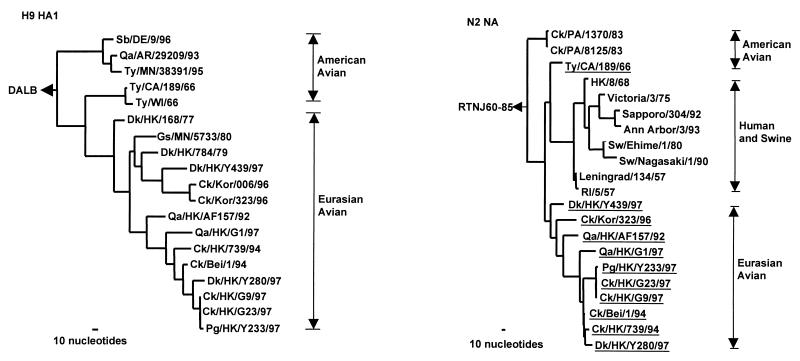
Figure 1.
Phylogenetic trees for the H9 HA1 and N2 NA genes of influenza A viruses. The nucleotide sequences of the HA1 and NA genes were analyzed with paup by using a maximum-parsimony algorithm. Nucleotides 43 to 1,047 (1,005 bp) of H9 HAs and nucleotides 21 to 1,393 (1,373 bp) of N2 NA were used for the phylogenetic analysis. The HA1 phylogenetic tree is rooted to duck/Alberta/60/76, DALB (H12N5). The N2 NA phylogenetic tree is rooted to A/ruddy turnstone/New Jersey/60/85, RTNJ60–85 (H4N9). The lengths of the horizontal lines are proportional to the minimum number of nucleotide differences required to join nodes. Vertical lines are for spacing branches and labels. Virus names and abbreviations are listed in Table 1, and the remaining sequences can be found in GenBank by using the following accession numbers: D90305, M12051, M11925, U42630, J02173, U42776, U43421, D00713, D29659, L37330, and M11205. All viruses underlined in the NA tree are H9N2 influenza viruses.