Abstract
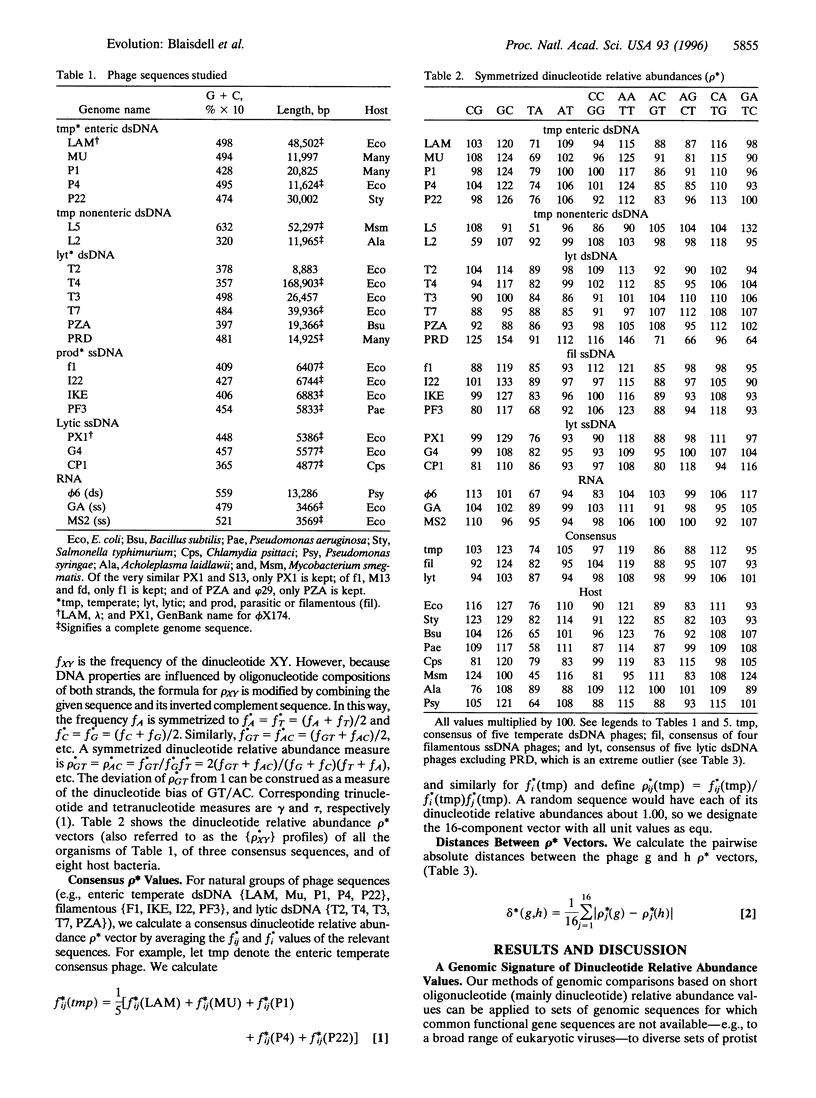
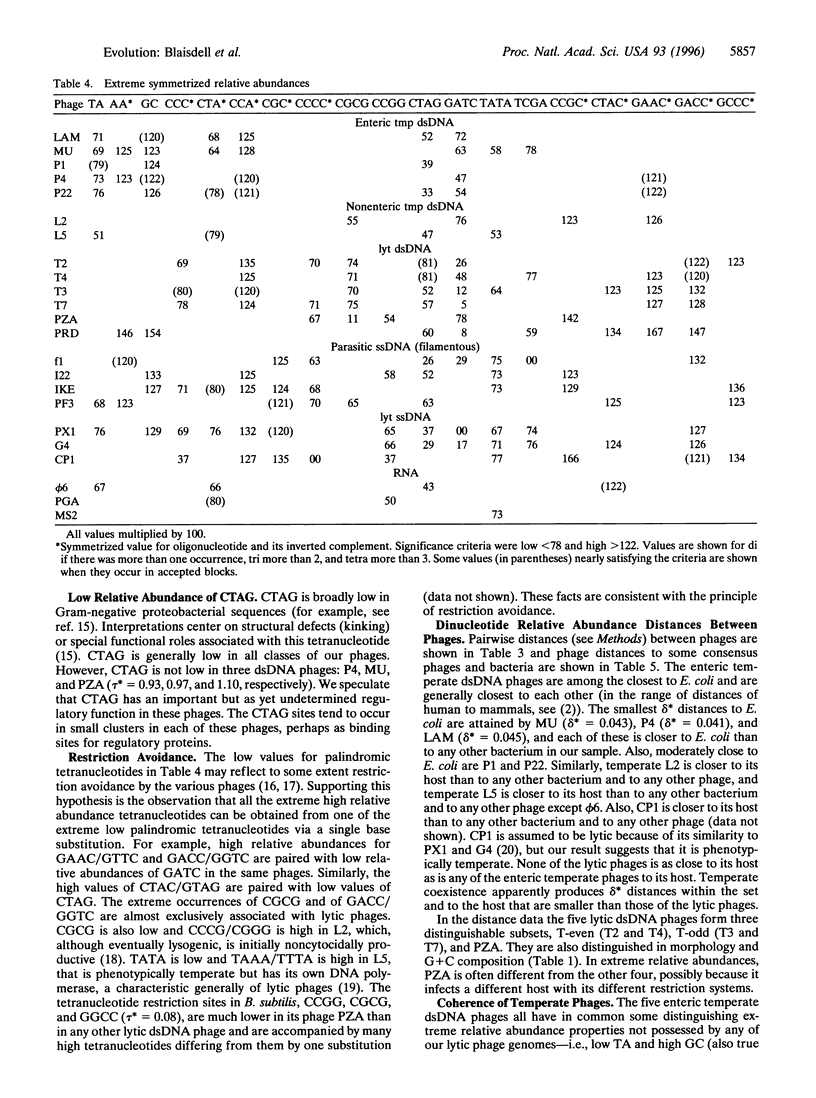
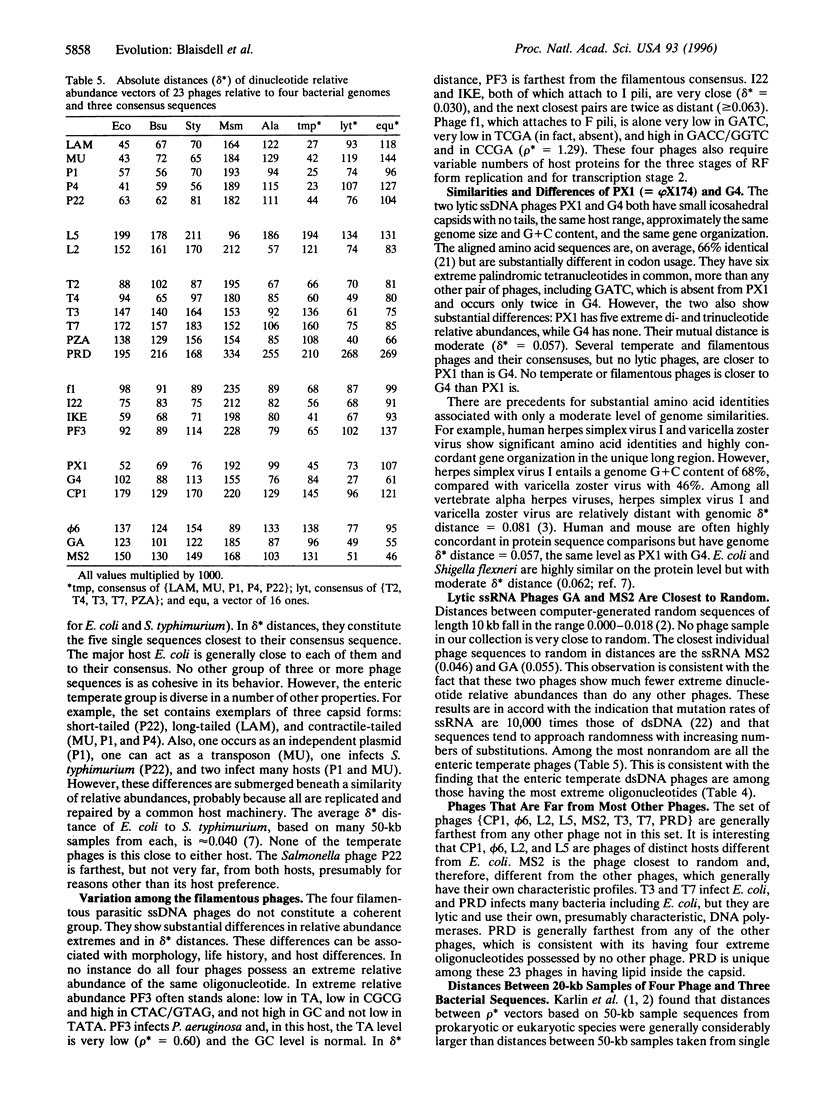
Genomic similarities and contrasts are investigated in a collection of 23 bacteriophages, including phages with temperate, lytic, and parasitic life histories, with varied sequence organizations and with different hosts and with different morphologies. Comparisons use relative abundances of di-, tri-, and tetranucleotides from entire genomes. We highlight several specific findings. (i) As previously shown for cellular genomes, each viral genome has a distinctive signature of short oligonucleotide abundances that pervade the entire genome and distinguish it from other genomes. (ii) The enteric temperate double-stranded (ds) phages, like enterobacteria, exhibit significantly high relative abundances of GpC = GC and significantly low values of TA, but no such extremes exist in ds lytic phages. (iii) The tetranucleotide CTAG is of statistically low relative abundance in most phages. (iv) The DAM methylase site GATC is of statistically low relative abundance in most phages, but not in P1. This difference may relate to controls on replication (e.g., actions of the host SeqA gene product) and to MutH cleavage potential of the Escherichia coli DAM mismatch repair system. (v) The enteric temperate dsDNA phages form a coherent group: they are relatively close to each other and to their bacteria] hosts in average differences of dinucleotide relative abundance values. By contrast, the lytic dsDNA phages do not form a coherent group. This difference may come about because the temperate phages acquire more sequence characteristics of the host because they use the host replication and repair machinery, whereas the analyzed lytic phages are replicated by their own machinery. (vi) The nonenteric temperate phages with mycoplasmal and mycobacterial hosts are relatively close to their respective hosts and relatively distant from any of the enteric hosts and from the other phages. (vii) The single-stranded RNA phages have dinucleotide relative abundance values closest to those for random sequences, presumably attributable to the mutation rates of RNA phages being much greater than those of DNA phages.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Burge C., Campbell A. M., Karlin S. Over- and under-representation of short oligonucleotides in DNA sequences. Proc Natl Acad Sci U S A. 1992 Feb 15;89(4):1358–1362. doi: 10.1073/pnas.89.4.1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell A. Comparative molecular biology of lambdoid phages. Annu Rev Microbiol. 1994;48:193–222. doi: 10.1146/annurev.mi.48.100194.001205. [DOI] [PubMed] [Google Scholar]
- Deschavanne P., Radman M. Counterselection of GATC sequences in enterobacteriophages by the components of the methyl-directed mismatch repair system. J Mol Evol. 1991 Aug;33(2):125–132. doi: 10.1007/BF02193626. [DOI] [PubMed] [Google Scholar]
- Haggård-Ljungquist E., Halling C., Calendar R. DNA sequences of the tail fiber genes of bacteriophage P2: evidence for horizontal transfer of tail fiber genes among unrelated bacteriophages. J Bacteriol. 1992 Mar;174(5):1462–1477. doi: 10.1128/jb.174.5.1462-1477.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatfull G. F., Sarkis G. J. DNA sequence, structure and gene expression of mycobacteriophage L5: a phage system for mycobacterial genetics. Mol Microbiol. 1993 Feb;7(3):395–405. doi: 10.1111/j.1365-2958.1993.tb01131.x. [DOI] [PubMed] [Google Scholar]
- JOSSE J., KAISER A. D., KORNBERG A. Enzymatic synthesis of deoxyribonucleic acid. VIII. Frequencies of nearest neighbor base sequences in deoxyribonucleic acid. J Biol Chem. 1961 Mar;236:864–875. [PubMed] [Google Scholar]
- Karlin S., Burge C. Dinucleotide relative abundance extremes: a genomic signature. Trends Genet. 1995 Jul;11(7):283–290. doi: 10.1016/s0168-9525(00)89076-9. [DOI] [PubMed] [Google Scholar]
- Karlin S., Cardon L. R. Computational DNA sequence analysis. Annu Rev Microbiol. 1994;48:619–654. doi: 10.1146/annurev.mi.48.100194.003155. [DOI] [PubMed] [Google Scholar]
- Karlin S., Ladunga I. Comparisons of eukaryotic genomic sequences. Proc Natl Acad Sci U S A. 1994 Dec 20;91(26):12832–12836. doi: 10.1073/pnas.91.26.12832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlin S., Mocarski E. S., Schachtel G. A. Molecular evolution of herpesviruses: genomic and protein sequence comparisons. J Virol. 1994 Mar;68(3):1886–1902. doi: 10.1128/jvi.68.3.1886-1902.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler C., Manta V. Specificity of restriction endonucleases and DNA modification methyltransferases a review (Edition 3). Gene. 1990 Aug 16;92(1-2):1–248. doi: 10.1016/0378-1119(90)90486-b. [DOI] [PubMed] [Google Scholar]
- Maniloff J., Kampo G. J., Dascher C. C. Sequence analysis of a unique temperature phage: mycoplasma virus L2. Gene. 1994 Apr 8;141(1):1–8. doi: 10.1016/0378-1119(94)90120-1. [DOI] [PubMed] [Google Scholar]
- Russell G. J., Walker P. M., Elton R. A., Subak-Sharpe J. H. Doublet frequency analysis of fractionated vertebrate nuclear DNA. J Mol Biol. 1976 Nov;108(1):1–23. doi: 10.1016/s0022-2836(76)80090-3. [DOI] [PubMed] [Google Scholar]
- Sharp P. M. Molecular evolution of bacteriophages: evidence of selection against the recognition sites of host restriction enzymes. Mol Biol Evol. 1986 Jan;3(1):75–83. doi: 10.1093/oxfordjournals.molbev.a040377. [DOI] [PubMed] [Google Scholar]
- Storey C. C., Lusher M., Richmond S. J. Analysis of the complete nucleotide sequence of Chp1, a phage which infects avian Chlamydia psittaci. J Gen Virol. 1989 Dec;70(Pt 12):3381–3390. doi: 10.1099/0022-1317-70-12-3381. [DOI] [PubMed] [Google Scholar]


