The assembly is viewed at lower magnification without individual nucleotides labeled. The colors indicating SNPs are now small tick marks. In the region shown, sequencing reads can be easily clustered on the basis of SNP frequency. Backgrounds colored in shades of brown (similar to F. acidarmanus) and blue (similar to Ferroplasma type II scaffolds) indicate these groupings. However, within each cluster there is some fine-scale variation. In B the reads in the brown cluster are grouped into two sequence types (one with SNPs and one without SNPs). One read, outlined in red, represents a recombination of the two types. The recombinant read can be compared to the reconstructed parent sequences, as shown in C. Thus, the divergence between the parent sequences can be calculated for that recombination event.