Abstract
Among bacteria, many species have synonymous codon usage patterns that have been influenced by natural selection for those codons that are translated more accurately and/or efficiently. However, in other species selection appears to have been ineffective. Here, we introduce a population genetics-based model for quantifying the extent to which selection has been effective. The approach is applied to 80 phylogenetically diverse bacterial species for which whole genome sequences are available. The strength of selected codon usage bias, S, is found to vary substantially among species; in 30% of the genomes examined, there was no significant evidence that selection had been effective. Values of S are highly positively correlated with both the number of rRNA operons and the number of tRNA genes. These results are consistent with the hypothesis that species exposed to selection for rapid growth have more rRNA operons, more tRNA genes and more strongly selected codon usage bias. For example, Clostridium perfringens, the species with the highest value of S, can have a generation time as short as 7 min.
INTRODUCTION
The frequency of use of alternative synonymous codons varies among species, and often also among genes from a single genome (1–3). The pattern of codon usage in any gene reflects a complex balance among biases generated by mutation, selection and random genetic drift (4–6). Among bacteria, genomic G+C content varies over a wide range, presumably reflecting variation in mutation biases (7), with a major impact on codon usage (8). In addition, three major factors have been found to contribute to codon usage variation among genes within a bacterial genome. First, mutation biases seem to differ between the leading and lagging strands of replication, since genes on the leading strand are often more G+T-rich (9,10). Second, in many species, there is evidence of natural selection on codon usage. Genes expressed at high levels exhibit a bias towards a subset of synonymous codons, which are those most accurately and/or efficiently recognized by the most abundant tRNA species, and the strength of this bias is correlated with the level of gene expression (2,11). Third, there is evidence of extensive horizontal gene transfer among bacteria (12), and genes recently acquired from sources other than close relatives have atypical codon usage. The extent or magnitude of all three factors varies greatly among species. Here, we focus on the manner in which selected codon usage bias varies among bacteria.
The first species in which codon usage was examined in detail, the bacterium Escherichia coli (13,14) and the yeast Saccharomyces cerevisiae (15,16), were both found to show strong evidence of natural selection on codon usage. Subsequently, it has often been assumed that such selection is ubiquitous, at least among unicellular organisms. However, there have been a number of reports of bacterial species exhibiting little or no evidence of selected codon usage bias. Some concern species with extremely A+T- or G+C-rich genomes (17–22), where mutational bias appears to swamp any selected bias. However, in other cases, there is no sign of selection, even though the genomic base composition is not extreme (23,24). In addition, there are species where codon selection has been detected, but the effect seems relatively minor (25–28). It would be useful to be able to quantify the strength of selected codon usage bias in such a way that the results can be compared between species. There are two particular difficulties. First, the extent of bias in the absence of selection varies among species due to mutational biases. Second, many of the codons favoured by selection vary between species, such that the nature of the bias within a set of synonyms for a particular amino acid can be quite different in different species.
To overcome the first of these problems, we use a population genetics model to assess the strength of selected codon usage bias (5), modifying it to take account of background mutation biases. To overcome the second problem, we focus on certain codons that are expected to be translationally advantageous in all bacterial species. For example, the two Phe codons (UUU and UUC) are recognized, through wobble, by a single species of tRNA with the anticodon sequence GAA. While the G at the wobble position may be modified [e.g. to 2′-O-methylguanosine in Bacillus subtilis (29)], it appears that the UUC codon is always better recognized and thus the translationally optimal codon (11).
The extent of selected synonymous codon usage bias might be expected to vary among species dependent on various factors. First, codons are thought to be selected for their effects on the efficiency and accuracy of translation, and ultimately for their effect on bacterial growth rate (30). Bacterial life styles vary markedly, with different species living within nutrient-rich eukaryotic cells but isolated from competitors, or as surface monocultures in oligotrophic external environments, or as complex mixed communities growing either in planktonic log phase within guts or as biofilms on rapidly cycling mucosal surfaces. Some species cross between diverse growth modes and rates, such as passing from terrestrial or aquatic environments to symbiotic relationships with eukaryotes. Thus, the relative importance of efficiency of rapid competitive growth as a component of fitness is likely to vary greatly among species. Second, the selection coefficient for a single synonymous mutation, in a genome with hundreds of thousands of synonymously variable sites, is expected to be extremely small. Then, although bacteria may have extremely large global population sizes, the population structure of the species may be such as to reduce the effective population size to the point where codon selection is less effective. Furthermore, the extent of recombination varies greatly among bacterial species (31), and in those with low recombination rates the linkage among numerous polymorphic synonymous sites on the bacterial chromosome may lead to interference in their selection (32). We consider these various factors in interpreting the variation in the strength of selected codon usage bias among species.
METHODS
Estimation of S
Following Bulmer (5), we can consider the case of an amino acid encoded by two synonyms, C1 and C2. The mutation rate from C1 to C2 is u; and from C2 to C1 is v. The selective difference between the two codons is s: the fitness of the optimal codon C1 is 1, while that of C2 is (1 − s). Under the combined effects of mutation, selection and random genetic drift, the equilibrium frequency (P) of C1 in a gene, or set of genes, is given by:
| 1 |
where S = 2 Ne s, U = 2 Ne u and V = 2 Ne v.
In genes where selection is strong enough to influence codon usage, the frequency of codons is determined by both the pattern of mutation and the strength of selection. The magnitude of S can be estimated from Equation 1:
| 2 |
where k = U/V.
In genes where selection is so weak as to be ineffective, the frequency of the codons is determined by the pattern of mutation between them:
| 3 |
This allows the estimation of k = (1 − P)/P for use in Equation 2 above.
This methodology was applied to codons for four amino acids (Phe, Tyr, Ile and Asn) where the nature of codon selection is expected to be the same in all species. For Tyr (codons UAU and UAC; anticodon GUA) and Asn (codons AAU and AAC; anticodon GUU), the situation is analogous to that for Phe described in the Introduction. For Ile, there are three synonyms, but one (AUA) is recognized by a distinct tRNA with the anticodon CAU; the other two synonyms (AUU and AUC) are recognized by a tRNA with anticodon GAU. Here, the AUA codon was ignored (it is often rare) and Ile was treated as if it were analogous to Phe, Tyr and Asn. There are no other amino acids for which it seems clear that the translationally optimal codon is the same in all species. S-values were calculated for each of the four amino acids: the overall value for a species was computed as the average weighted by the number of codons analysed for the highly expressed genes.
Sequence data
Complete genome sequences of bacterial species were obtained from GenBank release 136 (June 2003). Sequences were extracted using the ACNUC interface (33), and initial codon usage analyses performed using CodonW (34). Base composition statistics (GC3S and GT3S) were calculated as the frequency of these nucleotides at synonymously variable third positions of sense codons, i.e. excluding Met, Trp and termination codons.
In the case of species for which multiple strains have been sequenced, only one representative was selected. In addition, some other pairs of species are no more divergent than strains of a single species. To assess this, the average nucleotide sequence divergence across the genes rplA-C and rpsB-C was estimated. A criterion of at least 4% sequence divergence was used for inclusion of strains. This led to the exclusion of Mycobacterium bovis (0.05% different from Mycobacterium tuberculosis), Shigella flexneri (0.2% different from E.coli K12), Brucella suis (0.3% different from Brucella melitensis), Listeria innocua (1.5% different from L.monocytogenes) and Bacillus cereus (1.6% different from Bacillus anthracis). In contrast, Buchnera aphidicola strains Ap, Bp and Sg differed by 17–26%, and so all three were included. The least divergent pairs of species retained were Xanthomonas axonopodis and Xanthomonas campestris (4.0%) and E.coli and Salmonella enterica typhimurium (4.1%). With the exception of B.aphidicola, the 4% criterion would exclude all cases of multiple strains of a single species: the most divergent were Helicobacter pylori strains 26695 and J99 (3.2%) and Xylella fastidiosa strains 9a5c and Temecula (2.5%). Finally, Streptococcus mutans UA159 was excluded because several genes used in the analysis (see below) were incomplete or missing: the sequence has a deletion between the rplD and rpsS genes, truncating both and deleting the rplB and rplW genes that lie between rplD and rpsS in other Streptococcus species. The final data set included 80 different genomes (Table 1).
Table 1.
The 80 bacterial genome sequences analysed
| Species codea | Gene numbersb | GC contentc | Sd | Randome | Nf | Accession nosg | Species | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| rRNA | tRNA | ORF | i | ii | iii | ||||||
| Gamma proteobacteria | |||||||||||
| Esccol | 7 | 86 | 4289 | 51 | 54 | 48 | 1.488 | (0.308/−0.286) | 992 | U00096 | Escherichia coli K-12 |
| Salent | 7 | 84 | 4452 | 53 | 58 | 50 | 1.522 | (0.292/−0.254) | 993 | AE006468 | Salmonella enterica typhimurium |
| Yerpes | 6 | 68 | 4008 | 48 | 48 | 43 | 1.153 | (0.258/−0.243) | 991 | AL590842 | Yersinia pestis CO92 |
| BucaAp | 1 | 30 | 564 | 26 | 12 | 12 | −0.017 | (0.179/−0.228) | 1200 | BA000003 | Buchnera aphidicola Ap |
| BucaBp | 1 | 31 | 504 | 25 | 12 | 12 | −0.590 | (0.356/−0.448) | 1241 | AF492592 | Buchnera aphidicola Bp |
| BucaSg | 1 | 31 | 545 | 25 | 10 | 11 | −0.069 | (0.213/−0.265) | 1223 | AE013218 | Buchnera aphidicola Sg |
| Wigglo | 2 | 34 | 611 | 22 | 9 | 10 | 0.105 | (0.203/−0.247) | 1262 | BA000021 | Wigglesworthia glossinidia |
| Haeinf | 6 | 56 | 1709 | 38 | 27 | 24 | 1.492 | (0.330/−0.325) | 1001 | L42023 | Haemophilus influenzae |
| Pasmul | 6 | 56 | 2014 | 41 | 32 | 27 | 1.339 | (0.289/−0.282) | 1007 | AE004439 | Pasteurella multocida |
| Vibcho | 8 | 98 | 3828 | 47 | 47 | 37 | 1.725 | (0.294/−0.273) | 970 | AE003852* | Vibrio cholerae |
| Vibpar | 11 | 126 | 4832 | 45 | 44 | 33 | 1.886 | (0.336/−0.300) | 960 | BA000031* | Vibrio parahaemolyticus |
| Vibvul | 9 | 111 | 4537 | 47 | 47 | 34 | 1.950 | (0.296/−0.266) | 973 | AE016795* | Vibrio vulnificus CMCP6 |
| Sheone | 9 | 100 | 4630 | 46 | 45 | 37 | 1.377 | (0.313/−0.275) | 983 | AE014299 | Shewenella oneidensis |
| Pseaer | 4 | 62 | 5566 | 67 | 87 | 74 | −0.019 | (0.484/−0.507) | 940 | AE004091 | Pseudomonas aeruginosa |
| Pseput | 7 | 74 | 5350 | 62 | 77 | 64 | 0.917 | (0.360/−0.317) | 966 | AE015451 | Pseudomonas putida |
| Psesyr | 5 | 64 | 5566 | 58 | 71 | 58 | 0.701 | (0.255/−0.243) | 958 | AE016853 | Pseudomonas syringae |
| Xanaxo | 2 | 54 | 4312 | 65 | 80 | 80 | 0.636 | (0.273/−0.261) | 952 | AE008923 | Xanthomonas axonopodis |
| Xancam | 2 | 54 | 4181 | 65 | 81 | 80 | 0.607 | (0.292/−0.299) | 958 | AE008922 | Xanthomonas campestris |
| Xylfas | 2 | 49 | 2034 | 52 | 54 | 40 | −0.781 | (0.382/−0.324) | 990 | AE009442 | Xylella fastidiosa Temecula |
| Coxbur | 1 | 42 | 2009 | 43 | 38 | 43 | 0.175 | (0.170/−0.184) | 975 | AE016828 | Coxiella burnetii |
| Beta proteobacteria | |||||||||||
| Neimen | 4 | 58 | 2121 | 52 | 60 | 42 | −0.099 | (0.373/−0.346) | 1015 | AL157959 | Neisseria meningitidis Z2491 |
| Niteur | 1 | 41 | 2574 | 51 | 53 | 37 | −0.884 | (0.258/−0.253) | 1006 | AL954747 | Nitrosomonas europaea |
| Ralsol | 3 | 57 | 5120 | 67 | 87 | 80 | 0.024 | (0.451/−0.371) | 992 | AL646052* | Ralstonia solanacearum |
| Alpha proteobacteria | |||||||||||
| Agrtum | 4 | 53 | 4661 | 59 | 71 | 69 | 1.048 | (0.217/−0.202) | 1033 | AE008688* | Agrobacterium tumefaciens C58 (UW) |
| Sinmel | 3 | 54 | 6205 | 63 | 79 | 77 | 0.637 | (0.236/−0.225) | 1027 | AL591688 | Sinorhizobium meliloti |
| Brumel | 3 | 54 | 3198 | 57 | 66 | 67 | 0.896 | (0.237/−0.202) | 1037 | AE008917* | Brucella melitensis |
| Meslot | 2 | 52 | 6752 | 63 | 79 | 83 | 0.757 | (0.283/−0.245) | 1029 | BA000012 | Mesorhizobium loti |
| Brajap | 1 | 50 | 8317 | 64 | 82 | 86 | 0.741 | (0.312/−0.281) | 968 | BA000040 | Bradyrhizobium japonicum |
| Caucre | 2 | 51 | 3737 | 67 | 86 | 83 | 1.152 | (0.370/−0.310) | 970 | AE005673 | Caulobacter crescentus |
| Ricpro | 1 | 33 | 834 | 29 | 16 | 14 | −0.421 | (0.225/−0.243) | 1157 | AJ235269 | Rickettsia prowazekii |
| Riccon | 1 | 33 | 1374 | 32 | 21 | 17 | −0.410 | (0.234/−0.214) | 1135 | AE006914 | Rickettsia conorii |
| Epsilon proteobacteria | |||||||||||
| Camjej | 3 | 43 | 1654 | 31 | 17 | 16 | 0.486 | (0.300/−0.375) | 1119 | AL111168 | Campylobacter jejuni 11168 |
| Helpyl | 2 | 36 | 1491 | 39 | 41 | 42 | 0.016 | (0.184/−0.195) | 1138 | AE001439 | Helicobacter pylori J99 |
| Firmicutes (A+T-rich gram positives) | |||||||||||
| Bacsub | 10 | 86 | 4100 | 44 | 43 | 30 | 1.360 | (0.232/−0.224) | 1059 | AL009126 | Bacillus subtilis |
| Bacant | 11 | 95 | 5311 | 35 | 23 | 24 | 2.045 | (0.338/−0.316) | 1022 | AE016879 | Bacillus anthracis Ames |
| Bachal | 8 | 78 | 4066 | 44 | 40 | 34 | 0.999 | (0.166/−0.174) | 1046 | BA000004 | Bacillus halodurans |
| Oceihe | 7 | 69 | 3496 | 36 | 23 | 22 | 1.301 | (0.180/−0.197) | 1067 | BA000028 | Oceanobacillus iheyensis |
| Lismon | 6 | 67 | 2855 | 38 | 28 | 23 | 1.198 | (0.296/−0.288) | 1072 | AL591824 | Listeria monocytogenes EGD |
| Entfae | 4 | 67 | 3113 | 38 | 28 | 24 | 1.840 | (0.324/−0.287) | 1083 | AE016830 | Enterococcus faecalis |
| Lacpla | 5 | 72 | 3051 | 45 | 43 | 34 | 1.253 | (0.271/−0.268) | 1032 | AL935263 | Lactobacillus plantarum |
| Laclac | 6 | 62 | 2266 | 35 | 23 | 23 | 2.288 | (0.334/−0.321) | 1035 | AE005176 | Lactococcus lactis lactis |
| Straga | 7 | 80 | 2124 | 36 | 23 | 21 | 1.504 | (0.282/−0.252) | 1070 | AE009948 | Streptococcus agalactiae 2603V/R |
| Strpyo | 6 | 60 | 1696 | 39 | 30 | 24 | 1.759 | (0.299/−0.286) | 1081 | AE004092 | Streptococcus pyogenes M1 GAS SF370 |
| Strpne | 4 | 58 | 2043 | 40 | 34 | 26 | 1.720 | (0.380/−0.364) | 1074 | AE007317 | Streptococcus pneumoniae R6 |
| Staaur | 5 | 61 | 2593 | 33 | 20 | 18 | 1.564 | (0.248/−0.267) | 1084 | BA000018 | Staphlylococcus aureus N315 |
| Staepi | 5 | 58 | 2419 | 32 | 19 | 16 | 1.164 | (0.254/−0.243) | 1073 | AE015929 | Staphylococcus epididermis |
| Cloace | 11 | 73 | 3672 | 31 | 18 | 14 | 0.838 | (0.283/−0.286) | 856 | AE001437 | Clostridium acetobutylicum |
| Cloper | 10 | 95 | 2660 | 29 | 14 | 18 | 2.648 | (0.434/−0.420) | 838 | BA000016 | Clostridium perfringens |
| Clotet | 6 | 54 | 2373 | 29 | 14 | 13 | 1.004 | (0.244/−0.272) | 817 | AE015927 | Clostridium tetani |
| Theten | 4 | 55 | 2588 | 38 | 32 | 35 | 0.457 | (0.265/−0.266) | 842 | AE008691 | Thermoanaerobacter tengcongensis |
| Mycgen | 1 | 35 | 480 | 32 | 22 | 26 | 0.318 | (0.269/−0.310) | 1360 | L43967 | Mycoplasma genitalium |
| Mycpne | 1 | 36 | 688 | 40 | 41 | 43 | 0.324 | (0.206/−0.217) | 1307 | U00089 | Mycoplasma pneumoniae |
| Mycgal | 2 | 31 | 726 | 31 | 22 | 21 | 0.498 | (0.285/−0.391) | 1355 | AE015450 | Mycoplasma gallisepticum |
| Mycpen | 1 | 29 | 1037 | 26 | 12 | 11 | 0.496 | (0.237/−0.253) | 1379 | BA000026 | Mycoplasma penetrans |
| Mycpul | 1 | 28 | 782 | 27 | 13 | 12 | 0.380 | (0.235/−0.267) | 1235 | AL445566 | Mycoplasma pulmonis |
| Ureure | 1 | 29 | 611 | 26 | 11 | 10 | 0.401 | (0.232/−0.262) | 1223 | AF222894 | Ureaplasma urealyticum |
| Actinobacteria (G+C-rich gram positives) | |||||||||||
| Coreff | 5 | 56 | 2950 | 63 | 79 | 76 | 1.040 | (0.495/−0.395) | 1051 | BA000035 | Corynebacterium efficiens |
| Corglu | 6 | 60 | 3099 | 54 | 58 | 65 | 2.185 | (0.467/−0.381) | 1047 | BA000036 | Corynebacterium glutamicum |
| Myclep | 1 | 45 | 2720 | 58 | 64 | 73 | 0.515 | (0.224/−0.193) | 939 | AL450380 | Mycobacterium leprae |
| Myctub | 1 | 45 | 3918 | 66 | 79 | 83 | 0.452 | (0.256/−0.242) | 937 | AL123456 | Mycobacterium tuberculosis H37Rv |
| Strcoe | 6 | 63 | 7825 | 72 | 93 | 92 | 0.986 | (1.049/−0.618) | 921 | AL645882 | Streptomyces coelicolor |
| Strave | 6 | 68 | 7575 | 71 | 91 | 89 | 0.686 | (0.703/−0.501) | 937 | BA000030 | Streptomyces avermitilis |
| Trowhi | 1 | 50 | 808 | 46 | 41 | 46 | 0.014 | (0.189/−0.191) | 841 | AE014184 | Tropheryma whipplei Twist |
| Biflon | 4 | 56 | 1729 | 60 | 75 | 79 | 1.344 | (0.519/−0.449) | 999 | AE014295 | Bifidobacterium longum |
| Cyanobacteria | |||||||||||
| Nostoc | 4 | 67 | 5366 | 41 | 33 | 38 | 0.763 | (0.295/−0.271) | 1020 | BA000019 | Nostoc sp. PCC7120 |
| Theelo | 1 | 40 | 2475 | 54 | 57 | 56 | 0.178 | (0.306/−0.207) | 1018 | BA000039 | Thermosynechococcus elongatus |
| Syn680 | 2 | 41 | 3056 | 48 | 48 | 53 | 0.678 | (0.243/−0.253) | 1024 | BA000022 | Synechocystis PCC6803 |
| Spirochaetes | |||||||||||
| Borbur | 1 | 32 | 850 | 29 | 19 | 20 | −0.308 | (0.436/−0.579) | 1215 | AE000783 | Borrelia burgdorferi |
| Trepal | 2 | 45 | 1031 | 53 | 53 | 54 | −0.015 | (0.248/−0.255) | 956 | AE000520 | Treponema pallidum |
| Lepint | 1 | 37 | 4358 | 36 | 28 | 33 | 0.670 | (0.254/−0.258) | 1192 | AE010300* | Leptospira interrogans Lai |
| Chlamydiae | |||||||||||
| Chltra | 2 | 37 | 894 | 41 | 32 | 31 | 0.132 | (0.236/−0.247) | 974 | AE001273 | Chlamydia trachomatis |
| Chlmur | 1 | 37 | 904 | 40 | 31 | 30 | 0.145 | (0.244/−0.239) | 989 | AE002160 | Chlamydia muridarum |
| Chlcav | 1 | 38 | 998 | 39 | 30 | 28 | 0.113 | (0.224/−0.208) | 1028 | AE015925 | Chlamydophila caviae |
| Chlpne | 1 | 38 | 1110 | 41 | 33 | 26 | −0.065 | (0.223/−0.234) | 1027 | AE002161 | Chlamydophila pneumoniae AR39 |
| Fusobacteria | |||||||||||
| Fusnuc | 5 | 47 | 2067 | 27 | 10 | 10 | 1.244 | (0.242/−0.274) | 872 | AE009951 | Fusobacterium nucleatum |
| Bacteroidetes/Chlorobi | |||||||||||
| Bacthe | 5 | 70 | 4778 | 43 | 43 | 32 | 0.237 | (0.445/−0.418) | 1198 | AE015928 | Bacteroides thetaiotamicron |
| Chltep | 2 | 50 | 2252 | 57 | 72 | 65 | 0.069 | (0.301/−0.311) | 1072 | AE006470 | Chlorobium tepidum |
| Deinococci | |||||||||||
| Deirad | 3 | 49 | 2936 | 67 | 84 | 86 | 1.491 | (0.299/−0.280) | 990 | AE000513* | Deinococcus radiodurans |
| Thermotogae | |||||||||||
| Themar | 1 | 46 | 1846 | 46 | 51 | 48 | 0.365 | (0.281/−0.276) | 954 | AE000512 | Thermotoga maritima |
| Aquificae | |||||||||||
| Aquaeo | 2 | 43 | 1522 | 43 | 47 | 48 | 0.393 | (0.260/−0.273) | 837 | AE000657 | Aquifex aeolicus |
aThe species code (as used in Figure 1).
bThe numbers of rRNA operons, tRNA genes and putative protein-coding genes (ORFs).
cThe %G+C content of (i) the genome, or at synonymously variable third positions (ii) averaged over all genes and (iii) in the highly expressed gene data set.
dThe strength of selected codon usage bias (S) calculated for the highly expressed gene data set.
eThe 95% range of values of S among 1000 sets of randomly selected genes.
fThe number of codons used to estimate S, in the highly expressed gene data set.
gThe GenBank accession number for the genome sequence; asterisk indicates species with two chromosomes; the accession numbers for the two chromosomes are consecutive, except for D.radiodurans, where the second accession no. is AE001825.
To represent genes under the weakest selection, the codon usage of the entire genome was used, on the assumption that the number of genes expressed at high levels is a very small fraction of the genome as a whole. To represent genes where codon usage would be expected to be subject to strong translational selection, codon usage was summed across a set of 40 genes expected to be expressed constitutively at very high levels. This set included the genes encoding translation elongation factors Tu (tufA), Ts (tsf) and G (fusA), and 37 of the larger ribosomal proteins (encoded by genes rplA-rplF, rplI-rplT and rpsB-rpsT). No homologue of rplI was found in Mycoplasma penetrans; in this species rplU was added to the data set. Otherwise, the same 40 genes were used for all species. Many bacteria have two copies of the translation elongation factor Tu gene, although these are usually very similar due to concerted evolution (35), while some species have two or more homologues of fusA or certain ribosomal protein genes. In each case, the gene with the highest S-value was retained.
To assess whether the S-values observed were significantly greater than zero, for each species S-values were also calculated for 1000 sets of genes randomly selected from the genome. For each genome, the set of 40 highly expressed genes contained on average ∼1000 codons used in the analysis (Table 1). For the random data sets, genes were added until a total of at least 1000 codons were present for the four amino acids analysed. The range of S-values including 95% of these samples was recorded.
Phylogenetic analyses
The phylogenetic relationships of the 80 bacterial strains were estimated from a concatenated alignment of the proteins encoded by tuf, rplA-C and rpsB-C. Sequences were aligned using ClustalW (36), and sites with a gap in any sequence were removed. The tree was estimated by the Bayesian method implemented in MrBayesV3.0 (37), using the JTT model of protein evolution (38) with gamma distributed rates across sites. Phylogeny-independent correlations among species characters were estimated using the generalized least squares approach implemented in Continuous (39).
RESULTS
The strength of selected codon usage bias (S)
The strength of selected codon usage bias (S) was analysed for 80 genomes representing diverse major lineages of bacteria (Table 1 and Figure 1). S was estimated from the codon frequencies in a set of 40 genes expressed at very high levels compared with those in the genome as a whole, with the latter taken as an indication of the frequencies generated by mutation biases in the absence of selection. The analysis focused on four amino acids (Phe, Tyr, Ile and Asn), where the same codon is expected to be translationally advantageous in all species. The components of S for each of the four amino acids were highly correlated across species, and there was no clear indication that the U-ending codon is ever the optimal codon for any of the four amino acids.
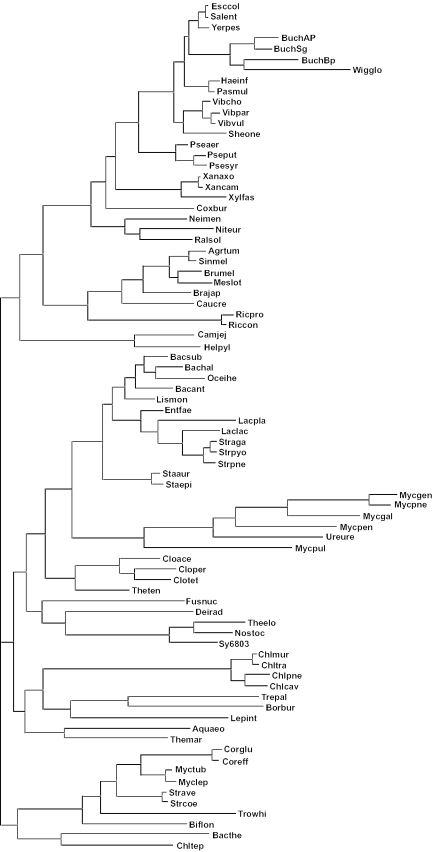
Figure 1.
Phylogenetic relationships of the 80 bacterial genomes analysed. Species codes are given in Table 1.
Some species have either two chromosomes (i.e. the three Vibrio species, Agrobacterium tumefaciens, Brucella melitensis, Leptospira interrogans and Deinococcus radiodurans) or one or more plasmids of larger than 1 Mb (Ralstonia solanacearum and Sinorhizobium meliloti). In each case, most (if not all) of the 40 genes expressed at high levels reside on just one of these chromosomes. Using the codon usage of genes from only this chromosome, rather than both, as the guide to mutational biases had only a minor impact on the S-values estimated: in all seven cases where both replicons are regarded as chromosomes the value of S was reduced by <3%. The effect was also minor in R.solanacearum, where S changed from 0.02 to −0.06, but more marked in S.meliloti, where the value decreased from 0.64 to 0.53, indicating a small difference in the overall codon usage between the plasmids and the chromosome in this species.
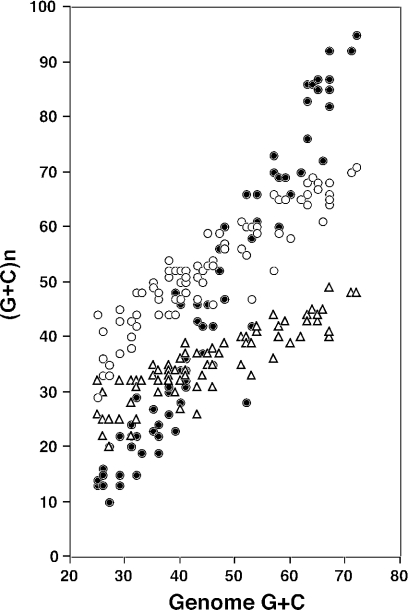
The species analysed here have genomic G+C contents ranging from 22 to 72%. Since bacterial genomes have little non-coding DNA, and the first two positions within codons are constrained by protein-coding requirements, most of the variation is due to the third position of codons [(8) and Figure 2]. Thus the overall G+C content at synonymously variable third positions (GC3S) ranged from 9 to 93% among the 80 genomes (Table 1). This base composition bias is so pervasive that it can be seen even when considering individual genes: e.g. for dnaA (a conserved gene with low selected codon usage bias), only one species (Xylella fastidiosa) showed a substantial deviation from the general trend, with a surprisingly low third position G+C content (28%) for a genome at 52% (Figure 2). This highlights the potential difficulty in estimating selected codon usage bias. The method used here for estimating S was explicitly designed to take account of genomic mutation biases, and indeed there was no correlation between S and the overall G+C content at synonymously variable third positions of codons (Figure 3). The optimal codons for the four amino acids analysed here are all C-ending, but there was no correlation between the S-value and the difference in GC3S values between the highly expressed gene data set and the genome as a whole; in fact, for 51 species the GC3S value for the highly expressed gene data set was the lower of the two (Table 1). This indicates that in species with high S-values many of the optimal codons for other amino acids are not C- or G-ending.
Figure 2.
G+C content at the three codon positions within the dnaA gene, compared with the G+C content of the genome as a whole, for 79 bacterial genomes (no dnaA homologue has been found in W.glossinidia). Positions 1, 2 and 3 are indicated by open circles, open triangles and filled circles, respectively. The third position is strongly influenced by G+C bias; the first two positions are also influenced, implying an effect on amino acid composition (68).
Figure 3.
Selected codon usage bias (S) and genomic G+C bias for 80 bacterial species. Genomic G+C bias is estimated by the overall GC3S. Open circles denote species where the S-value is not greater than found among randomly selected genes; filled triangles denote three Clostridium species.
The S-values showed a wide variation among species, ranging from −0.88 to 2.65 (Table 1). In most species, the 95% limits of the distribution of S-values for randomly selected genes were ∼0.2–0.3 either side of zero. For 24 species (i.e. 30% of the total), the S-value for the highly expressed genes was not as high as the upper 95% limit for the randomly selected genes, providing no immediate evidence that selection has affected codon usage in those genomes.
Negative S-values
The minimum S-values are expected to be around zero, but for five species the S-values were more highly negative than expected for randomly selected genes. This is surprising because the U-ending codons for the four amino acids analysed are unlikely to be translationally advantageous in any species, and the C-ending codons are not expected to be selected against in highly expressed genes. Two factors seem to contribute to these unexpectedly low S-values. First, in many species, there is a replication-dependent compositional skew between the leading and lagging strands, such that the leading strand is more G+T-rich, although the extent of this skew varies greatly among species (10). Most very highly expressed genes lie on the leading strand and so may have reduced frequencies of C-ending codons due to their location rather than because of selection. For example, in X.fastidiosa (S = −0.78), multivariate analysis of codon usage [following an approach outlined elsewhere (27)] found that the primary source of variation among genes was associated with this strand skew (40): the mean G+T contents (at synonymously variable third positions; GT3S) of leading and lagging strand genes are 0.61 and 0.40, respectively. Of the 40 highly expressed genes analysed here, 37 are encoded by the leading strand. When the highly expressed genes were compared with only those encoded on the leading strand, the S-value was much less highly negative (−0.43). Similarly, in Buchnera aphidicola strain Bp (S = −0.59), the average GT3S is 0.57 and 0.42 for genes on the leading and lagging strands, respectively; when the 34 highly expressed genes lying on the leading strand are compared with other leading strand genes, the S-value is −0.18. By comparison, in the other two B.aphidicola genomes (strains Ap and Sg), the skew between the two strands is much less pronounced, and the S-values are close to zero.
Second, many bacterial genomes contain regions (‘islands’) of unusual base composition, generally inferred to reflect horizontal gene transfer. In Nitrosomonas europaea (S = −0.88), where the average G+C content at synonymously variable third positions (GC3S) was 0.53 for the chromosome as a whole, many of the highly expressed genes lie within two islands with unusually low G+C content: 18 of the 40 genes in the highly expressed data set lie within a region encompassing 27 genes (rpsJ-rpoA, genes 400–426) where the average GC3S is 0.29, while 7 more lie in a cluster of 13 genes (rplL-NE2059, genes 2047–2059) with an average GC3S of 0.34. The S-value for these 25 genes is −1.36. The other 15 genes included in the set of 40 highly expressed genes are scattered around the genome, having an average GC3s of 0.45, and an S-value of −0.23. Horizontal transfer is thought to be rare for ‘informational’ genes, such as those encoding ribosomal proteins (41). However, since both regions include other genes, not expected to be highly expressed but with similarly low GC3S values, and since the highly expressed genes at other locations do not have such low GC3S values, the anomalously low S-values do not appear to be related to selection.
Correlation of selected codon usage bias with rRNA and tRNA gene numbers
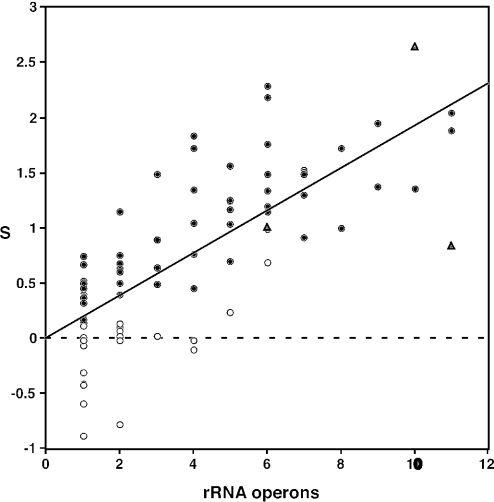
The strength of selection on synonymous codon usage is likely to be related to the degree to which speed and efficiency of growth and replication have been important during evolution. To investigate this, we have compared S-values with the numbers of rRNA operons and tRNA genes in each genome. Inter-specific variation in bacterial growth rate appears to be positively correlated with the number of rRNA operons (42). The abundance of different tRNAs is correlated with, and apparently largely determined by, gene copy number (11). The increased gene copy number, and consequent increased relative abundance, of particular tRNA species appears to be part of the strategy for optimizing translational efficiency (43,44). As expected, the numbers of rRNA and tRNA genes were found to be highly correlated in an analysis of 18 bacterial genomes (11). Among the 80 genomes analysed here, rRNA operon and tRNA gene copy numbers vary from 1 to 11, and 28 to 126, respectively (Table 1), and are very highly correlated (Figure 4).
Figure 4.
Ribosomal RNA operon copy number and tRNA gene number for 80 bacterial species. Symbols are as in Figure 3.
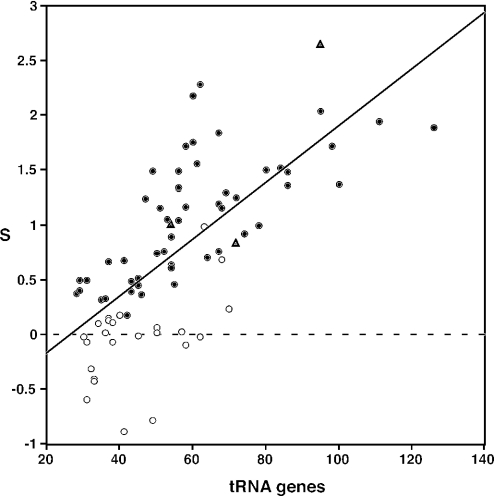
S-values are positively correlated with both rRNA operon and tRNA gene copy numbers (Figures 5 and 6). The highest S-value of all (2.65) was found in Clostridium perfringens, a genome with 10 rRNA operons and 95 tRNA genes. The species with the largest number of tRNA genes, Vibrio parahaemolyticus, is also among those with the largest number of rRNA operons, and has a high S-value (1.89). All species with >6 rRNA operons, and all species with >70 tRNA genes, have stronger codon usage bias in the highly expressed genes than in randomly selected genes. Among the 30 species with S-values >1, only two have fewer than four rRNA operons, and only two have fewer than 50 tRNA genes. Conversely, a majority of the species with only one rRNA operon, or <40 tRNA genes, show no evidence of selected codon usage bias.
Figure 5.
Selected codon usage bias (S) and ribosomal RNA operon copy number for 80 bacterial species. Symbols are as in Figure 3.
Figure 6.
Selected codon usage bias (S) and tRNA gene number for 80 bacterial species. Symbols are as in Figure 3.
The strengths of these correlations among rRNA operon numbers, tRNA gene copy numbers and S are overestimated by a simple analysis of the data as presented in Figures 4–6, due to the nonindependence of the data points. The 80 genomes are linked by a phylogenetic tree (Figure 1), and closely related species often share similar numbers of rRNA and tRNA genes, and have similar S-values, which may simply be due to their recent common ancestry. Using an approach to estimate the correlations after removing the effects of shared ancestry (39), the correlation coefficient for rRNA and tRNA gene copy numbers is 0.82, while the correlations between S and rRNA and tRNA gene copy numbers are 0.49 and 0.44, respectively (all values are highly statistically significant). While the phylogenetic relationships shown in Figure 1 are broadly consistent with those derived from analyses of other sequence data sets (45,46), there are some differences, such as Escherichia and Haemophilus being more closely related to each other than to Vibrio and Wigglesworthia lying within the radiation of Buchnera strains (47). However, we found that using alternative trees with such minor differences in topology had very little impact on the magnitude of the correlation coefficients.
DISCUSSION
Previous analyses of codon usage in bacteria have mostly focussed on the analyses of particular species, with no quantitative attempt to compare the strength of selected codon usage bias across different species (a recent exception is discussed below). Some analyses have started from the assumption that there is selected codon usage bias, without testing whether that is indeed the case (48), while others have concluded that ‘codon usage in most bacteria, if not all, is constrained by translation efficiency’ (11). Here, we have described a measure of the strength of selected codon usage bias, S, and a method for testing whether S is larger than expected by chance. The approach should be applicable to all species, and provides a means of comparing the strength of selected codon usage bias among them. We have applied this approach to 80 species. For 30% of these species, there was no evidence of selected codon usage bias, while among the others the value of S ranged widely.
Comparisons with previous analyses of individual species
The archetypal example of a species with strongly selected codon usage bias has been E.coli, where the selective pressure exerted via tRNA relative abundance and anticodon sequence was first elucidated (13,14). The S-value calculated here for E.coli is high (1.49), but 15 other species (∼20% of the species analysed) have even higher values, indicating more strongly selected codon usage bias. Among these 15 species are 8 members of the Firmicutes (A+T-rich gram positive bacteria), including C.perfringens with the highest value of 2.65. A recent analysis detected the selected codon usage bias in C.perfringens, and also noted that the bias was stronger than in Clostridium acetobutylicum (49); here, the latter species has an S-value of 0.84. In fact, with the exception of the Mollicutes and C.acetobutylicum, all of the Firmicutes have S-values >1.0 (Table 1). An early analysis of one of these species, B.subtilis, concluded (from 56 genes) that the selected codon usage bias was weaker than in E.coli (50), but here the S-value for B.subtilis is 1.36, not substantially different from E.coli.
An early analysis of M.tuberculosis (using 41 genes) reported weak but significant selected codon usage bias (25), and this is confirmed by the S-value of 0.45, compared with 0.26 as the upper limit of the 95% range for randomly selected genes in that species. Analysis of the genome of Thermotoga maritima detected selected codon usage in highly expressed genes, but found this to be a relatively minor source of variation among genes (28). No significant difference in the use of Tyr, Ile or Asn codons was found between genes expressed at high and low levels; and since these are three of the four amino acids used here, it is not surprising that the S-value is very low (0.37), and only just above the value (0.28) for randomly selected genes.
For two other species, where weak selected codon usage bias has been reported, the present analysis yields S-values within the range of randomly selected genes. In Chlamydia trachomatis, the major trend among genes in codon usage is related to strand skew (26). The average GT3S values for genes on the leading and lagging strands are 0.57 and 0.48, respectively. If the 40 genes are compared with only those on the leading strand, the S-value becomes 0.42, indicative of weak selection. Pseudomonas aeruginosa has an S-value close to zero (−0.02), providing no evidence for selection, whereas we previously found small but significant differences in codon usage between highly expressed and other genes (27). This discrepancy arises because the largest components of selected bias in this species relate to codons for Ser (especially UCC), Thr (ACC), Ala (GCU), Arg (CGU) and Gly (GGU), whereas frequencies of the C-ending codons for Phe, Tyr, Ile and Asn (used to calculate S) differ little between highly expressed genes and the genome as a whole (27).
Otherwise, few analyses have commented on the relative strength of selected codon usage bias, except in those cases where it appears to be absent. Evidence of a lack of selected codon usage bias has been reported for Helicobacter pylori (24), Rickettsia prowazekii (18), Treponema pallidum (23), Buchnera strains (21) and Wigglesworthia (22), all of which have S-values close to zero. In addition, an absence of selected codon usage bias has been reported in Borrelia burgdorferi (23,51) and Mycoplasma genitalium (19,20), but for these two species these conclusions have been questioned (52). For B.burgdorferi, there is no sign of selected codon usage bias in the present analysis, since the S-value is negative (−0.31). However, in this species, there is extremely pronounced skew between the chromosome strands: the average GT3S values for genes on the leading and lagging strands are 0.62 and 0.39, respectively. Of the 40 genes in the highly expressed data set, 38 lie on the leading strand, and when these are compared with genes from the leading strand only, the B.burgdorferi value is −0.04, still providing no evidence for selection. For M.genitalium, the possibility of selected codon usage bias was invoked on the grounds that highly expressed genes tend to use more G+C-rich codons (52). Indeed, here the S-value for M.genitalium (0.32) is slightly higher than expected for randomly selected genes. However, it has been shown that the major source of variation among M.genitalium genes is in G+C content, which varies systematically in a wave around the genome, seemingly affecting all genes irrespective of their expression level (19,20). A total of 29 of the 40 highly expressed genes used here lie within the most G+C-rich 40% of the genome. When these 29 genes are compared with the 192 genes in this region, the S-value is lower (0.17), and within the range of values for randomly selected genes from this region. This suggests that the minor difference in codon usage between highly expressed genes (in total) and the genome as a whole reflects compositional variation, and provides no evidence for selected codon usage bias in this species.
Streptomyces species are extremely G+C-rich, and this compositional bias was found to dominate codon usage in an early study (17). However, it was noted that tufA (the only unambiguously highly expressed gene sequence then available) had slightly different codon usage that might indicate the action of weak translational selection. Here, Streptomyces coelicolor has an S-value of 0.99. This value is close to that expected for a genome with 6 rRNA operons (Figure 5) and 63 tRNA genes (Figure 6), and all of these features are consistent with moderately strong translational selection. However, the difficulty in interpreting codon usage variation in this species is shown by the unusually broad range of values observed for randomly selected genes (Table 1). Among 1000 randomly selected S.coelicolor data sets, 28 had S-values as large as that for the highly expressed genes. For Streptomyces avermitilis, the S-value is lower (0.69), but again just within the range of values for 95% of randomly selected gene data sets. Overall, it appears that the codon selection in Streptomyces has been marginally effective in overcoming the very strong mutational bias.
Thus, the S-values obtained here are largely consistent with more detailed studies on individual species. However, because S is calculated from only four amino acids, where the choice is always between the translationally optimal C-ending codon and a U-ending codon, intragenomic variations in G+T content can impinge on the value obtained. Since most highly expressed genes lie on the leading (G+T-rich) strand this tends to reduce S, but the size of the effect, reflecting the extent of skew between the strands, varies substantially among species. For example, in E.coli the average GT3S values of genes on the leading and lagging strands are 0.55 and 0.51, respectively, and using only leading strand genes as the control for mutational bias leaves the S-value unaltered. It might be preferable to always only use genes on the leading strand as the control for mutational bias, but for many species this is impracticable because it is difficult to locate the origin and terminus of replication precisely. Furthermore, even closely related strains can show extensive genomic rearrangement [e.g. in the case of X.fastidiosa 9a5c compared with the Temecula strain analysed here (53,54)], which can confound comparisons of leading and lagging strand genes.
Intragenomic variations in G+C content can also impinge on the value of S. With the exception of M.genitalium (discussed above), intragenomic G+C variation mostly reflects ‘islands’ of atypical base composition. Typically, as many as half of the 40 highly expressed genes examined here are located in a single cluster, and we have noticed that in a number of species this cluster is more A+T-rich than the genome as a whole, tending to reduce the S-value. Islands of atypical base composition are usually explained as the result of horizontal gene transfer, but it is generally not expected that ribosomal protein genes undergo this process. Thus, the reason(s) for this base composition difference warrant further investigation.
These caveats regarding intragenomic variations in base composition serve to emphasise that any automated analysis of codon usage, without some detailed consideration of the variation among genes, may be prone to errors. However, the advantage of calculating S-values by the method described here is that a uniform approach can be used for all species, enabling comparisons among them.
Variation among bacteria in the strength of selected codon usage bias
At a biochemical level, the C-ending codons for Phe, Tyr, Ile and Asn are expected to be translationally optimal in all bacteria, but the wide range of S-values observed (Table 1) indicates that the strength and/or efficacy of selection for these optimal codons has varied considerably among species. The strength of selected codon usage bias, as estimated by S, is highly correlated with the number of rRNA operons and the number of tRNA genes. We expect that codon usage will have been more strongly selected in species which replicate fast. Information regarding the growth rate of bacteria in the wild is sparse, and so we have used the number of rRNA operons as a (very approximate) guide to the growth rate of species. Remarkably, C.perfringens, the species with the highest S-value (2.65) and 10 rRNA operons, can grow with a generation time under 7 min in specific laboratory conditions (55). In contrast, Mycobacterium species are renowned for their very slow growth: M.tuberculosis and M.leprae have generation times of ∼1 and 14 days, respectively. Both species have one rRNA operon and low S-values (∼0.5). These observations are consistent with the effects of selection for efficiency of translation under rapid and competitive growth conditions, and then the lack of selected codon usage bias in some species would reflect a relative unimportance of an exponential growth phase during their life history.
Alternatively, a lack of selected codon usage bias may reflect the greater impact of random genetic drift, due to a population structure with a low long-term effective population size and/or interference between linked synonymous sites due to a lack of recombination. For most species, it is difficult to know the long-term evolutionary effective population size relevant to codon usage. For example, M.tuberculosis currently infects many more people worldwide than M.leprae, such that the former is likely to have much the larger ongoing effective population size. However, M.tuberculosis exhibits little genetic diversity (56) and is thought to be a recently emerged clone from M.canetti (57); this evolutionary bottleneck would have reduced the effective population size of M.tuberculosis. But even this may have little relevance: in the same way that it is thought that the codon usage of horizontally transferred genes may take many millions of years to ameliorate to that of a new host genome (58), strongly selectively biased codon usage may take a very long time to decay after a reduction in effective population size, i.e. the codon usage bias currently observed may still be due in some part to evolutionary processes that occurred millions of years ago. The two Mycobacterium species currently have similar levels of selected codon usage bias.
Nevertheless, it seems clear that the life histories of some of the bacteria analysed are likely to lead to low effective population sizes. Many of the species with very low S-values are obligate intracellular parasites or endosymbionts: these include species in the genera Buchnera, Wigglesworthia, Coxiella, Rickettsia and Tropheryma, the Mollicutes (Mycoplasma plus Ureaplasma) as well as the four Chlamydiales. Among these 18 species, all have S-values <0.5, and only the Mollicutes have values >0.2, and marginally higher than expected from randomly selected genes. Most have reduced genome sizes (<1000 genes), all have only 1 or 2 rRNA operons, and most have <40 tRNA genes (Table 1). For example, Buchnera and Wigglesworthia are obligate endosymbionts of insects, with low effective population sizes (due to bottlenecks during their transmission) and limited recombination. It has been noted that, as well as an absence of selected codon usage bias, these species have rapid evolutionary rates, presumably reflecting the enhanced power of random genetic drift (21). In contrast, all of the bacteria with high S-values (say, >1.5) live outside host cells, typically in mixed environments, such as soil, water or the intestinal tracts of animals. Thus, this difference between an intracellular parasitic lifestyle and an extracellular existence appears to be a pervasive influence on S among the species included in this analysis.
A lack of recombination would be expected to impair the efficacy of selection on codon usage. Many of the intracellular parasitic species, noted above for their low S-values, are known or expected to be effectively clonal. Additionally, the primarily extracellular pathogenic spirochaete B.burgdorferi is extremely clonal (59) and has S near zero. In contrast, Streptococcus pneumoniae, Streptococcus pyogenes and Staphylococcus aureus all appear to have undergone high rates of recombination (60), and have high S-values (Table 1). However, E.coli and Haemophilus influenzae also have high S-values, despite apparently lower rates of recombination (60). It is clear that a high recombination rate alone is not enough to promote codon selection: H.pylori has perhaps the highest rate of recombination known among bacteria (61), and yet an S-value close to zero. In this case, the lack of selected codon usage bias has been interpreted as a consequence of the unimportance of competitive growth in the isolated acidic niche of this species (24).
Overall, it is difficult to disentangle the effects of low effective population size and a lack of recombination from the other aspects of these organisms' lifestyles discussed above. For example, among the spirochaetes, two (B.burgdorferi and T.pallidum) have S-values close to zero, whereas the third (L.interrogans) has a somewhat higher value (0.67). Both B.burgdorferi and T.pallidum are obligate parasites and grow slowly, whereas L.interrogans is a facultative parasite with many saprophytic relatives, is more metabolically versatile and can grow more rapidly. The stronger selected codon usage bias in L.interrogans appears to reflect this difference in lifestyles, although interestingly it is not accompanied by an increase in rRNA or tRNA gene number.
The correlations between S and rRNA and tRNA gene copy numbers are sufficiently strong that it is interesting to examine the outliers. For example, values for the three Clostridium species are highlighted in Figures 4–6. The S-value for C.acetobutylicum (0.84) is surprisingly low for a genome with 11 rRNA operons (Figure 5). It is similar to that of Clostridium tetani (1.00), with only 6 rRNA operons, but much lower than that of C.perfringens (2.65), a genome with 10 rRNA operons. However, the S-value for C.acetobutylicum is not unusual for a genome with 73 tRNA genes (Figure 6). Thus, it seems to be the high number of rRNA operons in C.acetobutylicum that is anomalous; this may reflect a very recent expansion in this gene family.
Perhaps the most surprising example of low codon usage bias is P.aeruginosa. This species can grow quite rapidly (doubling times <1 h) in laboratory planktonic cultures and is metabolically highly versatile. It is moderately recombinogenic via plasmid transfer, and there appear to be many horizontally transferred genes in its genome (27). The low selected bias was apparent in a full analysis of codon usage in this species (27), as well as the S-value calculated here. Selected codon usage bias is rather stronger in the two other Pseudomonas species analysed (Table 1). These paradoxical observations perhaps highlight our ignorance of the evolutionary history of even ‘well-known’ bacterial species.
Comparison with another estimate of S
Recently, another approach to estimating the strength of selected codon usage bias in a genome has been published by dos Reis and co-workers (62). These authors calculated two indices of codon usage bias. The first, based on the effective number of codons used in a gene (63), attempted to estimate the strength of general deviation from random codon usage in a gene. The second was a modification of the codon adaptation index, CAI (64), using tRNA gene copy number (as a surrogate for tRNA abundance) and the estimated strength of codon–anticodon interaction to assign fitness values to codons; the tRNA adaptation index for a gene was calculated as the average of these fitness values, as an attempt to estimate the adaptation of a gene's codon usage to the tRNA pool of the species. It was suggested that the strength of translationally selected codon usage bias, S (here termed St to distinguish it from S described above), could be estimated from the magnitude of the correlation between these two indices; the significance of St was estimated from a permutation test.
Dos Reis et al. (62) applied this methodology to 101 bacterial genomes, including 66 of those analysed here as well as another 20 genomes excluded here because of their close relationship to other strains. The St method found significant evidence for selection in only 26% of bacterial genomes analysed. Among the 66 species common to both analyses, S- and St-values are significantly correlated (coefficient = 0.46); 14 species were found to have significant evidence for selection in both analyses and 18 were found to lack such evidence in both analyses (Figure 7). However, 32 species found here to have significant S-values were not significant in the St analysis. These included a number of species where previous analyses have found clear evidence of selectively biased codon usage in highly expressed genes, such as B.subtilis (50,65), C.acetobutylicum (49) and Vibrio cholerae (40). Most strikingly, C.perfringens had the highest S-value among the 80 species analysed here, and yet was not significant in the St test; detailed analysis of codon usage in this species has revealed strongly selected bias in highly expressed genes (49).
Figure 7.
Comparison of two estimates of selected codon usage bias: x-axis values are taken from this paper, y-axis values from dos Reis et al. (62). Values significantly greater than zero in the dos Reis et al. analysis are shown as circles; values significantly greater than zero in our analysis are shown as filled symbols.
Interestingly, two species found here not to have significant S-values, Neisseria meningitidis and Bacteroides thetaiotamicron, were significant in the St test. Closer examination of these species [following an approach outlined elsewhere (27)] revealed that, in both, the primary trends in codon usage variation among genes were associated with leading versus lagging strand composition bias and G+C content, but there was evidence for weak selected codon usage bias in highly expressed genes. Overall, it appears that the estimation of S described here is generally much more effective than the St test at detecting translationally selected codon usage bias, even though S can sometimes be reduced by compositional biases. One difference between the two approaches should be noted. The method described here asks how strong the selected bias is in a specified set of very highly expressed genes, but not how many genes exhibit selected bias. The dos Reis et al. method aimed at quantifying the extent to which variation among genes across the genome as a whole can be explained as adaptation to the tRNA pool of the species. Given this difference, further comparison of the results of the two methods may shed additional light on the causes of selected codon usage bias.
Solving the riddle of codon usage preferences?
In their analysis dos Reis et al. included a small number of eukaryote genomes, as well as archaeal and bacterial species. They found that variation in the strength of codon usage bias among species was highly positively correlated with genome size and tRNA gene copy number (except in very large genomes), and concluded that these two factors ‘ultimately determine the action of natural selection’ on codon usage (62). They proposed a model whereby, from an ancestral bacterium with a small genome size, increases in genome size led to increases in tRNA gene copy number, which in turn led to selection for the optimization of codon usage. However, we find that genome size does not seem to cause tRNA gene copy number (among bacteria, at least), while it seems inappropriate to consider codon bias as the result of tRNA gene copy number. In contrast, we suggest that it is the biology of the organism (its ‘lifestyle’) that determines whether codon usage is affected by natural selection.
The overall results of dos Reis et al. were heavily influenced by the inclusion of eukaryote species, which contributed disproportionately to the variation in both genome size and tRNA gene number. Although there is a positive correlation between genome size and tRNA gene number among the 80 bacterial species examined here, this seems to be due only to species with small genomes. (Note that dos Reis et al. considered genome size in terms of DNA content, whereas we have used the estimated number of protein-coding genes; however, these two measures are extremely highly correlated among bacteria and so this difference should have no impact.) Among the larger bacterial genomes (e.g. the 42 species with >2500 genes), there is no significant correlation between genome size and tRNA copy number. For example, 10 of the 11 species with >5000 genes have <75 tRNA genes, while 10 of the 11 species with >75 tRNA genes have <5000 protein-coding genes; the single exception is B.anthracis with 5311 genes and 95 tRNA genes (Table 1). Thus, increases in genome size do not generally involve an increase in the number of tRNA genes. The forces that have led to reduced genome size (e.g. in Buchnera, Rickettsia and Mycoplasma species) may have impacted on tRNA gene copy number directly, but it seems more likely that these evolutionary pressures reflect the adoption of a lifestyle (typically intracellular parasitism), in which rapid replication was not advantageous (or perhaps even detrimental) and thus translational efficiency became less important, and additional tRNA genes became unnecessary.
It seems inappropriate to consider codon usage bias as simply being caused by tRNA abundances, since both factors are likely to co-evolve in response to selection for translational efficiency (44,66). Indeed, it is possible to consider circumstances where changes in codon usage bias, perhaps brought about by a change in the genome wide mutational bias, could select for a change in the tRNA pool (67). Thus, while we find correlations across species in the numbers of rRNA operons and tRNA genes, and the strength of selected codon usage bias, we do not invoke a causal relationship among any of these factors; rather, we take all three as indicative of the need for rapid and efficient bacterial growth.
Acknowledgments
We are very grateful to Michael Bulmer for discussion of his population genetic model of codon usage bias, and to Manolo Gouy and colleagues in Lyon for providing the ACNUC interface to GenBank. We also thank Mario dos Reis for discussion of his recent paper. This work was supported in part by studentships from the MRC (to R.J.G.) and the University of Nottingham (to J.F.P.). Funding to pay the Open Access publication charges for this article was provided by The University of Nottingham.
REFERENCES
- 1.Grantham R., Gautier C., Gouy M., Jacobzone M., Mercier R. Codon catalog usage is a genome strategy modulated for gene expressivity. Nucleic Acids Res. 1981;8:r43–r74. doi: 10.1093/nar/9.1.213-b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ikemura T. Codon usage and tRNA content in unicellular and multicellular organisms. Mol. Biol. Evol. 1985;2:13–34. doi: 10.1093/oxfordjournals.molbev.a040335. [DOI] [PubMed] [Google Scholar]
- 3.Sharp P.M., Cowe E., Higgins D.G., Shields D.C., Wolfe K.H., Wright F. Codon usage in Escherichia coli, Bacillus subtilis, Saccharomyces cerevisiae, Schizosaccharomyces pombe, Drosophila melanogaster and Homo sapiens; a review of the considerable within-species diversity. Nucleic Acids Res. 1988;16:8207–8211. doi: 10.1093/nar/16.17.8207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sharp P.M., Li W.-H. An evolutionary perspective on synonymous codon usage in unicellular organisms. J. Mol. Evol. 1986;24:28–38. doi: 10.1007/BF02099948. [DOI] [PubMed] [Google Scholar]
- 5.Bulmer M. The selection-mutation-drift theory of synonymous codon usage. Genetics. 1991;129:897–907. doi: 10.1093/genetics/129.3.897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sharp P.M., Stenico M., Peden J.F., Lloyd A.T. Codon usage: mutational bias, translational selection, or both? Biochem. Soc. Trans. 1993;21:835–841. doi: 10.1042/bst0210835. [DOI] [PubMed] [Google Scholar]
- 7.Sueoka N. On the genetic basis of variation and heterogeneity of DNA base composition. Proc. Natl Acad. Sci. USA. 1962;48:582–592. doi: 10.1073/pnas.48.4.582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Muto A., Osawa S. The guanine and cytosine content of genomic DNA and bacterial evolution. Proc. Natl Acad. Sci. USA. 1987;84:166–169. doi: 10.1073/pnas.84.1.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lobry J.R. Asymmetric substitution patterns in the two DNA strands of bacteria. Mol. Biol. Evol. 1996;13:660–665. doi: 10.1093/oxfordjournals.molbev.a025626. [DOI] [PubMed] [Google Scholar]
- 10.McLean M.J., Devine K.M., Wolfe K.H. Base composition skews, replication orientation, and gene orientation in 12 prokaryotic genomes. J. Mol. Evol. 1997;47:691–696. doi: 10.1007/pl00006428. [DOI] [PubMed] [Google Scholar]
- 11.Kanaya S., Yamada Y., Kudo Y., Ikemura T. Studies of codon usage and tRNA genes of 18 unicellular organisms and quantification of Bacillus subtilis tRNAs: gene expression level and species-specific diversity of codon usage based on multivariate analysis. Gene. 1999;238:143–155. doi: 10.1016/s0378-1119(99)00225-5. [DOI] [PubMed] [Google Scholar]
- 12.Ochman H., Lawrence J.G., Groisman E.A. Lateral gene transfer and the nature of bacterial innovation. Nature. 2000;405:299–304. doi: 10.1038/35012500. [DOI] [PubMed] [Google Scholar]
- 13.Post L.E., Nomura M. DNA sequences from the str operon of Escherichia coli. J. Biol. Chem. 1980;255:4660–4666. [PubMed] [Google Scholar]
- 14.Ikemura T. Correlation between the abundance of Escherichia coli transfer RNAs and the occurrence of the respective codons in its protein genes. J. Mol. Biol. 1981;146:1–21. doi: 10.1016/0022-2836(81)90363-6. [DOI] [PubMed] [Google Scholar]
- 15.Ikemura T. Correlation between the abundance of yeast transfer RNAs and the occurrence of the respective codons in protein genes. J. Mol. Biol. 1982;158:573–597. doi: 10.1016/0022-2836(82)90250-9. [DOI] [PubMed] [Google Scholar]
- 16.Bennetzen J.L., Hall B.D. Codon selection in yeast. J. Biol. Chem. 1982;257:3026–3031. [PubMed] [Google Scholar]
- 17.Wright F., Bibb M.J. Codon usage in the G+C-rich Streptomyces genome. Gene. 1992;113:55–65. doi: 10.1016/0378-1119(92)90669-g. [DOI] [PubMed] [Google Scholar]
- 18.Andersson S.G.E., Sharp P.M. Codon usage and base composition in Rickettsia prowazekii. J. Mol. Evol. 1996;42:525–536. doi: 10.1007/BF02352282. [DOI] [PubMed] [Google Scholar]
- 19.Kerr A.R.W., Peden J.F., Sharp P.M. Systematic base composition variation around the genome of Mycoplasma genitalium, but not Mycoplasma pneumoniae. Mol. Microbiol. 1997;25:1177–1179. doi: 10.1046/j.1365-2958.1997.5461902.x. [DOI] [PubMed] [Google Scholar]
- 20.McInerney J.O. Prokaryotic genome evolution as assessed by multivariate analysis of codon usage patterns. Microb. Comp. Genomics. 1997;2:1–10. [Google Scholar]
- 21.Wernegreen J.J., Moran N.A. Evidence for genetic drift in endosymbionts (Buchnera): analyses of protein-coding genes. Mol. Biol. Evol. 1999;16:83–97. doi: 10.1093/oxfordjournals.molbev.a026040. [DOI] [PubMed] [Google Scholar]
- 22.Herbeck J.T., Wall D.P., Wernegreen J.J. Gene expression level influences amino acid usage, but not codon usage, in the tsetse fly endomsymbiont Wigglesworthia. Microbiology. 2003;149:2585–2596. doi: 10.1099/mic.0.26381-0. [DOI] [PubMed] [Google Scholar]
- 23.Lafay B., Lloyd A.T., McLean M.J., Devine K.M., Sharp P.M., Wolfe K.H. Proteome composition and codon usage in spirochaetes: species-specific and DNA strand-specific mutational biases. Nucleic Acids Res. 1999;27:1642–1649. doi: 10.1093/nar/27.7.1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lafay B., Atherton J.C., Sharp P.M. Absence of translationally selected codon usage bias in Helicobacter pylori. Microbiology. 2000;146:851–860. doi: 10.1099/00221287-146-4-851. [DOI] [PubMed] [Google Scholar]
- 25.Andersson S.G.E., Sharp P.M. Codon usage in the Mycobacterium tuberculosis complex. Microbiology. 1996;142:915–925. doi: 10.1099/00221287-142-4-915. [DOI] [PubMed] [Google Scholar]
- 26.Romero H., Zavala A., Musto H. Codon usage in Chlamydia trachomatis is the result of strand-specific mutational biases and a complex pattern of selective forces. Nucleic Acids Res. 2000;28:2084–2090. doi: 10.1093/nar/28.10.2084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Grocock R.J., Sharp P.M. Synonymous codon usage in Pseudomonas aeruginosa PAO1. Gene. 2002;289:131–139. doi: 10.1016/s0378-1119(02)00503-6. [DOI] [PubMed] [Google Scholar]
- 28.Zavala A., Naya H., Romero H., Musto H. Trends in codon and amino acid usage in Thermotoga maritima. J. Mol. Evol. 2002;54:563–568. doi: 10.1007/s00239-001-0040-y. [DOI] [PubMed] [Google Scholar]
- 29.Arnold H.H., Keith G. The nucleotide sequence of phenylalanine tRNA from Bacillus subtilis. Nucleic Acids Res. 1977;4:2821–2829. doi: 10.1093/nar/4.8.2821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kurland C.G. Strategies for efficiency and accuracy in gene expression. 1. The major codon preference: a growth optimization strategy. Trends Biochem. Sci. 1987;12:126–128. [Google Scholar]
- 31.Maynard Smith J., Smith N.H., O'Rourke M., Spratt B.G. How clonal are bacteria? Proc. Natl Acad. Sci. USA. 1993;90:4384–4388. doi: 10.1073/pnas.90.10.4384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McVean G.A.T., Charlesworth B. The effects of Hill-Robertson interference between weakly selected mutations on patterns of molecular evolution and variation. Genetics. 2000;155:929–944. doi: 10.1093/genetics/155.2.929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gouy M., Gautier C., Attimonelli M., Lanave C., Di Paola G. ACNUC—a portable retrieval system for nucleic acid sequence databases: logical and physical design and usage. Comp. Appl. Biosci. 1985;1:167–172. doi: 10.1093/bioinformatics/1.3.167. [DOI] [PubMed] [Google Scholar]
- 34.Peden J.F. Analysis of codon usage. 1999. PhD Thesis, University of Nottingham, UK.
- 35.Sharp P.M. Determinants of DNA sequence divergence between Escherichia coli and Salmonella typhimurium: codon usage, map position and concerted evolution. J. Mol. Evol. 1991;33:23–33. doi: 10.1007/BF02100192. [DOI] [PubMed] [Google Scholar]
- 36.Thompson J.D., Higgins D.G., Gibson T.J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence-weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huelsenbeck J.P., Ronquist F. MRBAYES: Bayesain inference of phylogeny. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- 38.Jones D.T., Taylor W.R., Thornton J.M. The rapid generation of mutation data matrices from protein sequences. Comput. Appl. Biosci. 1992;8:275–282. doi: 10.1093/bioinformatics/8.3.275. [DOI] [PubMed] [Google Scholar]
- 39.Pagel M. Inferring the historical patterns of biological evolution. Nature. 1999;401:877–884. doi: 10.1038/44766. [DOI] [PubMed] [Google Scholar]
- 40.Grocock R.J. Evolution of codon usage among the gamma Proteobacteria. 2003. PhD Thesis, University of Nottingham, UK.
- 41.Jain R., Rivera M., Lake J.A. Horizontal gene transfer among genomes: the complexity hypothesis. Proc. Natl Acad. Sci. USA. 1999;96:3801–3806. doi: 10.1073/pnas.96.7.3801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Klappenbach J.A., Dunbar J.M., Schmidt T.M. rRNA operon copy number reflects ecological strategies of bacteria. Appl. Environ. Microbiol. 2000;66:1328–1333. doi: 10.1128/aem.66.4.1328-1333.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ehrenberg M., Kurland C.G. Costs of accuracy determined by a maximal growth rate constraint. Q. Rev. Biophys. 1984;17:45–82. doi: 10.1017/s0033583500005254. [DOI] [PubMed] [Google Scholar]
- 44.Berg O.G., Kurland C.G. Growth rate-optimised tRNA abundance and codon usage. J. Mol. Biol. 1997;270:544–550. doi: 10.1006/jmbi.1997.1142. [DOI] [PubMed] [Google Scholar]
- 45.Olsen G.J., Woese C.R., Overbeek R. The winds of (evolutionary) change—breathing new life into microbiology. J. Bacteriol. 1994;176:1–6. doi: 10.1128/jb.176.1.1-6.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Haubold B., Wiehe T. Comparative genomics: methods and applications. Naturwissenschaften. 2004;91:405–421. doi: 10.1007/s00114-004-0542-8. [DOI] [PubMed] [Google Scholar]
- 47.Wernegreen J.J., Degnan P.H., Lazarus A.B., Palacios C., Bordenstein S.R. Genome evolution in an insect cell: distinct features of an ant-bacterial partnership. Biol. Bull. 2003;204:221–231. doi: 10.2307/1543563. [DOI] [PubMed] [Google Scholar]
- 48.Karlin S., Mrazek J. Predicted highly expressed genes of diverse prokaryotic genomes. J. Bacteriol. 2000;182:5238–5250. doi: 10.1128/jb.182.18.5238-5250.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Musto H., Romero H., Zavala A. Translational selection is operative for synonymous codon usage in Clostridium perfringens and Clostridium acetobutylicum. Microbiology. 2003;149:855–863. doi: 10.1099/mic.0.26063-0. [DOI] [PubMed] [Google Scholar]
- 50.Shields D.C., Sharp P.M. Synonymous codon usage in Bacillus subtilis reflects both translational selection and mutational biases. Nucleic Acids Res. 1987;15:8023–8040. doi: 10.1093/nar/15.19.8023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.McInerney J.O. Replicational and transcriptional selection on codon usage in Borrelia burgdorferi. Proc. Natl Acad. Sci. USA. 1998;95:10698–10703. doi: 10.1073/pnas.95.18.10698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Perriere G., Thiolouse J. Use and misuse of correspondence analysis in codon usage studies. Nucleic Acids Res. 2002;30:4548–4555. doi: 10.1093/nar/gkf565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Simpson A.J.G., Reinach F.C., Arruda P., Abreu F.A., Acencio M., Alvarenga R., Alves L.M.C., Araya J.E., Baia G.S., Baptista C.S., et al. The genome sequence of the plant pathogen Xylella fastidiosa. Nature. 2000;406:151–159. doi: 10.1038/35018003. [DOI] [PubMed] [Google Scholar]
- 54.Van Sluys M.A., de Oliveira M.C., Monteior-Vitorello C.B., Miyaki C.Y., Furlan L.R., Camargo L.E.A., da Silva A.C.R., Moon D.H., Takita M.A., Lemos E.G.M., et al. Comparative analysis of the complete genome sequences of Pierce's disease and citrus variegated chlorosis strains of Xylella fastidiosa. J. Bacteriol. 2003;185:1018–1026. doi: 10.1128/JB.185.3.1018-1026.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Labbe R.G., Huang T.H. Generation times and modeling of enterotoxin-positive and enterotoxin-negative strains of Clostridium perfringens in laboratory media and ground beef. J. Food Prot. 1995;58:1303–1306. doi: 10.4315/0362-028X-58.12.1303. [DOI] [PubMed] [Google Scholar]
- 56.Sreevatsan S., Pan X., Stockbauer K.E., Connell N.D., Kreiswirth B.N., Whittam T.S., Musser J.M. Restricted structural gene polymorphism in the Mycobacterium tuberculosis complex indicates evolutionarily recent global dissemination. Proc. Natl Acad. Sci. USA. 1997;94:9869–9874. doi: 10.1073/pnas.94.18.9869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fabre M., Koeck J.-L., Le Fleche P., Simon F., Herve V., Vergnaud G., Pourcel C. High genetic diversity revealed by variable-number tandem repeat genotyping and analysis of hsp65 gene polymorphism in a large collection of “Mycobacterium canetti” strains indicates that the Mycobacterium tuberculosis complex is a recently emerged clone of “M. canetti”. J. Clin. Microbiol. 2004;42:3248–3255. doi: 10.1128/JCM.42.7.3248-3255.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lawrence J.G., Ochman H. Amelioration of bacterial genomes: rates of change and exchange. J. Mol. Evol. 1997;44:383–397. doi: 10.1007/pl00006158. [DOI] [PubMed] [Google Scholar]
- 59.Dykhuizen D.E., Polin D.S., Dunn J.J., Wilske B., Preac-Mursic V., Dattwyler R.J., Luft B.J. Borrelia burgdorferi is clonal: implications for taxonomy and vaccine development. Proc. Natl Acad. Sci. USA. 1993;90:10163–10167. doi: 10.1073/pnas.90.21.10163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Feil E.J., Holmes E.C., Bessen D.E., Chan M.-S., Day N.J.P., Enright M.C., Goldstein R., Hood D.W., Kalia A., Moore C.E., Zhou J., Spratt B.G. Recombination within natural populations of pathogenic bacteria: short-term empirical estimates and long-term phylogenetic consequences. Proc. Natl Acad. Sci. USA. 2001;98:182–187. doi: 10.1073/pnas.98.1.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Suerbaum S., Maynard Smith J., Bapumia K., Morelli G., Smith N.H., Kunstmann E., Dyrek I., Achtman M. Free recombination within Helicobacter pylori. Proc. Natl Acad. Sci. USA. 1998;95:12619–12624. doi: 10.1073/pnas.95.21.12619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dos Reis M., Savva R., Wernisch L. Solving the riddle of codon usage preferences: a test for translational selection. Nucleic Acids Res. 2004;32:5036–5044. doi: 10.1093/nar/gkh834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wright F. The ‘effective number of codons’ used in a gene. Gene. 1990;87:23–29. doi: 10.1016/0378-1119(90)90491-9. [DOI] [PubMed] [Google Scholar]
- 64.Sharp P.M., Li W.-H. The codon adaptation index—a measure of directional synonymous codon usage bias, and its potential applications. Nucleic Acids Res. 1987;15:1281–1295. doi: 10.1093/nar/15.3.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Moszer I., Rocha E.P.C., Danchin A. Codon usage and lateral gene transfer in Bacillus subtilis. Curr. Opin. Microbiol. 1999;2:524–528. doi: 10.1016/s1369-5274(99)00011-9. [DOI] [PubMed] [Google Scholar]
- 66.Bulmer M. Co-evolution of codon usage and transfer RNA abundance. Nature. 1987;325:728–730. doi: 10.1038/325728a0. [DOI] [PubMed] [Google Scholar]
- 67.Shields D.C. Switches in species-specific codon preferences: the influence of mutation biases. J. Mol. Evol. 1990;31:71–80. doi: 10.1007/BF02109476. [DOI] [PubMed] [Google Scholar]
- 68.Gu X., Hewett-Emmett D., Li W.-H. Directional mutational pressure affects the amino acid composition of proteins in bacteria. Genetica. 1998;102/103:383–391. [PubMed] [Google Scholar]