Summary
The emerged severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) Omicron subvariants BA.2.86 and JN.1 raise concerns regarding their potential to evade immune surveillance and spread globally. Here, we test sera from rhesus macaques immunized with 3 doses of wild-type SARS-CoV-2 receptor-binding domain (RBD)-Fc adjuvanted with the STING agonist CF501. We find that the sera can potently neutralize pseudotyped XBB.1.5, XBB.1.16, CH.1.1, EG.5, BA.2.86, and JN.1, with 50% neutralization titers ranging from 3,494 to 7,424. We also demonstrate that CF501, but not Alum, can enhance immunogenicity of the RBD from wild-type SARS-CoV-2 to improve induction of broadly neutralizing antibodies (bnAbs) with binding specificity and activity similar to those of SA55, BN03, and S309, thus exhibiting extraordinary broad-spectrum neutralizing activity. Overall, the RBD from wild-type SARS-CoV-2 also contains conservative epitopes. The RBD-Fc adjuvanted by CF501 can elicit potent bnAbs against JN.1, BA.2.86, and other XBB subvariants. This strategy can be adopted to develop broad-spectrum vaccines to combat future emerging and reemerging viral infectious diseases.
Keywords: coronavirus, SARS-CoV-2, JN.1, adjuvant, STING agonist, vaccine, conserved epitopes
Graphical abstract

Highlights
-
•
CF501/RBD-Fc induces potent bnAbs in macaques against JN.1 and BA.2.86 infection
-
•
Sera contain bnAbs targeting conserved epitopes on RBDs shared by SA55, BN03, and S309
-
•
RBDs of SARS-CoV-2 WT strain and JN.1 subvariant contain similar conserved epitopes
Liu et al. find that sera from macaques immunized with Fc-conjugated RBD from ancestral SARS-CoV-2 adjuvanted with a STING agonist, CF501, can potently neutralize BA.2.86, JN.1, and other Omicron variants, as the sera contains bnAbs targeting the conserved epitopes shared with the bnAbs SA55, BN03, and S309.
Introduction
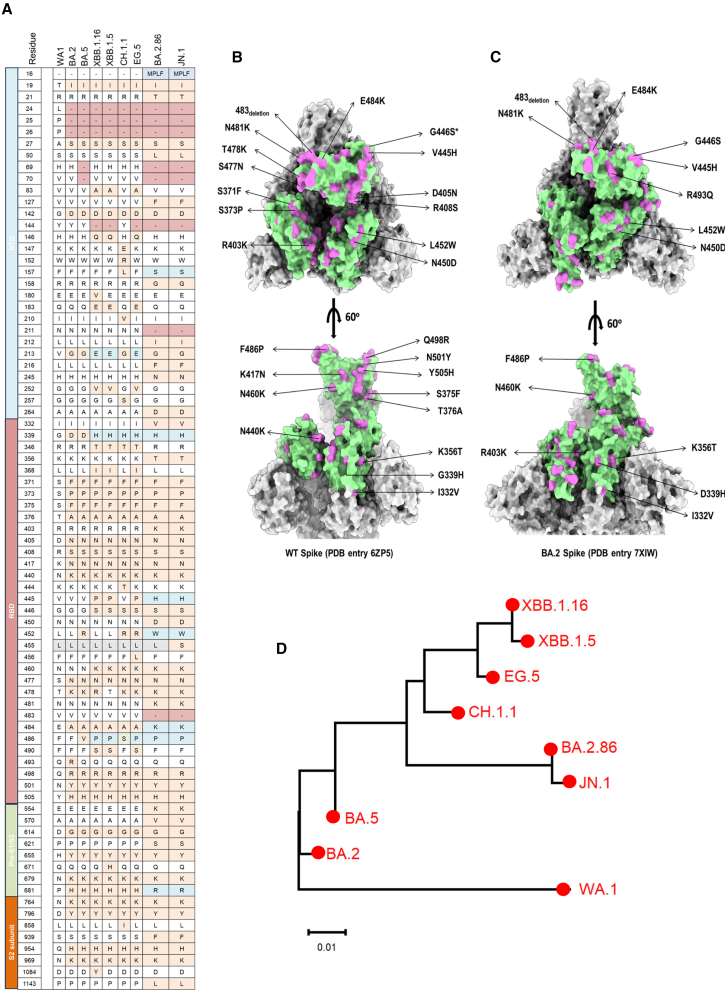
In 2021, the emergence of Omicron BA.1 with over 30 new mutations in its spike (S) protein relative to the original severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) strain has significantly influenced trends of the global pandemic.1,2 After that, a wide array of Omicron subvariants were continuously identified, such as BA.2, BA.5, and XBB. Most currently authorized COVID-19 vaccines such as parental mRNA vaccines and therapeutic antibodies showed a loss of efficacy against XBB owing to increased transmissibility and immune evasion.3,4,5,6 The XBB family has been the dominant variant globally for a long time. To combat the circulation of XBB, vaccine makers have updated the immunogen with XBB, e.g., Pfizer’s XBB.1.5-adapted monovalent COVID-19 vaccine. However, a new highly mutated Omicron subvariant, BA.2.86, has been recently identified in Israel and Denmark.7 Compared to the original SARS-CoV-2 and other strains such as XBB, Figure 1A illustrates the mutations on the S protein of BA.2.86. Numerous mutations have been found in BA.2.86, with 14 and 26 mutations in its receptor-binding domain (RBD) relative to parental BA.2 and wild-type (WT) SARS-CoV-2 strains, respectively (Figures 1A–1C). Up to now, the World Health Organization (WHO) has classified BA.2.86 as a variant under monitoring. Since the advent of BA.2.86, numerous reports have consistently demonstrated its status as one of the most immune-evasive variants to date.8,9,10 It exhibits the capability to evade the majority of monoclonal antibodies and sera from individuals immunized with mRNA vaccines.8,9,10 Recent studies have shown that pseudotyped BA.2.86 exhibits higher infectivity and cell-cell fusion rate in human lung cell line Calu-3 cells compared to BA.2 and XBB subvariants,11,12 implying that BA.2.86 might result in more severe COVID-19. Furthermore, Ho and colleagues reported that the receptor affinity of BA.2.86 was demonstrated to be higher than that of either BA.2 or XBB.1.5.13 Also, several antibodies able to neutralize XBB subvariants could not show neutralization activity against BA.2.86.13 Significantly, JN.1, the descendant strain of BA.2.86, contains an additional mutation (L455S) in the RBD region and has swiftly gained prevalence, emerging as the dominant strain in more than 40 countries, including the United States and France.14,15 The WHO has designated JN.1 as a variant of interest14 because JN.1 possesses a more robust immune escape capability compared to BA.2.86.15 Thus, the unique viral properties of JN.1 and BA.2.86 have raised concerns about their vaccine immune evasion potential.
Figure 1.
Sequence comparisons between BA.2.86, JN.1, and other representative Omicron subvariants
(A) Mutations in S proteins of BA.2, BA.5, XBB.1.5, XBB.1.16, CH.1.1, EG.5, BA.2.86, and JN.1 relative to original SARS-CoV-2 wild-type strain (WA1).
(B and C) Amino acid mutation sites in RBD of BA.2.86 compared with wild-type SARS-CoV-2 and BA.2. S trimers of BA.2 (PDB: 7XIW) and wild type (PDB: 6ZP5) are shown in a surface representation, colored in gray. The RBD region is colored in pale green. The mutations of BA.2.86 in the RBD, in comparison to BA.2 and the wild-type strain, are indicated and colored in orchid.
(D) Phylogenetic tree based on the RBD amino acid sequences shows the genetic distance between the SARS-CoV-2 wild-type strain (WA1) and its variants and subvariants, BA.2, BA.5, XBB.1.5, XBB.1.16, CH.1.1, EG.5, BA.2.86, and JN.1. The GeneBank accession numbers for their S protein amino acid sequences are GeneBank: YP_009724390.1 (WA1); GeneBank: WNO30888.1 (BA.2); GeneBank: WNH55069.1 (BA.5); GeneBank: WPU07054.1 (XBB.1.5); GeneBank: WNN93469.1 (XBB.1.16); GeneBank: WLS36136.1 (CH.1.1); GeneBank: WNK91259.1 (EG.5); GeneBank: WNK92404.1 (BA.2.86); and GeneBank: WQN42696.1 (JN.1).
Beginning with the outbreak of original SARS-CoV-2 (WA1), we have proposed a “non-changeable against changeable” strategy to combat the continuous evolution of SARS-CoV-2.16 The immunogen RBD-Fc from the original strain of SARS-CoV-2 was previously adjuvanted with the new immunopotentiator CF501, a STING agonist, and it elicited strong broadly neutralizing antibodies (bnAbs) across a broad spectrum against Delta, BA.2, BA.5, BQ.1.1, and even XBB in non-human primates.16,17,18 The distinct evolutionary direction between XBB subvariants and BA.2.86/JN.1 calls for further investigation of BA.2.86/JN.1. Specifically, we asked if BA.2.86/JN.1 exhibits stronger immune evasion compared to XBB and whether the antibodies in sera elicited by the pan-sarbecoviruses vaccine could strongly neutralize BA.2.86/JN.1 in the context of the constantly evolving SARS-CoV-2, a phenomenon that has now lasted for almost 4 years. For example, 26 mutations in the RBD of the BA.2.86 S protein have been identified compared to the RBD sequence of the WT SARS-CoV-2 S protein. Therefore, one key task is the discovery of conserved epitopes in the RBD of the WT SARS-CoV-2 S protein that have not yet mutated. This would provide a clue for understanding the limited mutation ability of the RBD, which would, in turn, guide broad-spectrum vaccine design. Here, we presented evidence that CF501/RBD-Fc (WT) vaccine-elicited sera could neutralize JN.1 and BA.2.86 in a manner that outperforms the neutralization of XBB.1.16, EG.5, and CH.1.1 by elevating the induction of the bnAbs with characteristics of the bnAbs that have been reported before, such as SA55,19 S309,20 and BN03.21 These results imply that some conserved epitopes in the WT RBD have never altered, despite the high number of mutations in the S proteins of JN.1 and BA.2.86. Thus, notwithstanding the evolution of WT SARS-CoV-2 over time, some conserved epitopes have remained unchanged up to now. Here, we demonstrate the use of adjuvants, namely CF501, to prime the induction of the bnAbs by targeting the latest Omicron variants carrying a high number of mutations, JN.1 and BA.2.86.
Results
JN.1 and BA.2.86 are genetically distant from XBB.1.16, BA.2, and WA1
Based on phylogenetic analysis, the genetic distance between BA.2.86/JN.1 and WA1 (WT) is further apart than that between XBB.1.16 and WA1 (Figure 1D). BA.2.86/JN.1 also showed a long genetic distance from XBB.1.16 (Figure 1D). We found that BA.2.86 exhibited several unique mutations in the RBD not found in other Omicron subvariants, such as I332V, K356T, R403K, V445H, N450D, L452W, N481K, Del 483, and E484K (Figures 1A–1C). These mutations might contribute to different infectivity rates and immune escape capacity.
Potency and durability of XBB.1.5-, XBB.1.16-, CH.1.1-, EG.5-, and BA.2.86-binding antibodies elicited by CF501/RBD-Fc in macaques
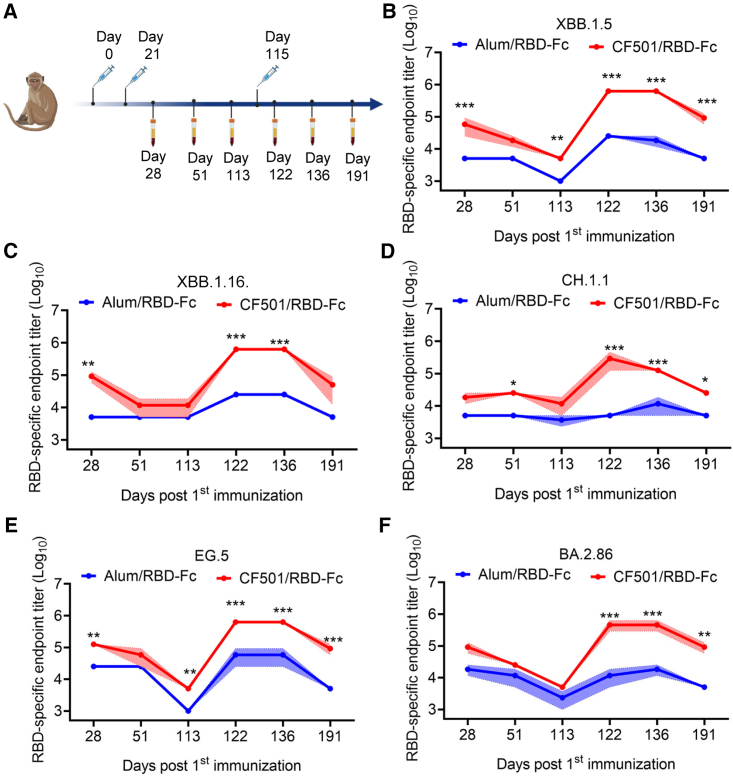
To determine antibody response against the recently circulating SARS-CoV-2 Omicron subvariants (XBB.1.5, XBB.1.16, CH.1.1, EG.5, BA.2.86, and JN.1), we collected sera at different time points from rhesus macaques previously immunized with CF501/RBD-Fc (n = 3) or Alum/RBD-Fc (n = 3).16 Then, we performed ELISA to evaluate RBD-specific immunoglobulin G (IgG) titers. Immunization and sample collection procedures are shown in Figure 2A. After two immunizations, RBD-specific IgG endpoint titers in the CF501/RBD-Fc group ranged from 18,333 to 125,000, about 4- to 18-fold higher than those in the Alum/RBD-Fc group (Figures 2B–2F). The endpoint titers at days 51 and 113 continuously decreased. However, when administered with a third immunization, the endpoint IgG titers (day 136) in the CF501/RBD-Fc group surged from 125,000 to 625,000, about 10- to 25-fold higher than titers in the Alum/RBD-Fc group (Figures 2B–2F). The titers of IgG for binding the RBDs of XBB.1.5, CH.1.1, EG.5, and BA.2.86 remained at a high level for a long time as judged by the endpoint titers at day 191 (76 days post-third immunization) in the sera of the CF501/RBD-Fc group, which were also significantly higher than those in the sera of the Alum/RBD-Fc group (Figures 2B–2F).
Figure 2.
Sera from CF501/RBD-Fc-immunized macaques showed high levels of RBD-binding IgG endpoint titers against XBB.1.5, XBB.1.16, CH.1.1, EG.5, and BA.2.86
(A) Vaccination and sample collection timeline in rhesus macaques (n = 3).
(B–F) The endpoint titers of RBD-specific antibodies against XBB.1.5 (B), XBB.1.16 (C), CH.1.1 (D), EG.5 (E), and BA.2.86 (F) in sera from macaques immunized with CF501/RBD-Fc or Alum/RBD-Fc from days 28 to 191 post-first immunization. Data are shown as mean ± SEM. The experiments were repeated a minimum of two times. Two-way ANOVA was used to compare endpoint titers between CF501/RBD-Fc and Alum/RBD-Fc. ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001.
Cross-neutralization activity against XBB.1.5, XBB.1.16, EG.5, CH.1.1, BA.2.86, and JN.1
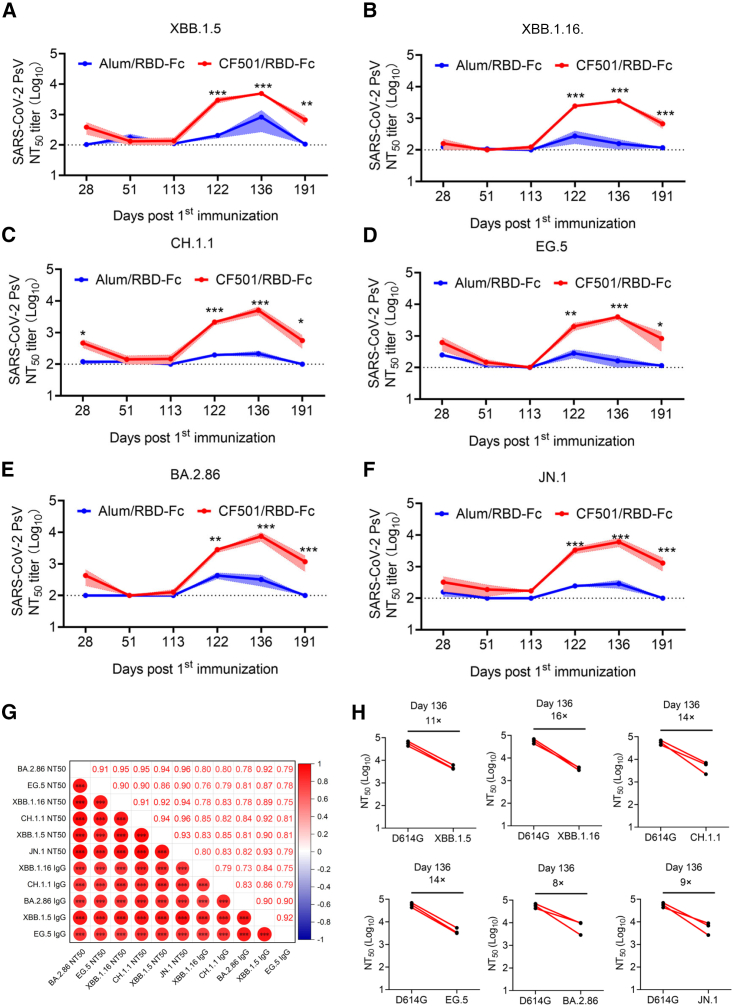
To investigate the neutralizing activities of sera, we conducted neutralization assays using pseudotyped viruses. Results revealed that the sera from the CF501/RBD-Fc group showed weak, but still slightly higher, neutralizing activity against XBB.1.5, XBB.1.16, CH.1.1, EG.5, BA.2.86, and JN.1 relative to the Alum/RBD-Fc group at day 28 (7 days post-second immunization) (Figures 3A–3F). Neutralizing titers in sera for both groups were below the limit of detection at days 51 and 113 post-first immunization (Figures 3A–3F). However, after the third immunization at day 115, the neutralizing titers of sera from macaques immunized with CF501/RBD-Fc saw a rapid surge, peaking at day 136 (14 days post-third immunization) (Figures 3A–3F). CF501/RBD-Fc-elicited sera could potently neutralize XBB.1.5, XBB.1.16, CH.1.1, EG.5, BA.2.86, and JN.1 with 50% neutralization titers (NT50s) of 4,924, 3,494, 5,032, 3,997, 7,424, and 6,025, respectively, at day 136 post-first immunization (Figures 3A–3F). In contrast, sera from the Alum/RBD-Fc group could scarcely neutralize XBB.1.5, XBB.1.16, CH.1.1, EG.5, BA.2.86, and JN.1 with NT50s of 819,157, 215, 162, 318, and 286, respectively, even at day 136 post-first immunization (Figures 3A–3F). Furthermore, the neutralizing effects of sera from the Alum/RBD-Fc group were transitory, and the neutralizing titers at day 191 post-first immunization were below the detection limit (Figures 3A–3F). Meanwhile, CF501/RBD-Fc elicited cross-neutralizing antibodies against XBB.1.5, XBB.1.16, CH.1.1, EG.5, BA.2.86, and JN.1 with NT50s of 669, 669, 560, 828, 1,173, and 1,294 respectively, at day 191 post-first immunization (Figures 3A–3F).
Figure 3.
CF501/RBD-Fc elicited potent and durable cross-neutralizing antibodies against XBB.1.5, XBB.1.16, CH.1.1, EG.5, BA.2.86, and JN.1 in sera of rhesus macaques
(A–F) Evaluation of the titers of cross-neutralizing antibodies against pseudotyped XBB.1.5 (A), XBB.1.16 (B), CH.1.1 (C), EG.5 (D), BA.2.86 (E), and JN.1 (F) in sera of CF501/RBD-Fc- or Alum/RBD-Fc-immunized rhesus macaques collected from days 28 to 191 post-first immunization. The dashed line represents the limit of detection, and 1:100 was defined as the limit of detection. Data are shown as mean ± SEM. The experiments were repeated a minimum of two times. Two-way ANOVA was used to compare NT50s between CF501/RBD-Fc and Alum/RBD-Fc. ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001.
(G) Correlation matrices showed Pearson correlations between any two parameters among the binding IgG titers and neutralizing titers against XBB.1.5, XBB.1.16, CH.1.1, EG.5, BA.2.86, and JN.1. The color and size of circle represent correlation values. ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001.
(H) Fold changes in NT50s against XBB.1.5, XBB.1.16, CH.1.1, EG.5, BA.2.86, and JN.1 compared to D614G for sera collected at day 136 post-first immunization.
Among these SARS-CoV-2 variants, JN.1 and BA.2.86 showed less capacity for immune evasion against CF501/RBD-Fc-elicited sera compared to the other XBB subvariants. Furthermore, sera from macaques immunized with 3 doses of CF501/RBD-Fc were effective at potently neutralizing most of the currently circulating Omicron subvariants tested18 (Figures 3A–3F). We also performed pairwise comparisons to evaluate the relationships between binding levels and neutralizing antibodies among these samples. The results demonstrated that any two parameters exhibited strong correlations (r: 0.73–0.95, p < 0.001) (Figure 3G).
Two doses of CF501/RBD-Fc administered to macaques elicited potent neutralizing antibodies against the D614G mutation in the SARS-CoV-2 S protein. However, less cross-neutralizing antibodies were raised against the detected SARS-CoV-2 variants, as demonstrated by reduced neutralizing antibodies against XBB.1.5, XBB.1.16, CH.1.1, EG.5, BA.2.86, and JN.1 by 55- to 212-fold relative to the titers against D614G (Figure S1). However, the second boost of CF501/RBD-Fc dramatically improved the titers of cross-neutralizing antibodies against XBB.1.5, XBB.1.16, CH.1.1, EG.5, BA.2.86, and JN.1, declining only 11-, 16-, 14-, 14-, 8-, and 9-fold compared to the titers against D614G, respectively (Figure 3H). We have also observed that there was no significant increase in the titer of neutralizing antibodies against the WT strain (D614G) in sera of rhesus macaques after the third immunization (day 136 post-first immunization) in comparison to the titers after two immunizations (day 28 post-first immunization). However, following the third immunization, cross-neutralizing antibody titers in sera of rhesus macaques against Omicron subvariants XBB.1.5, XBB.1.16, CH.1.1, EG.5, BA.2.86, and JN.1 significantly increased by 5- to 21-fold (Figure S2). These findings indicate that repeated vaccinations with the antigens from the same viral strain, in the presence of a potent adjuvant, do not yield a substantial increase in the titer of strain-specific neutralizing antibodies but resulted in a noteworthy elevation in the titer of cross-neutralizing antibodies against other strains.
Induction of bnAbs in macaques immunized with CF501/RBD-Fc
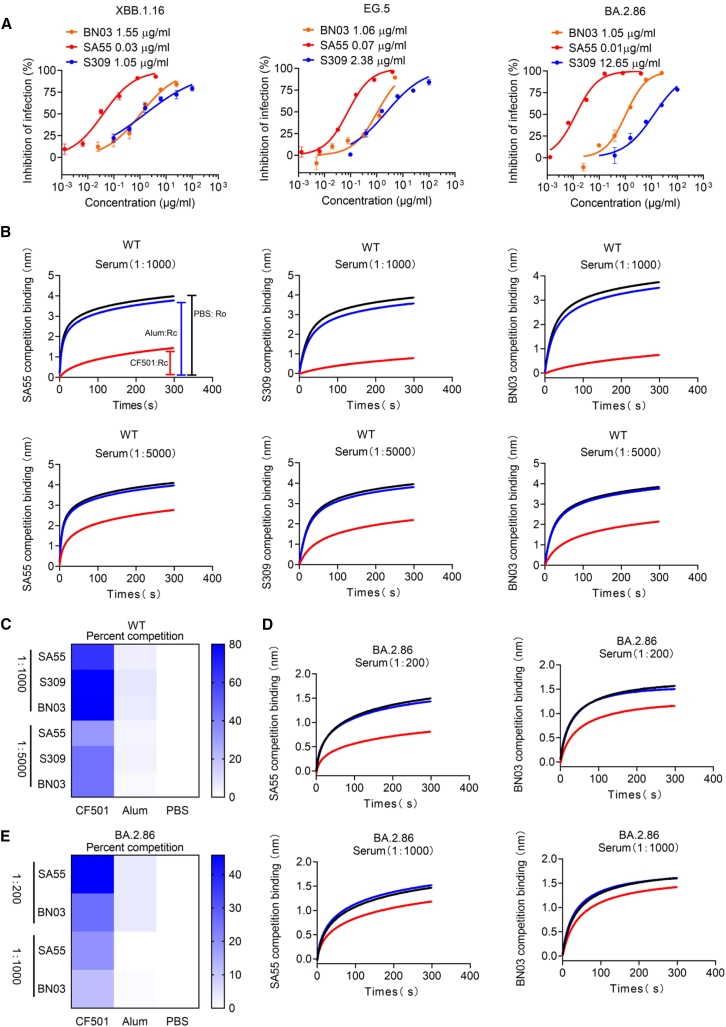
To assess whether XBB.1.16, EG.5, and BA.2.86 could escape antibodies that previously showed broadly neutralizing activities against Omicron subvariants, we selected three representative bnAbs, S309,22 SA55,19,23 and BN03,24 to test their neutralization effects. The epitopes recognized by these three antibodies on the RBD are distinct from each other (Figure S3A). BA.2.86 harbored two, three, and two mutations within the epitopes of S309, SA55, and BN03, respectively (Figures S3B–S3D). These mutations potentially influenced the neutralizing potency of the antibodies. When we performed an antibody neutralization assay, it was revealed that the neutralizing potency (NT50) of S309 against BA.2.86 was reduced by ∼12-fold compared to those against XBB.1.16 and EG.5, possibly owing to the mutations K356T and G339H (Figure 4A). However, BN03 could still neutralize XBB.1.16, EG.5, and BA.2.86 with NT50s of 1.55, 1.06, and 1.05 μg/mL (Figure 4A), respectively, whereas SA55 could potently neutralize all three variants (Figure 4A). Therefore, mutations in the region of BN03 and SA55 epitopes appear not to impact their neutralization activity.
Figure 4.
CF501 adjuvant enhances the immunogenicity of the epitopes in wild-type SARS-CoV-2 RBD protein to induce bnAbs with binding specificity and activity similar to those of S309, BN03, and SA55 bnAbs in the sera of macaques vaccinated with SARS-CoV-2 RBD-Fc protein
(A) Neutralizing activity of S309, BN03, and SA55 against XBB.1.16, EG.5, and BA.2.86. Data are shown as mean ± SEM. The experiments were repeated a minimum of two times.
(B) Pooled sera from macaques immunized with CF501/RBD-Fc or Alum/RBD-Fc to block binding of SA55, BN03, and S309 to RBD from wild-type SARS-CoV-2 strain. Ro represents the highest binding signals of SA55, BN03, and S309 when pooled sera are added from macaques administered with PBS. Rc represents the highest binding signals of SA55, BN03, and S309 tested with pooled sera from CF501/RBD-Fc- or Alum/RBD-Fc-vaccinated macaques. Red, blue, and black lines represent CF501/RBD-Fc, Alum/RBD-Fc, and PBS groups, respectively.
(C) Heatmap indicates the calculated percentage of competition. Results from (Ro − Rc) × 100/Ro represent competition levels.
(D) Pooled sera from macaques immunized with CF501/RBD-Fc or Alum/RBD-Fc to block binding of SA55 and BN03 to RBD from BA.2.86. Red, blue, and black lines represent CF501/RBD-Fc, Alum/RBD-Fc, and PBS groups, respectively.
(E) Heatmap indicates the calculated percentage of competition. Results from (Ro − Rc) × 100/Ro represent competition levels.
The induction of bnAbs is a critical factor impacting vaccine efficiency against the newly emerging Omicron subvariants. Therefore, we next asked why the antisera from macaques immunized with CF501/RBD-Fc exhibited potent and broad neutralizing activity. To address this question, we performed a serum competition assay based on bio-layer interferometry (BLI)25 to identify the antibodies in the CF501/RBD-Fc-immunized macaques with binding specificity and activity similar to those of S309, BN03, and SA55 bnAbs. We observed that the sera from macaques immunized with CF501/RBD-Fc effectively blocked the binding of S309, BN03, and SA55 to the RBD from the WT SARS-CoV-2 strain at dilutions of 1:1,000 and 1:5,000 (Figure 4B). By contrast, the sera from Alum/RBD-Fc showed almost no blocking effects (Figure 4B). Curve height Ro from the PBS group and competing binding curve height Rc from CF501/RBD-Fc or Alum/RBD-Fc were used for quantitative analysis. The competition levels of S309, BN03, and SA55 from sera in the CF501/RBD-Fc group were 10- to 21-fold higher than those elicited by Alum/RBD-Fc (Figure 4C). We also simultaneously tested the sera from two individuals in China who were infected during the BA.5/BF.7 outbreak. The results indicate that only serum 1 exhibits a marginal inhibition of the binding between SA55, S309, and BN03 and the RBD protein (Figure S4). In order to eliminate the possibility that antibodies targeting neighboring sites near these conservative epitopes may also affect the binding of these bnAbs, we further conducted a serum competition assay using the RBD in the BA.2.86 S protein. The results indicate that sera from the CF501/RBD-Fc-immunized rhesus macaques can still effectively inhibit the binding of SA55 and BN03 to the RBD of BA.2.86, while the blocking effect of antibodies in sera from the Alum/RBD-Fc-rhesus macaques is very weak (Figures 4D and 4E). We did not observe an effective binding between S309 and the RBD of BA.2.86, which is consistent with a notable decrease in the neutralizing activity of S309 against BA.2.86. These results indicated that adjuvant CF501, but not Alum, could effectively enhance the immunogenicity of the conserved epitopes shared with S309, BN03, and SA55 bnAbs.
Discussion
The recently emerging SARS-CoV-2 variants JN.1 and BA.2.86 carry over 30 new mutations, similar to the first emergent Omicron strain BA.1. Such evolutionary development might lead to completely different viral properties, such as infectivity and immune evasion. Thus, it is necessary to investigate the escape abilities of JN.1 and BA.2.86 against currently developed vaccine-elicited antibody immunity. When CF501/RBD-Fc was administered to macaques, we found that JN.1 and BA.2.86 were no more resistant than XBB.1.16, EG.5, and CH.1.1 to the elicited neutralizing activity of sera. With the continuous evolution of SARS-CoV-2, a high number of mutations have been found in the RBD. Yet, consensus has not settled the question of whether or not the WT RBD contains conserved epitopes that do not easily change with the progress of evolution. A previous study has indicated that natural infection with WT SARS-CoV-2 could still elicit RBD-targeting antibodies with broad-spectrum activity against Omicron subvariants.26 However, a recent study showed that repeated prototype-based booster vaccines did not improve neutralizing antibodies against Omicron subvariants.27 In contrast, our study provided solid evidence that the RBD from WT SARS-CoV-2 does contain some conserved epitopes recognized by antibodies such as SA55 and BN03, despite the acceleration of mutations in JN.1, BA.2.86, and XBB subvariants. Even with immune pressure, BA.2.86/JN.1 with over 30 mutations still did not change the conserved epitopes for the bnAbs such as SA55 and BN03, implying that the evolution of SARS-CoV-2 might not break through the completely conserved and unmutated epitopes, thus providing proof of concept for the strategy of “non-changeable against changeable” to develop universal COVID-19 vaccines. When we combined the adjuvant CF501, in comparison to Alum, with RBD-Fc, we found that CF501/RBD-Fc-elicited sera could effectively block the binding of S309, BN03, and SA55 to the RBD. This demonstrates that the non-nucleotide small-molecule STING agonist CF501, but not Alum, could remarkably enhance the immunogenicity of the conserved and well-exposed neutralizing epitopes in the RBD of the original SARS-CoV-2 strain presented in the RBD-Fc vaccine since the Alum/RBD-Fc could not elicit such a high titer of cross-neutralizing antibodies induced by CF501/RBD-Fc. Thus, the data revealed in the present study support the use of potent adjuvants to augment broad and durable antibody immunity against the ever-evolving WT SARS-CoV-2. Moreover, previous and current studies demonstrated that the bnAb BN03 maintained its neutralizing activity against all current SARS-CoV-2 variants, including BA.2.86, showing promise for clinical use.21,24
Boost immunization plays an important role in producing cross-neutralizing antibodies. Only 3 doses of CF501/RBD-Fc were required to elicit potent cross-neutralizing antibodies against the Omicron subvariants, while 2 doses did not. Several reports showed that extension of the boosting interval could significantly increase the magnitude of antibodies.28,29 In our study, the second boost vaccination was conducted at about 3 months post-first boost immunization, which may have also resulted in the high levels of neutralizing antibodies. Furthermore, beyond non-human primates, CF501/RBD-Fc has been shown to elicit significantly higher titers of neutralizing antibodies than Alum/RBD-Fc in mice and rabbits.16 While the CF501 adjuvant is currently in the preclinical studies, its evaluation in humans may be initiated in the near future.
Unlike most other reported COVID-19 vaccines, the pan-sarbecovirus does not need to replace the immunogen along with the newly emerged variants but just to increase the bnAbs to combat currently emerging SARS-CoV-2 variants. Once again, these data demonstrated the magnitude and breadth of neutralizing antibodies elicited by the pan-sarbecovirus vaccine. More importantly, our strategy for developing the pan-sarbecovirus vaccine using the first-generation immunogen containing conserved epitopes and the novel STING agonist could also be applied for the development of broad-spectrum vaccines against other viruses that may emerge or reemerge in the future.
Limitations of the study
In this study, the utilization of pseudotyped viruses rather than authentic viruses represents a limitation. However, it Is worth noting that we have previously validated our pseudoviral system alongside authentic SARS-CoV-2. Furthermore, SARS-CoV-2 pseudoviruses are widely employed in the field for assessing COVID-19 vaccines or examining the extent of immune evasion by newly emerging Omicron subvariants. Additionally, this study convincingly demonstrates, through serum competition assay, that sera from CF501/RBD-Fc-immunized rhesus macaques contain significantly higher titers of bnAbs with binding specificity and activity similar to those of SA55, BN03, and S309 than those from Alum/RBD-Fc-immunized rhesus macaques. Because of the technical constraints, a precise analysis of the quality and quantity of these bnAbs present in the sera could not be done currently but is warranted for future study.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Goat polyclonal Secondary Antibody to Monk–y IgG - H&L | Abcam | Cat#ab112764; RRID: AB_10861684 |
| SA55 mAb | Cao et al.23 | N/A |
| BN03 nanobody | Li et al.21 | N/A |
| S309 mAb | Pinto et al.20 | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| CF501 | Liu et al.16 | N/A |
| Imject™ Alum | Thermo Fisher Scientific | Cat# 77161 |
| Vigofect | Vigorous Biotechnology | Cat#T001 |
| EZ Trans transfection reagent | Life iLAB Bio Technology | Cat#AC04L082 |
| Bovine Serum Albumin | WeiAo Biotech, Shanghai | Cat#WH3044 |
| TMB | Sigma | Cat# T2885 |
| Fetal Bovine Serum | GEMINI | Cat#900-108 |
| Tween 20 | Sigma-Aldrich | Cat#P7949 |
| PBS | BI (Biological Industries) | Cat#02-023-1ACS |
| XBB.1.5 RBD protein | This study | N/A |
| BA.2.86 RBD protein | This study | N/A |
| XBB.1.16 RBD protein | This study | N/A |
| CH.1.1 RBD protein | This study | N/A |
| EG.5 RBD protein | This study | N/A |
| Critical commercial assays | ||
| Luciferase Assay System | Promega | Cat#E1501 |
| Experimental models: Cell lines | ||
| HEK293T cell line | ATCC | Cat# CRL-11268; RRID:CVCL_1926 |
| Huh-7 cell line | Fudan University30 | N/A |
| Expi293 Expression System | Thermo Fisher Scientific | Cat#A14635 |
| Recombinant DNA | ||
| pNL4-3.luc.RE | Fudan University30 | N/A |
| pcDNA3.1-JN.1-S | This study | N/A |
| pcDNA3.1-BA.2.86-S | This study | N/A |
| pcDNA3.1- CH.1.1-S | This study | N/A |
| pcDNA3.1- XBB.1.16-S | This study | N/A |
| pcDNA3.1- EG.5-S | This study | N/A |
| pcDNA3.1- XBB.1.5-S | This study | N/A |
| Pfuse-SARS-CoV-2 BA.2.86-RBD | This study | N/A |
| Pfuse-SARS-CoV-2 CH.1.1-RBD | This study | N/A |
| Pfuse-SARS-CoV-2 XBB.1.16-RBD | This study | N/A |
| Pfuse-SARS-CoV-2 EG.5-RBD | This study | N/A |
| Pfuse-SARS-CoV-2 XBB.1.5-RBD | This study | N/A |
| Software and algorithms | ||
| PRISM | GraphPad | https://www.graphpad.com |
| Biorender | biorender.com | N/A |
| Origin | Version 2022 | https://www.originlab.com/ |
| UCSF ChimeraX | Meng et al.31 | N/A |
| Experimental models: Organisms/strains | ||
| Rhesus macaques | Beijing Institute of Xieerxin Biology Resource | N/A |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Author Lu Lu.
Materials availability
All reagents generated in this study are available from the lead contact with a completed Materials Transfer Agreement.
Data and code availability
All data reported in this paper will be shared by the lead contact upon request. This paper does not report original code. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
Experimental model and subject details
Cell lines
HEK-293T cells and Huh-7 cells were obtained from the American Type Culture Collection (ATCC). HEK-293T cells and Huh-7 were maintained in Dulbecco-modified Eagle medium (DMEM) with supplementation of 10% inactivated fetal bovine serum (FBS). HEK293 serum-free OPM-293-CD05 medium (Sino Biological) was used to culture the Expi-293F cells.
Sera samples
The monkey anti-sera in this study were from our previous studies.16 The macaques (n = 3) were immunized with CF501/RBD-Fc or Alum/RBD-Fc respectively at days 0, 21 and 115. And the sera were collected as previously described. The RBD-Fc fusion protein was generated by fusing the RBD derived from the original SARS-CoV-2 strain (Residues 319–532, GenBank accession number: QHD43416.1) with the Fc region of human IgG1. This fusion construct was then cloned into the pFUSE-hIgG1-Fc2 plasmid and expressed through a eukaryotic expression system to obtain the protein. Human serum samples that have been infected BA.5/BF.7 were collected in Shanghai Fourth People’s Hospital and were approved by the ethics committee of Shanghai Fourth People’s Hospital (No. 2022098-001 and 2022185-001).
Method details
Protein expression and purification
RBD proteins of BA.2.86, XBB.1.16, EG.5 and CH.1.1 were expressed as previously reported.32 Briefly, EXPi293F cells were transfected by the expression plasmids of Pfuse-SARS-CoV-2-BA.2.86-RBD, Pfuse-SARS-CoV-2-XBB.1.16-RBD, Pfuse-SARS-CoV-2-CH.1.1-RBD, Pfuse-SARS-CoV-2-XBB.1.5-RBD and Pfuse-SARS-CoV-2-EG.5-RBD, respectively, using EZ Trans transfection reagents (Life iLAB Bio-Technology, China). At 5 days post-transfection, the supernatants of EXPi293F cells were harvested. Affinity chromatography was used to purify the RBDs using Ni-NTA (GE Health). Purified RBD proteins were quantified using the BCA protein assay kit.
ELISA
Binding antibodies against BA.2.86, XBB.1.16, XBB.1.5, CH.1.1, and EG.5 were evaluated using ELISA as previously reported.32 Briefly, 1 μg/mL of RBD was coated on wells of a 96-well ELISA plate and incubated at 4°C overnight. Sera from immunized rhesus macaques were serially diluted using PBST (PBS containing 0.05% Tween 20) and applied to the RBD-coated ELISA plates. After incubation for 45 min, the diluted sera were discarded. PBST was used to wash the plates 5 times. After that, a 1: 10,000 dilution of HRP-conjugated goat anti-monkey IgG (Abcam, UK) was incubated in wells of the microplate for an additional 45 min. After 5 washes with PBST, 3,3′,5,5′-tetramethylbenzidine (TMB) was conducted to visualize the reaction. After stopping the reaction using H2SO4, a microplate reader (InfiniteM200PRO, Switzerland) was used to detect the absorbance at 450 nm (A450). The highest dilution that showed A450 > 2.1-fold of background values was defined as the endpoint titers.
Pseuotyped virus neutralization
The production of JN.1, BA.2.86, XBB.1.16, XBB.1.5, CH.1.1, and EG.5 pseudoviruses was performed as previously reported.30 Briefly, the backbone plasmid of pNL4-3.Luc.R−E and one plasmid of spike, PcDNA3.1-BA.2.86-S (EPI_ISL_18138566), PcDNA3.1-XBB.1.5-S (EPI_ISL_16346200), PcDNA3.1-XBB.1.16-S (EPI_ISL_17958295), PcDNA3.1-EG.5-S (EPI_ISL_18052033), or PcDNA3.1-CH.1.1-S (EPI_ISL_18588371), were transfected into HEK293T cells using VigoFect. At 48 h post-transfection, the supernatant of HEK293T cells was collected to perform the neutralization assay. Huh-7 cells were used as the target cells. The sera and antibody neutralization assay were conducted as previously reported.33,34 Five thousand cells/well of Huh-7 were applied to wells of a 96-well plate. After 12 h, antisera from immunized macaques or antibodies were serially diluted and mixed with the pseudotyped JN.1, BA.2.86, XBB.1.16, XBB.1.5, CH.1.1, and EG.5 separately. The mixture of sera and pseudoviruses was added to Huh-7 cells and further cultured for 48 h. A Firefly Luciferase Assay Kit (Promega, USA) was finally used to calculate luciferase activity. NT50 was defined as the dilution that showed 50% of the luminescence units of the viral control.
Serum competition assay
BLI assay was used to evaluate serum competition as previously reported.25 A total of 10 μg/mL of RBD-His from wild-type SARS-CoV-2 or BA.2.86 were loaded on NI-NTA biosensors. The pooled sera from immunized macaques in Alum/RBD-Fc, CF501/RBD-Fc and PBS groups were diluted to different concentrations, respectively. The diluted pooled sera were further loaded into the RBD-captured NI-NTA biosensors. After saturating the RBD for 300 s, 250 nM SA55, S309 or BN03 were associated with the biosensors for 300 s. Ro represents the height of the saturated noncompeting binding curve. Rc represents the height of the saturated competing binding curve. The percent competition levels were calculated as (Ro-Rc)∗100/Ro.
Structural analysis and comparison
All protein structures used in this study were obtained from RCSB protein databank (PDB) (https://www.rcsb.org/). These structures included the spike trimer of BA.2 (PDB: 7XIW), the spike trimer of wild-type SARS-CoV-2 (PDB: 6ZP5), the RBD bound with S309 (PDB: 6WPS), the RBD bound with BN03 (PDB: 7WHI), and the RBD bound with SA55 (PDB: 7Y0W). Structural figures were generated using UCSF ChimeraX.31 To compare antibody epitopes, the RBD domains in the complex structures were aligned, and the epitopes were identified using the default parameters of the interface command in UCSF ChimeraX.
Quantification and statistical analysis
Statistical analysis in Figure 3G was conducted using Origin software. The other statistical analyses mentioned in the figure legends were performed using GraphPad Prism 9. Error bars in Figures 2B–2F, 3A-3F, and 4A represent means ± standard error. n represents the number of animals. To better approximate normality, log10 transformed NT50 values and the endpoint IgG titers were used for statistical significance analysis in Figures 2B–2F and 3A–3F using two-way ANOVA. Pearson correlation was employed for statistical significance analysis in Figure 3G.
Acknowledgments
This work was supported by the National Key Research and Development Program of China (2022YFC2305800 to Z.L., 2023YFC2307800 to S.J., and 2021YFC2300703 and 2022YFC2604102 to L.L.), the National Natural Science Foundation of China (82341036 to L.L. and 82202490 to Z.L.), and the Shanghai Municipal Science and Technology Major Project (ZD2021CY001 to L.L. and S.J.).
Author contributions
J.Z., W.W., Z.L., and K.Z. conducted the ELISA and pseudovirus neutralization assays. Z.L. performed the BLI assay. Y.Z. analyzed the protein structure. Z.L., J.Z., G.Z., Y.W., L.X., W.X., and Qian Wang analyzed the data. Qiao Wang and T.Y. provided the antibodies of SA55 and BN03. Z.L., L.L., and S.J. wrote the manuscript.
Declaration of interests
L.L., S.J., Z.L., J.Z., Q.W., and X.W. filed a patent application for the STING agonist.
Published: February 29, 2024
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xcrm.2024.101445.
Contributor Information
Zezhong Liu, Email: [email protected].
Shibo Jiang, Email: [email protected].
Lu Lu, Email: [email protected].
Supplemental information
References
- 1.Carreño J.M., Alshammary H., Tcheou J., Singh G., Raskin A.J., Kawabata H., Sominsky L.A., Clark J.J., Adelsberg D.C., Bielak D.A., et al. Activity of convalescent and vaccine serum against SARS-CoV-2 Omicron. Nature. 2022;602:682–688. doi: 10.1038/s41586-022-04399-5. [DOI] [PubMed] [Google Scholar]
- 2.Cao Y., Wang J., Jian F., Xiao T., Song W., Yisimayi A., Huang W., Li Q., Wang P., An R., et al. Omicron escapes the majority of existing SARS-CoV-2 neutralizing antibodies. Nature. 2022;602:657–663. doi: 10.1038/s41586-021-04385-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang Q., Iketani S., Li Z., Liu L., Guo Y., Huang Y., Bowen A.D., Liu M., Wang M., Yu J., et al. Alarming antibody evasion properties of rising SARS-CoV-2 BQ and XBB subvariants. Cell. 2023;186:279–286.e8. doi: 10.1016/j.cell.2022.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kurhade C., Zou J., Xia H., Liu M., Chang H.C., Ren P., Xie X., Shi P.Y. Low neutralization of SARS-CoV-2 Omicron BA.2.75.2, BQ.1.1 and XBB.1 by parental mRNA vaccine or a BA.5 bivalent booster. Nat. Med. 2023;29:344–347. doi: 10.1038/s41591-022-02162-x. [DOI] [PubMed] [Google Scholar]
- 5.Uraki R., Ito M., Furusawa Y., Yamayoshi S., Iwatsuki-Horimoto K., Adachi E., Saito M., Koga M., Tsutsumi T., Yamamoto S., et al. Humoral immune evasion of the omicron subvariants BQ.1.1 and XBB. Lancet Infect. Dis. 2023;23:30–32. doi: 10.1016/S1473-3099(22)00816-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang X., Chen L.L., Ip J.D., Chan W.M., Hung I.F.N., Yuen K.Y., Li X., To K.K.W. Omicron sublineage recombinant XBB eutralizingalising antibodies in recipients of BNT162b2 or CoronaVac vaccines. Lancet. Microbe. 2023;4:e131. doi: 10.1016/S2666-5247(22)00335-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rasmussen M., Møller F.T., Gunalan V., Baig S., Bennedbæk M., Christiansen L.E., Cohen A.S., Ellegaard K., Fomsgaard A., Franck K.T., et al. First cases of SARS-CoV-2 BA.2.86 in Denmark, 2023. Euro Surveill. 2023;28 doi: 10.2807/1560-7917.ES.2023.28.36.2300460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Uriu K., Ito J., Kosugi Y., Tanaka Y.L., Mugita Y., Guo Z., Hinay A.A., Jr., Putri O., Kim Y., Shimizu R., et al. Transmissibility, infectivity, and immune evasion of the SARS-CoV-2 BA.2.86 variant. Lancet Infect. Dis. 2023;23:e460–e461. doi: 10.1016/S1473-3099(23)00575-3. [DOI] [PubMed] [Google Scholar]
- 9.Yang S., Yu Y., Jian F., Song W., Yisimayi A., Chen X., Xu Y., Wang P., Wang J., Yu L., et al. Antigenicity and infeceutralizingionisation of SARS-CoV-2 BA.2.86. Lancet Infect. Dis. 2023;23:e457–e459. doi: 10.1016/S1473-3099(23)00573-X. [DOI] [PubMed] [Google Scholar]
- 10.Sheward D.J., Yang Y., Westerberg M., Öling S., Muschiol S., Sato K., Peacock T.P., Karlsson Hedestam G.B., Albert J., Murrell B. Sensitivity of the SARS-CoV-2 BA.2.86 variant to preveutralizingalising antibody responses. Lancet Infect. Dis. 2023;23:e462–e463. doi: 10.1016/S1473-3099(23)00588-1. [DOI] [PubMed] [Google Scholar]
- 11.Qu P., Xu K., Faraone J.N., Goodarzi N., Zheng Y.M., Carlin C., Bednash J.S., Horowitz J.C., Mallampalli R.K., Saif L.J., et al. Immune evasion, infectivity, and fusogenicity of SARS-CoV-2 BA.2.86 and FLip variants. Cell. 2024;S0092–8674 doi: 10.1016/j.cell.2023.12.026. 01400-01409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang L., Jiao F., Jiang H., Yang Y., Huang Z., Wang Q., Xu W., Zhu Y., Xia S., Jiang S., Lu L. Fusogenicity of SARS-CoV-2 BA.2.86 subvariant and its sensitivity to the prokaryotic recombinant EK1 peptide. Cell Discov. 2024;10:6. doi: 10.1038/s41421-023-00631-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang Q., Guo Y., Liu L., Schwanz L.T., Li Z., Nair M.S., Ho J., Zhang R.M., Iketani S., Yu J., et al. Antigenicity and receptor affinity of SARS-CoV-2 BA.2.86 spike. Nature. 2023;624:639–644. doi: 10.1038/s41586-023-06750-w. [DOI] [PubMed] [Google Scholar]
- 14.Looi M.K. Covid-19: WHO adds JN.1 as new variant of interest. BMJ. 2023;383:p2975. doi: 10.1136/bmj.p2975. [DOI] [PubMed] [Google Scholar]
- 15.Yang S., Yu Y., Xu Y., Jian F., Song W., Yisimayi A., Wang P., Wang J., Liu J., Yu L., et al. Fast evolution of SARS-CoV-2 BA.2.86 to JN.1 under heavy immune pressure. Lancet Infect. Dis. 2024;24:e70–e72. doi: 10.1016/S1473-3099(23)00744-2. [DOI] [PubMed] [Google Scholar]
- 16.Liu Z., Zhou J., Xu W., Deng W., Wang Y., Wang M., Wang Q., Hsieh M., Dong J., Wang X., et al. A novel STING agonist-adjuvanted pan-sarbecovirus vaccine elicits potent and durable neutralizing antibody and T cell responses in mice, rabbits and NHPs. Cell Res. 2022;32:269–287. doi: 10.1038/s41422-022-00612-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu Z., Chan J.F.W., Zhou J., Wang M., Wang Q., Zhang G., Xu W., Chik K.K.H., Zhang Y., Wang Y., et al. A pan-sarbecovirus vaccine induces highly potent and durable neutralizing antibody responses in non-human primates against SARS-CoV-2 Omicron variant. Cell Res. 2022;32:495–497. doi: 10.1038/s41422-022-00631-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu Z., Zhou J., Wang X., Xu W., Teng Z., Chen H., Chen M., Zhang G., Wang Y., Huang J., et al. A pan-sarbecovirus vaccine based on RBD of SARS-CoV-2 original strain elicits potent neutralizing antibodies against XBB in non-human primates. Proc. Natl. Acad. Sci. USA. 2023;120 doi: 10.1073/pnas.2221713120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cao Y., Jian F., Wang J., Yu Y., Song W., Yisimayi A., Wang J., An R., Chen X., Zhang N., et al. Imprinted SARS-CoV-2 humoral immunity induces convergent Omicron RBD evolution. Nature. 2023;614:521–529. doi: 10.1038/s41586-022-05644-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pinto D., Park Y.J., Beltramello M., Walls A.C., Tortorici M.A., Bianchi S., Jaconi S., Culap K., Zatta F., De Marco A., et al. Cross-neutralization of SARS-CoV-2 by a human monoclonal SARS-CoV antibody. Nature. 2020;583:290–295. doi: 10.1038/s41586-020-2349-y. [DOI] [PubMed] [Google Scholar]
- 21.Li C., Zhan W., Yang Z., Tu C., Hu G., Zhang X., Song W., Du S., Zhu Y., Huang K., et al. Broad neutralization of SARS-CoV-2 variants by an inhalable bispecific single-domain antibody. Cell. 2022;185:1389–1401.e18. doi: 10.1016/j.cell.2022.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Addetia A., Piccoli L., Case J.B., Park Y.J., Beltramello M., Guarino B., Dang H., de Melo G.D., Pinto D., Sprouse K., et al. Neutralization, effector function and immune imprinting of Omicron variants. Nature. 2023;621:592–601. doi: 10.1038/s41586-023-06487-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cao Y., Jian F., Zhang Z., Yisimayi A., Hao X., Bao L., Yuan F., Yu Y., Du S., Wang J., et al. Rational identification of potent and broad sarbecovirus-neutralizing antibody cocktails from SARS convalescents. Cell Rep. 2022;41 doi: 10.1016/j.celrep.2022.111845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hao A., Song W., Li C., Zhang X., Tu C., Wang X., Wang P., Wu Y., Ying T., Sun L. Defining a highly conserved cryptic epitope for antibody recognition of SARS-CoV-2 variants. Signal Transduct. Targeted Ther. 2023;8:269. doi: 10.1038/s41392-023-01534-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kang Y.F., Sun C., Zhuang Z., Yuan R.Y., Zheng Q., Li J.P., Zhou P.P., Chen X.C., Liu Z., Zhang X., et al. Rapid Development of SARS-CoV-2 Spike Protein Receptor-Binding Domain Self-Assembled Nanoparticle Vaccine Candidates. ACS Nano. 2021;15:2738–2752. doi: 10.1021/acsnano.0c08379. [DOI] [PubMed] [Google Scholar]
- 26.Ju B., Zhang Q., Wang Z., Aw Z.Q., Chen P., Zhou B., Wang R., Ge X., Lv Q., Cheng L., et al. Infection with wild-type SARS-CoV-2 elicits broadly neutralizing and protective antibodies against omicron subvariants. Nat. Immunol. 2023;24:690–699. doi: 10.1038/s41590-023-01449-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang H., Xue Q., Zhang H., Yuan G., Wang X., Sheng K., Li C., Cai J., Sun Y., Zhao J., et al. Neutralization against Omicron subvariants after BA.5/BF.7 breakthrough infection weakened as virus evolution and aging despite repeated prototype-based vaccination(1) Emerg. Microb. Infect. 2023;12 doi: 10.1080/22221751.2023.2249121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Payne R.P., Longet S., Austin J.A., Skelly D.T., Dejnirattisai W., Adele S., Meardon N., Faustini S., Al-Taei S., Moore S.C., et al. Immunogenicity of standard and extended dosing intervals of BNT162b2 mRNA vaccine. Cell. 2021;184:5699–5714.e11. doi: 10.1016/j.cell.2021.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhao X., Li D., Ruan W., Chen Z., Zhang R., Zheng A., Qiao S., Zheng X., Zhao Y., Dai L., et al. Effects of a Prolonged Booster Interval on Neutralization of Omicron Variant. N. Engl. J. Med. 2022;386:894–896. doi: 10.1056/NEJMc2119426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xia S., Liu M., Wang C., Xu W., Lan Q., Feng S., Qi F., Bao L., Du L., Liu S., et al. Inhibition of SARS-CoV-2 (previously 2019-nCoV) infection by a highly potent pan-coronavirus fusion inhibitor targeting its spike protein that harbors a high capacity to mediate membrane fusion. Cell Res. 2020;30:343–355. doi: 10.1038/s41422-020-0305-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Meng E.C., Goddard T.D., Pettersen E.F., Couch G.S., Pearson Z.J., Morris J.H., Ferrin T.E. UCSF ChimeraX: Tools for structure building and analysis. Protein Sci. 2023;32 doi: 10.1002/pro.4792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu Z., Xu W., Xia S., Gu C., Wang X., Wang Q., Zhou J., Wu Y., Cai X., Qu D., et al. RBD-Fc-based COVID-19 vaccine candidate induces highly potent SARS-CoV-2 neutralizing antibody response. Signal Transduct. Targeted Ther. 2020;5:282. doi: 10.1038/s41392-020-00402-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu Z., Xu W., Chen Z., Fu W., Zhan W., Gao Y., Zhou J., Zhou Y., Wu J., Wang Q., et al. An ultrapotent pan-beta-coronavirus lineage B (beta-CoV-B) neutralizing antibody locks the receptor-binding domain in closed conformation by targeting its conserved epitope. Protein Cell. 2022;13:655–675. doi: 10.1007/s13238-021-00871-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhou Y., Liu Z., Li S., Xu W., Zhang Q., Silva I.T., Li C., Wu Y., Jiang Q., Liu Z., et al. Enhancement versus neutralization by SARS-CoV-2 antibodies from a convalescent donor associates with distinct epitopes on the RBD. Cell Rep. 2021;34 doi: 10.1016/j.celrep.2021.108699. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data reported in this paper will be shared by the lead contact upon request. This paper does not report original code. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.