Abstract
We analyzed near-complete population (composite) genomic sequences for coexisting acidophilic iron-oxidizing Leptospirillum group II and III bacteria (phylum Nitrospirae) and an extrachromosomal plasmid from a Richmond Mine, Iron Mountain, CA, acid mine drainage biofilm. Community proteomic analysis of the genomically characterized sample and two other biofilms identified 64.6% and 44.9% of the predicted proteins of Leptospirillum groups II and III, respectively, and 20% of the predicted plasmid proteins. The bacteria share 92% 16S rRNA gene sequence identity and >60% of their genes, including integrated plasmid-like regions. The extrachromosomal plasmid carries conjugation genes with detectable sequence similarity to genes in the integrated conjugative plasmid, but only those on the extrachromosomal element were identified by proteomics. Both bacterial groups have genes for community-essential functions, including carbon fixation and biosynthesis of vitamins, fatty acids, and biopolymers (including cellulose); proteomic analyses reveal these activities. Both Leptospirillum types have multiple pathways for osmotic protection. Although both are motile, signal transduction and methyl-accepting chemotaxis proteins are more abundant in Leptospirillum group III, consistent with its distribution in gradients within biofilms. Interestingly, Leptospirillum group II uses a methyl-dependent and Leptospirillum group III a methyl-independent response pathway. Although only Leptospirillum group III can fix nitrogen, these proteins were not identified by proteomics. The abundances of core proteins are similar in all communities, but the abundance levels of unique and shared proteins of unknown function vary. Some proteins unique to one organism were highly expressed and may be key to the functional and ecological differentiation of Leptospirillum groups II and III.
To understand how microorganisms contribute to biogeochemical cycling, it is necessary to determine the roles of uncultivated as well as cultivated groups and to establish how these roles vary during ecological succession and when environmental conditions change. Shotgun genomic sequencing (metagenomics) has opened new opportunities for culture-independent studies of microbial communities. Examples include investigations of acid mine drainage (AMD) biofilm communities (4, 43, 75), symbiosis in a marine worm involving sulfur-oxidizing and sulfate-reducing bacteria (85), and enhanced biological phosphorous removal by sludge communities (32). From these genomic data sets, it has been possible to reconstruct aspects of the metabolism of individual organisms (32) and coexisting community members (29, 75) and to identify which organisms contribute community-essential functions (75). An interesting question relates to how differences in metabolic potential between organisms from the same lineage allow them to occupy distinct niches. Identification of potentially adaptive traits in closely related organisms is also important from an evolutionary perspective.
Genomic data do not reveal how organisms alter their metabolisms in response to the presence of other organisms or environmental conditions. Proteomics methods for analysis of metabolic responses of isolates (16, 17, 42, 80, 81) have been extended to analyze the functioning of the dominant members of natural consortia (56, 69), with strain-level resolution (43, 82). In these studies, peptides are separated by liquid chromatography (LC) and identified by tandem mass spectrometry (MS-MS) through reference to appropriate genomic databases. Proteomic analysis is possible even if the genome sequences are not identical to those of the organisms present (24); however, missing sequence information reduces the resolution of such proteogenomic studies.
Due to dominance by a few organism types, chemoautotrophic microbial AMD biofilms from Richmond Mine, Iron Mountain, CA, are tractable model systems used to develop cultivation-independent metagenomic and proteogenomic methods for analysis of community structure, function, and ecology (13). Acidophilic Leptospirillum bacteria dominate this AMD system (15), other AMD systems (54), and bioleaching systems used for recovery of metals (19, 53, 86). These bacteria play pivotal roles in sulfide mineral dissolution because they are iron oxidizers (53, 75), and ferric iron drives sulfide oxidation, leading to formation of metal-rich sulfuric acid solutions. According to a recent microscopy-based study (83), Leptospirillum group II are the first colonists in AMD biofilm communities whereas Leptospirillum group III generally appear later, sometimes partitioned within biofilm interiors. Because only Leptospirillum group III appear to be able to fix nitrogen, they may be keystone species in AMD ecosystems (75). This observation enabled the isolation of one representative, “Leptospirillum ferrodiazotrophum” (76). In prior work, we reported near-complete genome sequences of two Leptospirillum group II types (43, 65), but detailed functional annotations and metabolic analyses have not been published. Genomic data have been used to explore the metabolism of Leptospirillum bacteria in one biofilm community (56), but proteomic and genomic analyses of the same biofilm community have not been performed.
Here, we report a near-complete genomic sequence for Leptospirillum group III, derived from a biofilm obtained from the UBA site within the Richmond Mine, Iron Mountain, CA; a detailed functional annotation of the genomes of Leptospirillum groups II and III; and a genomic and proteomic comparison of them. In addition, we report the sequence of an extrachromosomal plasmid associated with these organisms. This study represents the first comprehensive genomics-based analysis of the metabolism of bacteria in the Nitrospirae phylum and the first environmental community proteogenomic study where the genomic and proteomic data were derived from the same sample. We compared the proteomic profiles of three different biofilm communities to evaluate the importance of shared and unique genes and pathways in environmental adaptation.
MATERIALS AND METHODS
Samples.
Biofilm samples were collected underground within Richmond Mine, Iron Mountain, CA (Fig. 1). The UBA biofilm was collected from the surface of a slowly draining ∼0.5-cm-deep pool in a stream with a pH of 1.1 and a temperature of 41°C in the A drift in June 2005 (see Fig. S1 in the supplemental material). The thin (a few tens of micrometers thick, as estimated by microscopy on similar biofilms) (see Fig. S1 in the supplemental material) floating ABend biofilm was collected from the surface of a deeper pool in the AB drift in January 2004. Geochemical data and other information were reported by Ram et al. (56). Briefly, the pH of the solution was 1.07 and the temperature 43°C. The ABfront biofilm was also collected in the AB drift, about 2 m from the ABend location, in June 2004 (Fig. 1). The ABfront sample is inferred to be a much more mature biofilm than the ABend sample on the basis of its thickness (∼200 μm) (see Fig. S1 in the supplemental material). At the time of sampling, the pH at the ABfront location was 0.99 and the temperature 39°C.
FIG. 1.
Map showing sampling locations.
Assembly.
Total DNA recovered from the UBA biofilm was cloned and sequenced (∼3-kb library), as reported previously (43). Briefly, 100 Mb of sequence was obtained from the UBA site (Fig. 1), sequences were assembled (Phred/Phrap), and contigs were manually curated to correct misassemblies and remove errors such as coassembly of fragments from different organism types (identified based on misplacement of mate-paired sequences). Misassembles due to repetitive sequences were either resolved based on surrounding unique sequences by using mate pair information or allowed to terminate scaffolds. Contig fragmentation due to multiple genome paths for different individuals within a single population (strain variation) was identified so that larger scaffolds could be established. Contig editing was done using Consed (33).
The very near-complete, deeply sampled (∼25×) genome of Leptospirillum group II recovered from the UBA genomic data set (Leptospirillum group II UBA) is in seven composite scaffolds (43). The population is genomically distinct from a Leptospirillum ferriphilum-like strain (Leptospirillum group II 5-way CG), previously derived from the 5-way CG site in the Richmond Mine (Fig. 1) (65, 75). The two genomically characterized types (UBA and 5-way CG) share 99.7% 16S rRNA gene sequence identity and ∼94% DNA sequence similarity for orthologs (38).
After manual curation of the assembly, contigs of Leptospirillum group III from the UBA genomic data set were separated from archaea on the basis of GC content and from low-abundance bacteria (average depth, ∼2×) on the basis of sequence depth (average depth, ∼10×). Binning was verified using tetranucleotide sequence signatures (1) analyzed using emergent, self-organizing maps (G. J. Dick, A. F. Andersson, B. J. Baker, S. L. Simmons, B. C. Thomas, A. P. Yelton, and J. F. Banfield, unpublished data).
We reconstructed what we believe to be a near-complete composite sequence (∼250 kb) for a large extrachromosomal plasmid. Fragments were clustered using emergent, self-organizing maps, with a strong distance structure that definitively separated the sequences from any others in the community (Dick et al., unpublished). The numerous small contigs were manually curated into 10 scaffolds.
Annotation.
Gene predictions for Leptospirillum groups II and III were made using a combination of FgenesB (Softberry, Inc.), CRITICA (11), and Glimmer (21). Automated function predictions were generated by searching for all predicted peptides in the TIGRFAMs (with trusted HMMPfam cutoffs), PRIAM (with the rpsblast 1e−30 cutoff), PFAM (with trusted HMMPfam cutoffs), InterPro (with default interproscan cutoffs), and COGs (with the rpsblast 1e−10 cutoff) databases. Proteins that failed to return a definitive result with the aforementioned profile searches were annotated on the basis of BLASTP searches in the KEGG and SwissProt-TREMBL (1e_5 cutoff) peptide databases. The tRNAScanSE tool (44) was used to find tRNA genes, while ribosomal RNAs were found using BLASTn searches in the 16S and 23S rRNA databases. Other “standard” structural RNAs (e.g., 5S rRNA, rnpB, tmRNA, and SRP RNA) were found using covariance models with the Infernal search tool (25).
Manual curation of gene annotation.
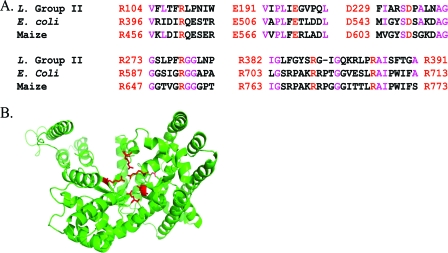
The automatic gene annotations were manually curated. Product descriptions were assigned when scores for matches to protein families in the PRIAM database were e−30 or less. Functions were also inferred based on the TIGRFAM or PFAM assignments as long as the protein had an ortholog in the public databases with >70% identity over >70% of the length of the protein alignment. “Putative” was added to product descriptions for proteins with PFAM assignments and an ortholog in the public databases with between 30% and 70% identity and alignments involved over 70% of the protein length. “Probable” was added to product descriptions for predicted proteins with >30% identity to proteins in the SwissProt database. For these cases, BLAST (3) matches in the NCBI nonredundant sequence database (http://blast.ncbi.nlm.nih.gov/Blast.cgi) were also considered. The term “conserved protein of unknown function” was used when the predicted protein was a conserved hypothetical protein identified by proteomics in the current study or validated in previous studies. Similarly, “protein of unknown function” was used when the predicted protein was a hypothetical protein identified by proteomics in the current study or in prior studies of these AMD biofilm communities. “Conserved hypothetical protein” was used when the predicted protein had an alignment of >70% with one or more hypothetical proteins and >30% identity with these. The term “hypothetical protein” was used when there was no good alignment with predicted proteins in the NCBI nonredundant sequence database. In the specific case of a possible phosphoenolpyruvate (PEP) carboxylase, the protein structure of Leptospirillum group II was modeled after the Maize (Protein Data Bank entry 1jqo_A) and Escherichia coli (Protein Data Bank entry 1jqn_A) crystal structures (87) (K. E. Wheeler, A. Zemla, D. S. Aliaga Goltsman, Y. Jiao, J. F. Banfield, and M. P. Thelen, unpublished data).
Both Leptospirillum group II and group III genomes are composites in that they generally report a single genome path, although multiple paths exist in some regions. Due to the strain variation and gaps present in both genomes, a subset of sequencing reads, potentially carrying important genes, were not brought into the composite sequences. Consequently, analysis of gene content included consideration of the read databases as well as composite sequences and strain variant paths.
Proteomics.
Proteomic data were obtained from the same UBA biofilm samples used for genomic library construction as well as two other samples, the ABfront biofilm and the ABend biofilm. Complete descriptions of the ABend biofilm preparation and analyses were published previously (43, 56). The UBA and ABfront proteomes were prepared and analyzed via comparable methods. Briefly, proteins in the biofilms were released via sonication and fractionated based on cellular location (membrane, soluble, whole cell, and extracellular). Proteome fractions (∼3 mg total protein per fraction) were denatured, reduced, digested using sequencing grade trypsin (Promega, Madison, WI), desalted, concentrated, and frozen until analyses.
Two-dimensional nano LC-electrospray ionization-MS-MS analyses of all samples were performed with a linear ion trap mass spectrometer (LTQ; Thermo Fisher, San Jose, CA) as previously described (16, 56). Four different fractions were analyzed using the same methodology on the same LC-MS system, with three technical replicates for each sample. The samples were loaded (∼500 μg starting material) onto a split-phase column (packed in-house with a C18 reverse-phase column and strong cation exchange chromatographic resin) (48) placed behind a 15-cm C18 analytical column (packed in-house). Both were situated in front of a Proxeon nanospray source (Odense, Denmark) on the LTQ device. Flow was provided via an Ultimate high-performance-LC pump (LC Packings, a division of Dionex, San Francisco, CA), with an initial flow rate of ∼100 μl/min that was split precolumn to obtain a flow rate of ∼300 nL/min at the nanospray tip; a voltage of 3.8kV was applied on the waste line. Chromatographic separation of the tryptic peptides was conducted over a 22-h period of increasing (0 to 500 mM) pulses of ammonium acetate salt, followed by a 2-h aqueous-to-organic-solvent gradient. The LTQ device was operated in a data-dependent manner with two microscan full scans (400 to 1,700 m/z) and two microscan MS-MS scans (top five most abundant), and dynamic exclusion was set at 1 (16, 56).
MS-MS spectra from all individual 24-h two-dimensional LC-MS-MS runs were searched using the SEQUEST algorithm (28) in a global database created from proteins predicted from AMD genomic sequences. The database was concatenated with a list of common contaminants (trypsin and keratin, etc.). All searches were run with the following settings: enzyme type, trypsin; parent mass tolerance, 3.0; fragment ion tolerance, 0.5; up to 4 missed cleavages allowed; and fully tryptic peptides only (no posttranslational modifications were considered for this study). The output data files from all searches were filtered and sorted with the DTASelect algorithm (73), using the following parameters: fully tryptic peptides only; delCN values of at least 0.08; and cross-correlation scores of at least 1.8 (+1), 2.5 (+2), and 3.5 (+3). Identification of at least two peptides within the same 24-h run was required in order for a protein to be deemed identified. From the DTASelect output files, the total numbers of proteins, peptides, and spectra and percent sequence coverage for each protein as well as the numbers of unique peptides per protein were extracted. The cross-correlation values used in the current study have been rigorously tested and typically give maximum false-positivity rates of 1 to 2% for both bacterial isolates (16) and microbial communities (43, 56, 82). All databases, peptide and protein results, MS-MS spectra, and supplementary tables for all database searches have been archived and made available as open access via the link http://compbio.ornl.gov/comparative_genomics_proteomics_of_leptospirillum/.
Proteomic analyses.
Given that our goal was to compare Leptospirillum group II to group III at a functional level, we combined the spectral counts (number of unique counts for each type + number of nonunique counts shared by the two types) for the two Leptospirillum group II genomic types (UBA and 5-way CG [43, 65]). Similarly, nonunique and unique spectral counts were combined for Leptospirillum group III proteins. Virtually no cross-identification is expected for proteins sharing <85% sequence identity (24) (Leptospirillum groups II and III share ∼55% average sequence identity). Although analysis of similar amounts of protein for each fraction (∼3 mg) could potentially lead to an overrepresentation of proteins in the less abundant extracellular fraction, relative comparisons between samples should not be affected.
For comparison of protein abundance levels for Leptospirillum groups II and III and the extrachromosomal plasmid, proteomics data were normalized using the normalized spectral abundance factor (NSAF) method (30, 89). This method estimates protein abundance by first dividing the spectral count for each protein by the protein length and then dividing this number by the sum of all length-normalized spectral counts for each organism and multiplying by 100. For each sample, we summed the spectral counts from each fraction and combined the spectral counts of each protein over the three technical replicates prior to calculation of the NSAF value. The NSAF value for a Leptospirilllum group II protein thus estimates the percentage of the total Leptospirillum group II protein pool that each protein represents.
Circular comparative genomic and proteomic representations were made using the Circos version 0.46 software program (http://mkweb.bcgsc.ca/circos/). Heat maps of the protein abundance levels were based on log2-transformed NSAF values.
The NSAF values were clustered using Cluster version 3.0 (26) (http://bonsai.ims.u-tokyo.ac.jp/∼mdehoon/software/cluster/software.htm#ctv) and visualized as heat maps with TreeView software (61). For the clustering analyses, we considered only proteins identified in more than half of the six data sets (two organisms in three environmental samples). To check the robustness of the results, we also performed the analysis by (i) considering only proteins identified in five or more of the six data sets and (ii) considering proteins identified in one or more data sets. For a single organism in a single sample, the median protein NSAF value was determined, and proteins with higher or lower values were classified as over- or underrepresented in the proteome (indicated by shades of red and green, respectively). This is referred to as median centering by organism. In some cases, simultaneously with median centering by organism, the abundances of all proteins were compared among the six organism data sets, the median value for each protein was determined, and the protein levels were assigned values indicating under- or overrepresentation. This is referred to as median centering by protein and organism data set. For both forms of median centering, the six organism data sets were clustered based on the resulting patterns by using average linkage and Kendall's Tau distance matrix.
Nucleotide sequence accession numbers.
The Whole Genome Shotgun project containing the assembly and annotation of Leptospirillum sp. group III has been deposited in DDBJ/EMBL/GenBank under accession number ACNP00000000. The version described in this paper is the first version, ACNP01000000. The Whole Genome Shotgun project containing the assembly and annotation of Leptospirillum sp. group II was previously deposited, and the manually curated annotation is available under accession number AAWO00000000.
RESULTS
Genomics statistics and overall proteomic results.
We reconstructed a near-complete composite genome for a Leptospirillum group III population closely related (99.8% 16S rRNA gene sequence identity) to the Leptospirillum ferrodiazotrophum strain previously isolated from the same AMD system (76) (Fig. 2). The Leptospirillum group III genomic data set comprises 39 scaffolds (average depth, ∼10×). An isolate with 100% 16S rRNA sequence identity has been recovered and is available for further characterization (strain UBA:5) (Fig. 2).
FIG. 2.
Phylogenetic tree based on 16S rRNA genes of Leptospirillum spp. (maximum-likelihood method). Statistically supported bootstrap values are labeled at the nodes. The scale bar represents 0.10 changes per site, or 10%. Filled squares indicate isolates, while filled circles indicate composite genomes.
The genome annotations of Leptospirillum group II and Leptospirillum group III are reported in Tables S1 and S2 in the supplemental material, respectively, and the annotation files have been deposited in GenBank. Table S2 in the supplemental material also reports three strain variant contigs within the Leptospirillum group III population. While these organisms share only 92% 16S rRNA gene sequence identity (Fig. 2), more than 60% of the genes in Leptospirillum groups II and III are orthologs (55% average amino acid identity) and ∼78% of the proteins in each organism identified by proteomics are orthologs (Table 1). Table 1 also summarizes other basic statistics.
TABLE 1.
Basic statistics of the Leptospirillum group II and III genomes
| Category | No. (%) of genes in Leptospirillum group
|
||||
|---|---|---|---|---|---|
| Group II
|
Group III
|
||||
| Total | Detected in all samples | Total | Detected in all samples | Predicted level (%)a | |
| Predicted genes | 2,625 | 1,696 (65) | 2,659 | 1,201 (45) | 64 |
| Proteins detected in UBA sample only | 1,384 (53) | 998 (37) | 53 | ||
| Genes with annotation | 1,717 (65) | 1,258 (48) | 1,716 (65) | 934 (35) | 50 |
| Orthologs | 1,257 | 1,037 (40) | 1,257 | 789 (30) | 42 |
| Unique to each species | 460 | 221 (8) | 459 | 145 (5) | 8 |
| Genes without annotation | 908 | 438 (17) | 943 | 267 (10) | 14 |
| Orthologs | 445 | 280 (11) | 444 | 157 (6) | 8 |
| Unique to each species | 463 | 158 (6) | 499 | 110 (4) | 6 |
| Hypothetical and conserved hypothetical proteins | 455 | 538 | |||
| Subset unique to Leptospirillum groups II and III | 149 | 149 | |||
| Hypothetical proteins unique to each species | 306 | 389 | |||
Values corrected to show abundance levels in samples in which Leptospirillum groups II and III are present at the same concentration (observed values corrected on the basis of abundance ratios of Leptospirillum groups II and III in the biofilm samples).
Representations of the Leptospirillum group II and III genomes illustrating synteny between orthologs are shown in Fig. 3 and Fig. S2 in the supplemental material. Proteomic data from ABend and ABfront biofilms were included alongside results for the UBA sample to provide insight into site-to-site variation. Syntenous regions primarily encode core metabolic functions (Fig. 3; see also Tables S1 and S2 in the supplemental material) and tend to have similar protein abundance patterns across the three biofilms (compare the heat map rings within each figure; see also Fig. S3 in the supplemental material), and between organisms (Fig. 3; see also Fig. S2 in the supplemental material). Regions of consistently high protein abundance correspond to ribosomal proteins, RNA polymerase, and proteins involved in energy metabolism. The genomic regions where the gene content shared between Leptospirillum groups II and III is lower than average correspond primarily to an integrated plasmid.
FIG. 3.
Diagram of the genome of Leptospirillum group II (outer circle) and orthologs in Leptospirillum group III (inner circle, color coded by scaffold). A histogram of percent identity between orthologs is shown by black bars on the inner ring. Heat maps of protein identification values (NSAF) are given by six middle rings (gray, no identification).
Clustering of NSAF values, median centered by organism data set (see Fig. S3 in the supplemental material) and by organism data set and protein (Fig. 4), yielded two clusters, one containing all three data sets for Leptospirillum group II and the other containing all three data sets for Leptospirillum group III. The same results were obtained independent of the filtering level and when clustering was done with four organism data sets (two organisms each in two of the three available environmental samples; data not shown). Many protein subclusters are apparent in Fig. 4. Although ribosomal proteins are highly abundant in all data sets (Fig. 3; see also Fig. S2 and S3 in the supplemental material), they are generally less abundant in Leptospirillum group III than in group II (e.g., clusters A and B in Fig. 4). Vitamin and cofactor biosyntheses (within cluster A) are generally more abundant in Leptospirillum group II than in group III, and transport and secretion proteins (in cluster B) are overrepresented in all three biofilm samples in Leptospirillum group II, whereas proteins involved in chemotaxis and energy generation (in clusters A and C) are overrepresented in Leptospirillum group III.
FIG. 4.
Protein abundance values (NSAF) for Leptospirillum group II and III orthologs in UBA, ABfront (AB-F), and ABend (AB-E) samples. Red, overrepresented; green, underrepresented; black, median; gray, no identification. The functional categories for the most abundant proteins are in three clusters. Cluster A (only part shown) represents transcription, translation, ribosomal structure, and biogenesis (7 proteins); coenzyme transport and metabolism (6 proteins); transport and secretion (5 proteins); energy production and conversion (3 proteins); and other functions (22 proteins). Cluster B represents transport and secretion (10 proteins); translation, ribosomal structure, and biogenesis (5 proteins); posttranslational modification, protein turnover, and chaperones (4 proteins); lipid transport and metabolism (2 proteins); and other functions (25 proteins). Cluster C represents energy production and conversion (10 proteins); cell motility (10 proteins); amino acid transport and metabolism (6 proteins); transcription, translation, ribosomal structure, and biogenesis (6 proteins); and other functions (24 proteins).
Proteins of unknown function in Leptospirillum group II are generally located in genomic regions with consistently high protein abundance, whereas hypothetical proteins not identified by proteomics tend to occur in genomic regions where few proteins are identified. Several proteins of unknown function that are unique to Leptospirillum group II or Leptospirillum group III are highly abundant, whereas others show notable intersample protein abundance variation (Fig. 5).
FIG. 5.
Inferred abundances (NSAF) of proteins of unknown function in Leptospirillum groups II and III. (A) Orthologs; (B) proteins unique to Leptospirillum group II; (C) proteins unique to Leptospirillum group III. Red, overrepresented; green, underrepresented; black, median; gray, no identification.
There are fewer conserved proteins of unknown function than proteins of unknown function in Leptospirillum groups II and III, and these appear to be spread across the genomes. Many conserved proteins of unknown function were found in operons with one or more genes with functional predictions, and may have related roles. Examples occur in operons with genes for the proteasome, flagella, transport and secretion, plasmid functions, folate metabolism, and t-RNA synthetases (see Tables S1 and S2 in the supplemental material).
Comparative genomics. (i) Energy metabolism.
(a) Electron transport chain. The conserved motif typical of c-type cytochromes (CXXCH) was found in 34 predicted Leptospirillum group II proteins. After in-depth analysis, 13 were annotated as c-type cytochromes (Table 2). Seven of the putative Leptospirillum group II cytochromes have an ortholog in Leptospirillum group III on the basis of reciprocal best hit. An independent survey of the Leptospirillum group III genome did not uncover any predicted c-type cytochromes without a homolog in the Leptospirillum group II genome. Two Leptospirillum group II c-type cytochromes, cytochrome 579 (Cyt579) and cytochrome 572, isolated directly from Richmond Mine biofilms, were biochemically characterized recently (36, 67). Although the composite Leptospirillum group III genome fragments lack an ortholog of Cyt579, sequence reads not brought into the assembly indicate that Leptospirillum group III has a gene for Cyt579.
TABLE 2.
c-type cytochromes in Leptospirillum groups II and III
| Leptospirillum group II protein | Predicted no. of heme binding sites | Annotation | Group II proteomics
|
Leptospirillum group III protein (% amino acid identity) | Group III proteomics
|
||||
|---|---|---|---|---|---|---|---|---|---|
| UBA NSAF | ABfront NSAF | ABend NSAF | UBA NSAF | ABfront NSAF | ABend NSAF | ||||
| 8062_147 | 1a | Cyt579 | 2.149420 | 0.962943 | 1.881661 | Noneb | |||
| 8062_372 | 1a | Cyt579 | 3.530970 | 1.898338 | 2.700427 | Noneb | |||
| 8241_149 | 1a | Cyt572 | 0.079349 | 0.075519 | 0.084001 | 9627_29 (58) | 0.280592 | 0.290043 | 0.499131 |
| 8524_197 | 1 | Putative cytochrome c, class I | 0.071612 | 0.079448 | 0.060592 | 9545_121 (45) | |||
| 8524_235 | 1 | Putative cytochrome c, class I | 0.058754 | 0.093354 | 0.078289 | 7442_12 (64) | 0.235543 | 0.204500 | 0.021282 |
| 7931_87 | 1 | Putative cytochrome c, class I | 0.162992 | 0.106659 | 0.199494 | 9545_121 (38)c | |||
| 7931_111 | 2 | Putative cytochrome | 0.052524 | 0.022980 | 0.056351 | 7442_67 (59) | 0.023914 | 0.031897 | 0.039541 |
| c, class I | 9453_106 (68)c | 0.021120 | 0.017754 | 0.045729 | |||||
| 7931_112 | 2 | Putative cytochrome | 0.054150 | 0.044047 | 0.042870 | 7442_66 (64) | 0.003507 | 0.024767 | |
| c, class I | 9453_105 (60)c | 0.003564 | |||||||
| 7931_121 | 2 | Probable cytochrome c, class I | 0.001323 | 9453_105 (58)c | 0.003564 | ||||
| 7931_122 | 2 | Probable cytochrome c, class I | 0.004426 | 0.002643 | 9453_106 (62)c | 0.021120 | 0.017754 | 0.045729 | |
| 7931_123 | 2 | Probable cytochrome | 0.003873 | 0.002056 | 9453_106 (49)c | 0.021120 | 0.017754 | 0.045729 | |
| c, class I | 7442_66 (43)c | 0.003507 | 0.024767 | ||||||
| 8524_98 | 4 | Probable cytochrome c 554 | 0.005953 | 0.003950 | 9595_5 (63) | ||||
| 8062_150 | 4 | Probable cytochrome c NapC/NirT | 7442_35 (64) | ||||||
Visible-light spectroscopy indicates that these proteins have modified c-type heme groups (67).
Unassembled reads contain a sequence with high degrees of similarity to C-terminal 77-amino-acid sequences for 8062_147 (79%) and 8062_372 (89%).
Not orthologs, based on reciprocal best hit.
Genes encoding a putative bc1 complex have been identified in Leptospirillum groups II and III (see Table S3 in the supplemental material). Cytochrome b/b6 protein is bifurcated, and the c1 component contains binding sites for four-heme prosthetic groups. Proteins with sequence similarity to two subunits of a cytochrome cbb3 oxidase were predicted to occur in Leptospirillum groups II and III, and both subunits are duplicated (see Table S3 in the supplemental material). Only Leptospirillum group II cytochrome cbb3 oxidase gene products were identified by proteomics. Subunits of a cytochrome bd oxidase were also predicted to occur in both Leptospirillum genomes (see Table S3 in the supplemental material); however, peptides for these proteins were not observed in any proteomic data sets.
A conserved cluster of 14 NADH dehydrogenase genes is present in Leptospirillum groups II and III (see Table S4 in the supplemental material), and all of the corresponding proteins were identified by proteomics. In addition, there are several extra copies of different subunits of NADH dehydrogenase, two copies of NADPH-quinone reductase, and other energy genes (cytochromes) scattered around the genomes, usually in plasmid/phage regions.
(b) CO2 fixation. Both Leptospirillum group II and III isolates from the Richmond Mine can be grown without sources of fixed carbon, and all previously characterized Leptospirillum species grow only chemoautotrophically (43, 60). Therefore, it is very likely that the Leptospirillum species in the biofilm utilize CO2 as their sole carbon source. The most common autotrophic pathway is the Calvin-Benson cycle; however, both Leptospirillum group II and group III lack ribulose-5-phosphate kinase, a key enzyme in the pathway. Leptospirillum groups II and III have two and three copies of a ribulose-bisphosphate carboxylase-like protein (RuBisCO-like), respectively, but none were predicted to have carboxylase and oxygenase activity, based on phylogenetic classification with the Form IV Rubisco-like group (10). Thus, neither Leptospirillum group II nor group III appears to use the Calvin-Benson cycle for CO2 fixation. The RuBisCO-like proteins were identified by proteomics and are likely involved in sulfur metabolism (10).
It is unlikely that Leptospirillum groups II and III fix carbon via the Wood-Ljungdahl pathway, which converts CO2 to acetyl-coenzyme A (CoA) through the bifunctional enzyme carbon monoxide dehydrogenase (CODH)/acetyl-CoA synthase. Although both groups have paralogous genes with sequence similarity to CODH, the predicted proteins lack the active site for CODH and acetyl-CoA synthase on the basis of multiple alignments. It is interesting, however, that one of the candidate CODH genes is in the second cluster of pyruvate-ferredoxin oxidoreductase (PFOR) instead of the epsilon subunit in Leptospirillum group II. All copies of annotated CODH proteins were identified at various levels by proteomics.
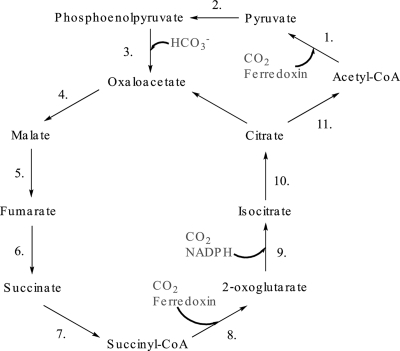
The most likely pathway for carbon fixation is via the reductive tricarboxylic acid (rTCA) cycle (Fig. 6). Several proteins involved in the rTCA cycle, especially PFOR and isocitrate dehydrogenase, are identified at high levels in proteomic data sets. A key component in the rTCA cycle, PEP carboxylase, could not be initially located in either species. However, on the basis of a TIGRFam domain and protein structure prediction, we annotated a protein of unknown function as PEP carboxylase (see Tables S1 [8241_GENE_507] and S2 [7952_GENE_51] in the supplemental material) (Fig. 7). ATP citrate lyase and 2-oxoglutarate-ferredoxin oxidoreductase were not annotated for Leptospirillum groups II and III. Recent work with Hydrogenobacter thermophilus TK-6, an aerobic hydrogen-oxidizing bacterium, has demonstrated a novel citrate-cleaving reaction catalyzed by two enzymes, citryl-CoA synthetase and citryl-CoA lyase (6, 7). The first shares similarity with succinyl-CoA synthetase, and both Leptospirillum species contain two different copies of this enzyme. H. thermophilus citryl-CoA lyase shares strong similarity with Leptospirillum citrate synthase. Both enzymes were identified by proteomics and may be involved in citrate cleavage. Additionally, both genomes have duplicated operons for PFOR, one of which may carboxylate 2-oxoglutarate instead of pyruvate (5). These results are in agreement with reverse transcriptase PCR results suggesting CO2 fixation via the rTCA pathway in a strain related to L. ferriphilum for which the complete genome sequence has not been deposited (41).
FIG. 6.
Proposed CO2 fixation pathway (rTCA) for Leptospirillum groups II and III. 1, PFOR; 2, PEP synthase; 3, PEP carboxylase (PEPC); 4, malate dehydrogenase; 5, fumarate hydratase; 6, fumarate reductase; 7, succinyl-CoA synthetase; 8. PFOR (second copy); 9, isocitrate dehydrogenase; 10, aconitate hydratase; 11, succinyl-CoA synthetase (second copy) and citrate synthase.
FIG. 7.
(A) Alignment of PEP carboxylase regions containing conserved active site residues (in red) from Maize and E. coli (47) and from Leptospirillum group II. Other, identical residues are shown in purple. (B) Predicted protein structure of PEP carboxylase in Leptospirillum group II, with active site residues shown in red.
The first part of the rTCA cycle is shared with the pathway for CO2 incorporation recently described to occur in the archaeon Igniococcus hospitalis (34, 35), where acetyl-CoA is carboxylated to pyruvate by PFOR. I. hospitalis has been shown to regenerate acetyl-CoA through a complex pathway, with 4-hydroxybutyryl-CoA as a central intermediate (34); however, Leptospirillum groups II and III lack the genes for this route. If this pathway operates in Leptospirillum, a novel pathway for regeneration of acetyl-CoA is required.
(c) TCA cycle. Leptospirillum group II has genes for most steps in the TCA cycle. The dihydrolipoamide dehydrogenase subunit of the oxoglutarate dehydrogenase complex was predicted and identified by proteomics (intriguingly, three copies are present), but the other two subunits of this complex were not found. The oxoglutarate dehydrogenase complex is similar to the pyruvate dehydrogenase complex, which Leptospirillum group II lacks (likely, the carboxylation step catalyzed by PFOR reverses to break down pyruvate to acetyl-CoA). Incomplete TCA cycles have been shown to occur in chemoautotrophs as biosynthetic rather than energy generation pathways (40).
Leptospirillum group III may have a complete TCA cycle. The uncertain component is also the oxoglutarate dehydrogenase complex. Leptospirillum group III has three subunits that could serve this function or could alternatively be a pyruvate dehydrogenase complex, otherwise lacking in Leptospirillum group III. The key E2 component has a relatively high enzyme-specific profile score for oxoglutarate dehydrogenase (EC 2.3.1.61), while the component E1 could belong to either oxoglutarate or pyruvate dehydrogenase (EC 1.2.4.1). As in Leptospirillum group II, there are three copies of dihydrolipoamide dehydrogenase at different locations in the genome; none are in proximity to the other putative oxoglutarate dehydrogenase complex components. The putative E1 and E2 proteins and all dihydrolipoamide dehydrogenase proteins were identified by proteomics.
(d) Gluconeogenesis, glycolysis, and sugar metabolism. Leptospirillum groups II and III have all the enzymes needed for gluconeogenesis.
Leptospirillum groups II and III do not appear to metabolize glucose through the Entner-Doudoroff pathway. Most enzymes in glycolysis (Embden-Meyerhoff) are present in Leptospirillum groups II and III. A key enzyme, phosphofructokinase, has not been identified in either species. A hexokinase is also missing in both organisms; however, one of several carbohydrate kinase family proteins may confer this function. Pyruvate kinase, the last energy-generating step in glycolysis, was found only in Leptospirillum group III. Thus, Leptospirillum group III may carry out glycolysis, but this function is apparently not possible for group II. All the proteins identified as potentially involved in gluconeogenesis/glycolysis in Leptospirillum groups II and III were identified by proteomics.
Leptospirillum group III has two copies of glucoamylase, an enzyme that degrades starch to glucose, and both copies were identified by proteomics. Leptospirillum group II lacks glucoamylase but has genes for degradation of extracellular maltose. It is possible that Leptospirillum group III uses glucoamylase for mobilization through the biofilm.
(ii) Nitrogen and sulfur metabolism.
(a) Nitrogen metabolism genes. Leptospirillum group III carries all the genes for nitrogen fixation (75, 76). These are next to a cluster of molybdenum uptake genes necessary for nitrogen fixation (22). Notably, in our current analysis, the proteins involved in nitrogen fixation were not identified in any of the samples (see Table S2 in the supplemental material).
Leptospirillum group III has four nitrogen-regulatory transduction PII proteins involved in nitrogen regulation or sensing of α-ketoglutarate (52). Two associated with the nitrogen fixation region are not identified by proteomics and thus probably regulate the nitrogen fixation. Of the two identified by proteomics, one is clustered with an ammonium transporter and is likely regulating ammonium uptake and the other is clustered with redox enzymes.
Although Leptospirillum group II does not fix nitrogen, it does harbor various nitrogen metabolism genes. There are three ammonium transporters (all identified by proteomics) clustered with nitrogen-regulatory PII proteins. This gene organization is very conserved among other organisms and suggests that the regulatory proteins are related to ammonium uptake (18, 52). Once inside, the ammonium is assimilated by the glutamine synthase/glutamate synthase pathway (88). Ammonium uptake proteins present in Leptospirillum group II were identified at high levels by proteomics. One copy of nitrogen-regulatory protein PII is located close to a transcriptional regulator, NifA (Fis family), and shows high protein coverage. Although nitrogen fixation proteins, including NifL, are not present in Leptospirillum group II, NifA may still be involved in nitrogen sensing.
In addition to acquiring ammonium via uptake, Leptospirillum group II may form ammonium from nitrite. A cytochrome c NapC/NirT family protein involved in respiratory nitrite ammonification (66) was found in Leptospirillum group II, but the gene for the catalytic subunit for this route has not been identified. Leptospirillum groups II and III have two genes for nitrite/sulfite reductase (ferredoxin) required in assimilatory nitrite ammonification and could use these to directly reduce nitrite to ammonium for amino acid biosynthesis (20, 66). Only the gene products for the assimilatory route are identified by proteomics.
The finding of an ammonia monooxygenase subunit, amoA, in both Leptospirillum group II and group III genomes is intriguing. AmoA is one of three subunits required for oxidation of ammonia and contains the active site for substrate oxidation (9); however, the lack of other subunits in Leptospirillum prevents us from inferring this functionality. AmoA was not identified by proteomics for either Leptospirillum species. Ammonia monooxygenase may also be involved in methane oxidation and hydrocarbon degradation (9).
(b) Sulfur metabolism. Leptospirillum group II and Leptospirillum group III have a complete assimilatory pathway for sulfate reduction. Interestingly, APS reductase, which is present as two subunits in Leptospirillum species, lacks a conserved motif described by Valdes et al. (78). The genes for sulfate assimilation are next to a region that contains Fe-S accessory proteins and a cysteine desulfurase in Leptospirillum group III but clustered in a plasmid/phage region in Leptospirillum group II.
Leptospirillum groups II and III could oxidize hydrogen sulfide with a siroheme-like enzyme, rhodanese-like proteins, or a sulfide-quinone reductase. The sulfide-quinone reductase is duplicated in both organisms, and one is clustered with the cytochrome bd operon, suggesting sulfur oxidation for energy generation. Siroheme-like protein could also be involved in nitrite oxidation (49). Siroheme and rhodanese-like proteins were identified by proteomics in Leptospirillum group II only, whereas sulfide-quinone reductase was identified by proteomics in both species.
(iii) Biosynthesis and degradation pathways.
(a) Cofactor biosynthesis. Biotin is a predicted cofactor for several biotin-dependent carboxylases and decarboxylases (59), and both Leptospirillum group II and group III have the five genes required for the bacillus-type pathway using BioW. All biotin biosynthesis gene products were identified by proteomics in Leptospirillum group II, but only BioA was identified in Leptospirillum group III.
Biosynthesis of riboflavin and flavin adenine dinucleotide in both Leptospirillum group II and group III is organized in two operons, and all of the gene products were identified by proteomics.
All the genes needed for thiamine biosynthesis are present in Leptospirillum groups II and III, and all of the gene products were identified by proteomics. Two copies of thiS and thiF are clustered with genes involved in biosynthesis of methionine, cysteine, and molybdopterin. This gene organization may reflect the common need for sulfur.
Cobalamin biosynthesis can occur via aerobic and anaerobic pathways (57). Leptospirillum groups II and III are probably able to synthesize cobalamin by using an anaerobic pathway. They carry the genes for 19 of the 20 steps required, including cbiX, the gene for the second and characteristic step for the anaerobic pathway. Although Leptospirillum groups II and III lack the genes cbiJ and cobK, this function could be complemented at very low levels by alternative nonspecific reactions as in Methanococcus maripaludis (37). It is possible that both organisms could also produce cobalamin through the aerobic pathway; most of the steps are shared between pathways, and they contain cobB, the aerobic alternative to cbiA. Most of the gene products putatively involved in cobalamin biosynthesis were identified by proteomics.
(b) Fatty acid and lipid biosynthesis. Both species contain the complete pathway for fatty acid biosynthesis. Most of the genes are arranged in clusters, and all proteins were identified by proteomics. Only Leptospirillum group II contains a fatty acid desaturase, an enzyme involved in converting saturated bonds to double bonds (2).
In addition to large clusters of genes involved in lipopolysaccharide biosynthesis, there are genes that may indicate production of glycosphingolipids (e.g., ceramide glucosyltransferase) and other membrane components (e.g., squalene/hopene) that may play roles in membrane stabilization and/or acid resistance.
(c) Degradation of aromatic compounds. Two putative extradiol (LigB) for cleavage of aromatic rings (part of cathecol dioxygenase) and a carboxymuconolactone decarboxylase were found in Leptospirillum group II (in a mobile element region) and in Leptospirillum group III and were identified by proteomics (at low abundance). These enzymes are part of the β-ketoadipate pathway, which degrades protocatechuate, an intermediate product of aromatic compound breakdown (46). However, LigA and other pathway steps were not found.
5-Carboxymethyl-2-hydroxymuconate δ-isomerase, an enzyme that generates an intermediary for the production of oxaloacetate in the benzoate degradation via a hydroxylation pathway, is present and identified by proteomics in both species. In addition, carboxymethyl butenolidase (dienelactone hydrolase), an enzyme that generates 2-maleylacetate in the metabolic pathway of chloroaromatic compounds (51), was identified at high levels by proteomics in Leptospirillum groups II and III. The presence of unique enzymes involved in aromatic compound metabolism suggests that Leptospirillum species are degrading aromatic compounds, but the sources of the aromatic substrates and the final products remain unclear.
(d) Cellulose biosynthesis. Leptospirillum group II has the genes for biosynthesis of cellulose, cellobiose, and starch/amylose. The genes for synthesis of cellulose are clustered. The regulatory subunit is identified at very low levels, while the catalytic subunit and subunit C were not identified by proteomics. A second copy of cellulose synthase subunit C, in cluster with an endoglucanase and a peptidoglycan glycosyl transferase, was identified by proteomics at high levels. Both copies of cellulose synthase subunit C are much shorter than the sequences with which they share similarity. Although cellulose synthase and cellulase genes were not found in the Leptospirillum group III genome, some unassembled reads suggest that this function might be present.
(e) Proteasomes. Within the bacteria, proteasomes involved in proteolysis have previously been found only in Actinobacteria (23); however, Leptospirillum group II (23) and Leptospirillum group III contain two gene clusters for this pathway, and all of the gene products are identified by proteomics. Within the community genomic data set, there are multiple contigs carrying proteasome genes, including an actinobacterial contig with a cluster of four proteasome genes. The Leptospirillum-type, Actinobacteria-type, and other unassigned bacterial proteasomes from the AMD data set cluster together in a gene tree (see Fig. S4 in the supplemental material), suggesting acquisition of proteasomes via lateral gene transfer.
(iv) Signal transduction and information processing.
(a) Signal transduction. For most genes encoding signal transduction histidine kinase proteins in Leptospirillum group II, we found a syntenous ortholog in Leptospirillum group III. These include chemotaxis-specific (CheA) genes, osmosensitive K+ channel-specific genes, and genes for proteins with PAS/PAC sensor domains. Several transcriptional regulators are encoded by Leptospirillum groups II and III, including LysR, ArsR, Fis, LuxR, MerR, and other families.
Leptospirillum group II encodes 29 diguanylate cyclase/phosphodiesterase proteins, whereas 39 are encoded by the Leptospirillum group III genome. One of the only four orthologs and 20 of the 25 proteins lacking orthologs were identified by proteomics in Leptospirillum group II. Similarly, in Leptospirillum group III, two of the four orthologs and 20 of the 35 Leptospirillum group III-specific proteins were identified by proteomics.
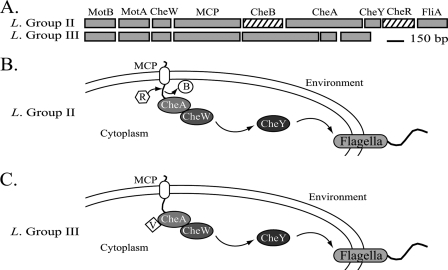
(b) Chemotaxis. Leptospirillum group III lacks cheB and cheR (Fig. 8), which are involved in methylation-dependent adaptation of the receptor in chemotactic and aerotactic sensing pathways (70, 72). Both are present and identified by proteomics in Leptospirillum group II. Leptospirillum group III contains CheV, a methyl-independent adaptation protein that is believed to interact directly with CheA (72), also present in Leptospirillum group II. Overall, however, Leptospirillum group III has many more genes for methyl-accepting chemotaxis sensory transducer-like proteins than Leptospirillum group II, most spread across the genome and identified by proteomics.
FIG. 8.
(A) Diagram of the chemotaxis gene cluster in Leptospirillum groups II and III. Orthologs are shown in gray, and unique proteins are shown in a black pattern. MCP, methyl-accepting chemotaxis sensory transducer. Cartoons show predicted methyl-dependent (B) and methyl-independent (C) chemotaxis systems in Leptospirillum groups II and III, respectively. Adaptation chemotaxis proteins: R, CheR; B, CheB; and V, CheV.
(c) DNA polymerases. Leptospirillum groups II and III have a gene for DNA polymerase I and five subunits of DNA polymerase III. These are spread around the genome and have low normalized spectral count values. DNA polymerase family B is likely the product of lateral transfer from archaea, given its strong similarity only with archaeal proteins. Although detected, this protein is not abundant, based on proteomics data.
Leptospirillum groups II and III contain various genes involved in DNA repair and recombination, including ruvABC, organized in an operon and highly expressed (43).
(d) RNA polymerase. Leptospirillum group II and III genomes encode the subunits of the typical bacterial RNA polymerase complex. Both carry sigma-70 (RpoD), sigma-28 (RpoF; in the chemotaxis cluster and close to the flagellar genes), and sigma-54 (RpoN).
Of particular interest is the sigma factor RpoD, which has a fused adenine phosphoribosyltransferase (APRT) domain in Leptospirillum group II (see Fig. S5 in the supplemental material). In-depth analysis ruled out an assembly error, a missed stop codon, or an insertion/deletion that could have generated a frameshift mutation between the two genes. The genes are adjacent to each other in Leptospirillum group III but not fused. The fused sigma factor shows extensive protein coverage, and at least one peptide maps between normal RpoD and APRT coding regions (see Fig. S5 in the supplemental material), confirming that the whole protein is translated. The APRT domain protein may have a regulatory function, perhaps connected to nucleotide synthesis. Another interesting feature of RpoD in both species is the lack of region 1.1, shown to be important in initiation of transcription in E. coli (84).
Leptospirillum groups II and III lack the sigma-32 factor, which responds to extracytoplasmatic stress, for example, to induce heat shock genes. All the heat shock gene products are highly abundant based on proteomics, suggesting that the niche for these organisms could require the constitutive expression of otherwise conditionally induced functions. Alternatively, an unknown heat shock sigma factor might play the role.
(v) Stress and transport.
(a) Oxidative stress. Leptospirillum groups II and III have the complete pathway for synthesis of phytoene and carotene, and most of the gene products were identified. Carotenoids can act as antioxidants (77), and synthesis of carotenoids by the Leptospirillum species could be related to radical detoxification.
Leptospirillum group II contains the genes for rubrerythrin and peroxiredoxin, and both are very highly expressed. Leptospirillum group III carries an alkylhydroperoxidase not found in Leptospirillum group II, the gene products of which were identified at high levels by proteomics.
(b) Ectoine and trehalose. Leptospirillum groups II and III can synthesize trehalose by using three of four known pathways (27). Leptospirillum group II has the complete pathway for ectoine biosynthesis, another compatible solute for tolerance in high-salinity and high-temperature environments (31). The genes are arranged in an operon (ectABCD), and a transporter for ectoine is located upstream of the biosynthetic operon; Leptospirillum group III does not have a specific ectoine transporter or genes for synthesis of ectoine. All of the protein products in trehalose and ectoine pathways were identified by proteomics. Some of the genes for the ectoine and trehalose biosynthesis pathways were previously documented for Leptospirillum ferrooxidans (54); however, this is the first time the complete pathway for biosynthesis of ectoine and hydroxyectoine has been described for an acidophilic bacterium.
(c) Transport and acquisition. Leptospirillum groups II and III contain several transporters for citrate, potassium, phosphate (Pst in cluster with a phosphate uptake regulator, PhoU), sulfate-transporting ATPases, and transporters related to antibiotic resistance. Most secretion proteins are next to metal efflux pumps or membrane efflux proteins. In addition, Leptospirillum group II contains copper-translocating ATPases, whereas Leptospirillum group III lacks them. Leptospirillum groups II and III contain an iron permease, many ferric uptake regulators (Fur family proteins), and several TonB-dependent receptors. Most of these proteins were identified by proteomics. Interestingly, Fig. 4 shows that some Leptospirillum group II transport proteins are overrepresented in all three samples relative to transport proteins in Leptospirillum group III.
(d) Metal and antibiotic resistance. Arsenic resistance genes, such as those encoding transcriptional regulators (ArsR) and the arsenite transporter (ArsB), are present in Leptospirillum groups II and III. In addition, Leptospirillum group III contains the genes encoding ArsA (an arsenite-activated ATPase), ArsD (arsenical resistance operon trans-acting regulatory protein), and ArsC (arsenate reductase) arranged in an operon, and the products of these genes were identified by proteomics. These genes may be key to the ability of Leptospirillum to thrive in solutions that can contain mM concentrations of arsenic.
Leptospirillum groups II and III contain a mercuric reductase (MerA) and a mercuric transcriptional regulator in a cluster with an ion channel. A phage/plasmid region in Leptospirillum group II contains a probable mercuric transporter; however, this protein was not identified by proteomics. Reduction of mercury is typically favored in anaerobic organisms (50). It is interesting that some strains of Leptospirillum group II have a mercuric reductase frame shifted (insertions/deletions in multiple reads), suggesting that this capability may not be necessary for Leptospirillum group II in the current AMD environment, where mercury levels are very low.
Several antibiotic resistance genes, such as beta-lactamase, acriflavin, fusaric acid, and glyoxalase family proteins, are present in Leptospirillum groups II and III. Most of the gene products were identified by proteomics.
(vi) Mobile elements.
(a) Plasmid regions and extrachromosomal plasmid. Regions of an integrated plasmid are present in Leptospirillum group II (scaffold 8692) and Leptospirillum group III (scaffold 4481). The shared gene content and organization, as well as the comparability to other, nonplasmid regions in terms of product amino acid identity and GC content, indicate that the blocks were acquired long ago, perhaps before the species diverged.
Many conjugal transfer proteins (type II and IV secretion system components) from the integrated plasmid region share ∼30% average amino acid sequence identity with proteins from an extrachromosomal plasmid (see Table S5 in the supplemental material). Consequently, the plasmid may be associated with Leptospirillum. The presence of conjugation systems in the integrated and nonintegrated plasmids suggests that both Leptospirillum types can transfer plasmids. Interestingly, only a few proteins encoded by genes in the integrated plasmid regions of Leptospirillum groups II and III and none of the proteins encoded by genes in the conjugative transfer region were identified by proteomics. In contrast, many of the conjugative transfer proteins encoded by genes in the extrachromosomal plasmid were identified by proteomics (see Table S5 in the supplemental material).
Other mobile regions in Leptospirillum group II encode copper-, arsenic-, and mercury-transporting ATPase and secretion proteins; glycosyltransferases; metal-related transcriptional regulators; and proteins of the β-ketoadipate pathway. NADPH-quinone reductase, NADH dehydrogenase subunits, and cytochromes are associated with other phage/plasmid regions.
Clusters of toxin-antitoxin system proteins are present in Leptospirillum groups II and III. These systems are known to retain bacterial plasmids during segregation, and some have been suggested to arrest growth during nutritional stress (8). Only one antitoxin protein in Leptospirillum group II was identified by proteomics in the UBA sample.
Leptospirillum groups II and III contain two copies of reverse transcriptase genes, probably associated with group II introns (58), and all show protein coverage. A gene tree of the reverse transcriptase genes in Leptospirillum groups II and III and other organisms places these genes in the chloroplast-like group II intron class (74). The evolutionary mechanisms and functions of group II introns are still unknown.
A mobile element on scaffold 8524 in Leptospirillum group II encodes a putative defect in organelle-trafficking lipoproteins (Dot) and intracellular-multiplication (Icm) proteins. Only some orthologs for Icm and Dot proteins are present in Leptospirillum group III, and the surrounding proteins include many methyl-accepting chemotaxis sensory transducers (all identified by proteomics) without orthologs in Leptospirillum group II. Icm and Dot proteins were not identified by proteomics in any sample.
Two copies of a gene encoding a methyltransferase of the FkbM family occur in a plasmid-like region, and another copy is found among a large cluster of Leptospirillum group II genes for biosynthesis, export, and reconfiguration of sugar/polysaccharides (only the product of the last of these genes was identified by proteomics), while Leptospirillum group III lacks these genes.
(b) CRISPR. Clustered, regularly interspaced, short palindromic repeats (CRISPRs) and CRISPR-associated (CAS) genes are involved in a recently described viral and plasmid defense mechanism found in Bacteria and Archaea (14, 45). Leptospirillum group II carries a cluster of Cas proteins (Cas2/1/3/5/4/2/1/3). Orthologs (mostly syntenous) occur in one of the multiple Cas clusters of Leptospirillum group III. Most proteins in the orthologous clusters were identified by proteomics, and Cas protein abundance levels vary significantly among biofilms. Another CRISPR region, also carrying this repeat, occurs at a different genomic locus and in several different mobile elements (without genes encoding identifiable Cas proteins; data not shown). A second Leptospirillum group III CAS cluster encodes Cas and Csm proteins, and transposases interrupt the CRISPR region and Cas1 protein (see Table S2 in the supplemental material). This CRISPR locus reconstructed from the population genomic data set is essentially clonal (unlike almost all other CRISPR loci in the AMD data sets), and none of the spacers match any viral sequences. These observations suggest that this second locus is inactive. However, despite the interruption of Cas1 by a transposase, three of the Csm family proteins were identified by proteomics. This second Leptospirillum group III CRISPR locus is next to an integrase and is interspersed by transposases in a genomic region with many hypothetical proteins; thus, it is possibly part of an integrated mobile element. The repeat in this second cluster has a region of similarity to a repeat in a CRISPR locus carried by the extrachromosomal plasmid (locus 13 in scaffold 15659) (see Table S5 in the supplemental material). The CRISPR spacer and repeat sequences carried by plasmid-like contigs were reported previously (4).
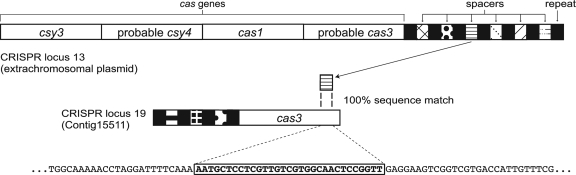
Spacers at the CRISPR loci of the plasmid-like population match plasmid-like contigs, perhaps indicating that CRISPR spacers are involved in competition among mobile elements. Some spacers match only regions encoding Cas proteins of other plasmids, potentially indicating CRISPR silencing of acquired resistance. Specifically, a spacer at CRISPR locus 13 (on the extrachromosomal plasmid) targets (based on nucleotide identity) the Cas3 helicase encoded by CRISPR locus 19 (contig 15511) (Fig. 9). Another spacer, at CRISPR locus 13, encodes a product that matches a hypothetical protein encoded between the Cas-cys3 gene and a Cas-cys2 gene in plasmid-like contig 12113, whereas a spacer at CRISPR locus 11 (plasmid-like contig 11387) targets the Cas-cys3 gene.
FIG. 9.
Diagram of CRISPR/CAS loci associated with the extrachromosomal plasmid. Black bars show repeat sequences, while bars between the repeats represent spacer sequences. (While cas genes are displayed accurately, the sets of spacers are shown schematically.) A spacer at CRISPR locus 13 (extrachromosomal plasmid) targets a cas3 gene at CRISPR locus 19 (plasmid-like contig 15511). The inset displays a portion of the cas3 gene targeted by the spacer shown bold.
Some spacers at plasmid-borne CRISPR loci also target non-Cas genes. For example, a spacer at CRISPR locus 13 encodes a product that matches a hypothetical protein encoded by the same extrachromosomal plasmid (contig 11623). A DNA polymerase encoded by plasmid-like contig 15498 is also targeted by a spacer at CRISPR locus 13.
DISCUSSION
Leptospirillum groups II and III are the first organisms from the Nitrospirae lineage for which extensive (near-complete) genomic data and detailed functional annotations are available. For Leptospirillum group II, there are two composite sequences from the Richmond Mine, the UBA type (43) and a 5-way CG type related to L. ferriphilum (65). In addition, there are several isolates that are similar to these genomically characterized populations (based on multilocus sequence typing [43]). We refer to the now extensively characterized UBA subgroup as “Leptospirillum rubarum”. The genomes of L. rubarum and L. ferriphilum are syntenous, but there are major rearrangements relative to Leptospirillum group III. Leptospirillum groups II and III share most of their core metabolic pathways, and the organization of key genes is maintained despite their divergence.
The significant evolutionary distance between the leptospirilli and other organisms for which extensive genomic and biochemical information is available somewhat limits physiological interpretations. Gaps in generally well-known metabolic pathways likely reflect the paucity of information for closely related lineages, although the incomplete nature of both genomic data sets makes any firm conclusion about missing genes impossible. For this reason, proteomic identification of enzymes characteristic of specific pathways provides key indications of the associated functionality. It is important to note that, although there are information gaps, the physiological, ecological, and evolutionary insights obtained here were achieved for natural populations without cultivation.
Studies of the Leptospirillum genus support assignment of its members as obligate Fe(II) oxidizers (62). Therefore, Fe(II) oxidation and electron transport are key functions in the maintenance of Leptospirillum cellular metabolism. A bifurcated cytochrome b/b6 protein has previously been found only in gram-positive bacteria (68). This unusual structure for a bc1 complex, along with tetraheme cytochrome as the c1 component, is also observed in the genome of “Candidatus Kuenenia stuttgartensis,” a planctomycete that performs anaerobic ammonium oxidation and generates reductant for autotrophic growth by reverse electron transport (71). The high abundance of now well-studied cytochromes in Leptospirillum group II (and group III) supports the key role of iron oxidation in the growth of these organisms.
For both organisms, ribosomal proteins are generally very abundant, consistent with high levels of activity. Other proteins identified at high levels in all three proteomic samples in Leptospirillum group II are cold shock proteins and other molecular chaperones; histone-like proteins; cytochromes, such as Cyt579; and translation elongation factors. Although it is possible that high levels of cold and heat shock proteins are artifacts of sample preparation, preliminary proteomics data on the same biological sample subjected to different freezing regimes suggest that this is not the case (V. J. Denef, R. S. Mueller, M. B. Shah, R. L. Hettich, and J. F. Banfield, unpublished data). An alternative explanation is that protein-modifying, RNA-binding, and nucleic acid-binding histone-like proteins perform an alternative function, possibly related to cell stabilization in the extreme environment (79).
Some proteins with clear functional annotations were not identified by proteomics. Although an absence of peptide identification could reflect low rather than no abundance or a variety of experimental problems, a failure to detect all predicted peptides in whole groups of proteins in large complexes is notable. For example, Leptospirillum species carry formate-hydrogen lyase clusters found in microaerophilic/anaerobic organisms (63), but none of the gene products were identified in the samples studied. This complex may convert formate to hydrogen and CO2 or play a role in carbon fixation. Its presence in both genomes is consistent with the capability for anaerobic growth. Carbon fixation via the rTCA pathway, anaerobic cobalamin pathway genes, and highly abundant PFOR have been found only in known anaerobes, suggesting that Leptospirillum groups II and III may grow in anaerobic as well as microaerophilic and aerobic environments.
On the basis of functional predictions, proteomic information, and data from studies of isolated species, both organisms are likely capable of carbon fixation and most core metabolic functions. Both face the same environmental challenges, particularly the very low pHs and high concentrations of toxic metals. Notable are the identification of multiple pathways for production of compatible solutes that presumably provide a response to surrounding high-ionic-strength solutions, membrane stabilization molecules, and metal efflux pumps.
Icm proteins may be involved in pathogenesis (64), conjugation (64), and resistance to eukaryotic predation (55). In Leptospirillum group II, Icm, Dot, and FkbM proteins may protect against grazing by protists and from fungi that proliferate in some higher-developmental-stage biofilms (12, 83). Both organisms appear to have functional CRISPR/Cas loci for viral defense. To date, we have only detected evidence suggestive of silencing of one plasmid's CRISPR locus by another. However, the same approach would be effective for silencing the host resistance system (and is likely if lateral transfer from mobile elements is in fact the major source of bacterial and archaeal CRISPR loci).
Differences in gene complement point to important physiological distinctions that may have been key to the likely sympatric divergence of the Leptospirillum groups. Leptospirillum group II is better equipped than Leptospirillum group III to deal with osmotic challenges associated with the near-molar FeSO4 solutions and produce potentially key polymers for establishment of floating biofilms (e.g., cellulose, cellobiose, and starch/amylose). Leptospirillum group III is apparently better optimized for energy generation (given a possible complete glycolysis and TCA pathways) and nitrogen fixation. These findings are consistent with the identifications of Leptospirillum group II as the early colonist and Leptospirillum group III as a member in biofilms at late developmental stages (83).
Intriguingly, the complements of signal transduction and chemotaxis genes in Leptospirillum group II and Leptospirillum group III are quite different, as are many regulatory genes, pointing to adaptation to different microenvironments (e.g., with specific levels of oxygen, redox potential, and availability of fixed nitrogen). Genomic and proteomic data suggest that signal transduction, motility, and chemotaxis are more important in Leptospirillum group III than in group II (Fig. 4). Biofilm characterization studies place Leptospirillum group III as dispersed cells and microcolonies in interior regions of biofilms (83), where geochemical gradients are expected to be pronounced. This distribution, in combination with the inferred metabolic characteristics, may point to Leptospirillum group III as a microaerophile that locates in nutrient-poor regions of biofilms, where its ability to fix nitrogen may be key. The distribution of Leptospirillum group II at the bases of some biofilms, where oxygen availability is almost certainly low, may indicate an optional but as yet incompletely defined anaerobic metabolism (for example, making use of a within-biofilm nitrogen cycle).
Nitrogen fixation proteins of Leptospirillum group III were not identified by proteomics. Both the late arrival of this organism during biofilm development (83) and the near absence of evidence for nitrogen fixation are at odds with the simplest ecological model in which this organism is the keystone species and first colonist. An important observation is that the biofilms studied here form at the confluence of drainage streams with sources throughout the biologically active and probably highly productive subsurface ecosystem. Thus, a significant load of fixed nitrogen in influent solutions is not surprising (L. Kalnejais, unpublished data). Furthermore, biofilm recycling occurs periodically when biofilms sink. Thus, it is perhaps expected that Leptospirillum group III bacteria invest little energy in nitrogen fixation in the biofilms studied here. Likely, this function is important when these bacteria grow in microenvironments not yet studied (e.g., in association with pyrite surfaces in the sediment). Alternatively, nitrogen fixation may occur below detection levels in anaerobic regions of thicker biofilms where fixed nitrogen has been depleted by surrounding organisms.
The identification of an extrachromosomal plasmid with relatively high proteome coverage indicates that the physiology of both Leptospirillum types cannot be described on the basis of their core metabolic potential alone. The effect of plasmids on the metabolism of these bacteria is difficult to deduce because most proteins identified have no known function. It is interesting that the few proteomically identified proteins from integrated plasmids (a subset of which is common to both Leptospirillum types and may predate lineage divergence) are not involved in conjugation but that the conjugation apparatus of the extrachromosomal plasmids may contribute to ongoing plasmid transfer.
The genomic and proteomic data sets provide evidence of interesting new biochemical functionalities. For example, the identification of a sigma factor with the fused APRT points to potentially novel aspects of genome regulation. In addition to the indications of functional differentiation noted above, comparative proteogenomic analyses highlight many proteins of unknown function that are unique and, in some cases, expressed at high levels. These are obvious targets for future functional screening and crystallography studies (39). Some of these species-specific proteins are relatively abundant in only a subset of biofilm communities, suggesting roles in microenvironmental adaptation. Expression profiles of proteins shared in Leptospirillum species strongly cluster by organism rather than environment, suggesting that the organism is more important in determining protein expression pattern than environmental parameters. Proteins that are shared by both Leptospirillum types but are otherwise unique likely reflect Nitrospirae lineage adaptations to the low-pH, metal-rich environments.
Conclusion.
This study represents the first in-depth functional (simultaneous proteomic and genomic) analysis of closely related species as members of communities in the same natural environment and the first detailed genome-based functional analysis of members of the Nitrospirae lineage. Given that they consistently coexist in acidic, metal-rich environments, Leptospirillum groups II and III most likely underwent sympatric divergence. Documented differences in their genotypes and protein expression patterns (proteins that cluster by organism, not environment) may largely account for differences in ecological behavior, such as the early predominance of Leptospirillum group II and different partitioning of Leptospirillum groups II and III (83). Specifically, we highlighted important differences in the complements of chemosensory genes and in the levels of their protein products, differences in metabolic potential, distinct expression patterns for both orthologous and unique proteins of unknown function, and notable differences in the complements of signal transduction proteins. This study demonstrates the power of combining comparative, cultivation-independent genomics and community proteomics to study closely related organisms within their natural environment.
Supplementary Material
Acknowledgments
We thank T. W. Arman, President, Iron Mountain Mines, Inc., and R. Sugarek for access to the Richmond Mine and R. Carver for on-site assistance. E. Watkin provided genotype information for a Leptospirillum group III colony 5 isolate (UBA:5). P. Abraham is thanked for his assistance with UBA proteomic measurements. DNA sequencing was carried out at The Joint Genome Institute.
D.S.A.G. acknowledges support from an NSF Graduate Research Fellowship. Oak Ridge National Laboratory is managed by University of Tennessee-Battelle, LLC, for the Department of Energy under contract DOE-AC05-00OR22725. This research was supported by The U.S. Department of Energy, Office of Biological and Environmental Research, Genomics (Genomes to Life Program).
Footnotes
Published ahead of print on 8 May 2009.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Abe, T., S. Kanaya, M. Kinouchi, Y. Ichiba, T. Kozuki, and T. Ikemura. 2003. Informatics for unveiling hidden genome signatures. Genome Res. 13:693-702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aguilar, P. S., and D. de Mendoza. 2006. Control of fatty acid desaturation: a mechanism conserved from bacteria to humans. Mol. Microbiol. 62:1507-1514. [DOI] [PubMed] [Google Scholar]
- 3.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 4.Andersson, A. F., and J. F. Banfield. 2008. Virus population dynamics and acquired virus resistance in natural microbial communities. Science 320:1047-1050. [DOI] [PubMed] [Google Scholar]
- 5.Aoshima, M., and Y. Igarashi. 2006. A novel oxalosuccinate-forming enzyme involved in the reductive carboxylation of 2-oxoglutarate in Hydrogenobacter thermophilus TK-6. Mol. Microbiol. 62:748-759. [DOI] [PubMed] [Google Scholar]
- 6.Aoshima, M., M. Ishii, and Y. Igarashi. 2004. A novel enzyme, citryl-CoA lyase, catalysing the second step of the citrate cleavage reaction in Hydrogenobacter thermophilus TK-6. Mol. Microbiol. 52:763-770. [DOI] [PubMed] [Google Scholar]
- 7.Aoshima, M., M. Ishii, and Y. Igarashi. 2004. A novel enzyme, citryl-CoA synthetase, catalysing the first step of the citrate cleavage reaction in Hydrogenobacter thermophilus TK-6. Mol. Microbiol. 52:751-761. [DOI] [PubMed] [Google Scholar]
- 8.Arcus, V. L., P. B. Rainey, and S. J. Turner. 2005. The PIN-domain toxin-antitoxin array in mycobacteria. Trends Microbiol. 13:360-365. [DOI] [PubMed] [Google Scholar]
- 9.Arp, D. J., and L. Y. Stein. 2003. Metabolism of inorganic N compounds by ammonia-oxidizing bacteria. Crit. Rev. Biochem. Mol. Biol. 38:471-495. [DOI] [PubMed] [Google Scholar]
- 10.Ashida, H., A. Danchin, and A. Yokota. 2005. Was photosynthetic RuBisCO recruited by acquisitive evolution from RuBisCO-like proteins involved in sulfur metabolism? Res. Microbiol. 156:611-618. [DOI] [PubMed] [Google Scholar]
- 11.Badger, J. H., and G. J. Olsen. 1999. CRITICA: coding region identification tool invoking comparative analysis. Mol. Biol. Evol. 16:512-524. [DOI] [PubMed] [Google Scholar]
- 12.Baker, B. J., M. A. Lutz, S. C. Dawson, P. L. Bond, and J. F. Banfield. 2004. Metabolically active eukaryotic communities in extremely acidic mine drainage. Appl. Environ. Microbiol. 70:6264-6271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Banfield, J. F., N. C. Verberkmoes, R. L. Hettich, and M. P. Thelen. 2005. Proteogenomic approaches for the molecular characterization of natural microbial communities. OMICS 9:301-333. [DOI] [PubMed] [Google Scholar]
- 14.Barrangou, R., C. Fremaux, H. Deveau, M. Richards, P. Boyaval, S. Moineau, D. A. Romero, and P. Horvath. 2007. CRISPR provides acquired resistance against viruses in prokaryotes. Science 315:1709-1712. [DOI] [PubMed] [Google Scholar]
- 15.Bond, P. L., G. K. Druschel, and J. F. Banfield. 2000. Comparison of acid mine drainage microbial communities in physically and geochemically distinct ecosystems. Appl. Environ. Microbiol. 66:4962-4971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brown, S. D., M. R. Thompson, N. C. Verberkmoes, K. Chourey, M. Shah, J. Zhou, R. L. Hettich, and D. K. Thompson. 2006. Molecular dynamics of the Shewanella oneidensis response to chromate stress. Mol. Cell. Proteomics 5:1054-1071. [DOI] [PubMed] [Google Scholar]
- 17.Callister, S. J., M. A. Dominguez, C. D. Nicora, X. Zeng, C. L. Tavano, S. Kaplan, T. J. Donohue, R. D. Smith, and M. S. Lipton. 2006. Application of the accurate mass and time tag approach to the proteome analysis of sub-cellular fractions obtained from Rhodobacter sphaeroides 2.4.1. Aerobic and photosynthetic cell cultures. J. Proteome Res. 5:1940-1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Conroy, M. J., A. Durand, D. Lupo, X. D. Li, P. A. Bullough, F. K. Winkler, and M. Merrick. 2007. The crystal structure of the Escherichia coli AmtB-GlnK complex reveals how GlnK regulates the ammonia channel. Proc. Natl. Acad. Sci. USA 104:1213-1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Coram, N. J., and D. E. Rawlings. 2002. Molecular relationship between two groups of the genus Leptospirillum and the finding that Leptospirillum ferriphilum sp. nov. dominates South African commercial biooxidation tanks that operate at 40°C. Appl. Environ. Microbiol. 68:838-845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Curdt, I., B. B. Singh, M. Jakoby, W. Hachtel, and H. Bohme. 2000. Identification of amino acid residues of nitrite reductase from Anabaena sp. PCC 7120 involved in ferredoxin binding. Biochim. Biophys. Acta 1543:60-68. [DOI] [PubMed] [Google Scholar]
- 21.Delcher, A. L., D. Harmon, S. Kasif, O. White, and S. L. Salzberg. 1999. Improved microbial gene identification with GLIMMER. Nucleic Acids Res. 27:4636-4641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Delgado, M. J., A. Tresierra-Ayala, C. Talbi, and E. J. Bedmar. 2006. Functional characterization of the Bradyrhizobium japonicum modA and modB genes involved in molybdenum transport. Microbiology 152:199-207. [DOI] [PubMed] [Google Scholar]
- 23.De Mot, R. 2007. Actinomycete-like proteasomes in a Gram-negative bacterium. Trends Microbiol. 15:335-338. [DOI] [PubMed] [Google Scholar]
- 24.Denef, V. J., M. B. Shah, N. C. Verberkmoes, R. L. Hettich, and J. F. Banfield. 2007. Implications of strain- and species-level sequence divergence for community and isolate shotgun proteomic analysis. J. Proteome Res. 6:3152-3161. [DOI] [PubMed] [Google Scholar]
- 25.Eddy, S. R. 2002. A memory-efficient dynamic programming algorithm for optimal alignment of a sequence to an RNA secondary structure. BMC Bioinformatics 3:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Eisen, M. B., P. T. Spellman, P. O. Brown, and D. Botstein. 1998. Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. USA 95:14863-14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Empadinhas, N., and M. S. da Costa. 2006. Diversity and biosynthesis of compatible solutes in hyper/thermophiles. Int. Microbiol. 9:199-206. [PubMed] [Google Scholar]
- 28.Eng, J. K., A. L. McCormack, and J. R. Yates. 1994. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J. Am. Soc. Mass Spectrom. 5:976-989. [DOI] [PubMed] [Google Scholar]
- 29.Eppley, J. M., G. W. Tyson, W. M. Getz, and J. F. Banfield. 2007. Genetic exchange across a species boundary in the archaeal genus ferroplasma. Genetics 177:407-416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Florens, L., M. J. Carozza, S. K. Swanson, M. Fournier, M. K. Coleman, J. L. Workman, and M. P. Washburn. 2006. Analyzing chromatin remodeling complexes using shotgun proteomics and normalized spectral abundance factors. Methods 40:303-311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.García-Estepa, R., M. Argandoña, M. Reina-Bueno, N. Capote, F. Iglesias-Guerra, J. J. Nieto, and C. Vargas. 2006. The ectD gene, which is involved in the synthesis of the compatible solute hydroxyectoine, is essential for thermoprotection of the halophilic bacterium Chromohalobacter salexigens. J. Bacteriol. 188:3774-3784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.García Martín, H., N. Ivanova, V. Kunin, F. Warnecke, K. W. Barry, A. C. McHardy, C. Yeates, S. He, A. A. Salamov, E. Szeto, E. Dalin, N. H. Putnam, H. J. Shapiro, J. L. Pangilinan, I. Rigoutsos, N. C. Kyrpides, L. L. Blackall, K. D. McMahon, and P. Hugenholtz. 2006. Metagenomic analysis of two enhanced biological phosphorus removal (EBPR) sludge communities. Nat. Biotechnol. 24:1263-1269. [DOI] [PubMed] [Google Scholar]
- 33.Gordon, D. 2003. Viewing and editing assembled sequences using Consed. Curr. Protoc. Bioinformatics 2003(August):Unit 11.2. [DOI] [PubMed] [Google Scholar]
- 34.Huber, H., M. Gallenberger, U. Jahn, E. Eylert, I. A. Berg, D. Kockelkorn, W. Eisenreich, and G. Fuchs. 2008. A dicarboxylate/4-hydroxybutyrate autotrophic carbon assimilation cycle in the hyperthermophilic Archaeum Ignicoccus hospitalis. Proc. Natl. Acad. Sci. USA 105:7851-7856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jahn, U., H. Huber, W. Eisenreich, M. Hugler, and G. Fuchs. 2007. Insights into the autotrophic CO2 fixation pathway of the archaeon Ignicoccus hospitalis: comprehensive analysis of the central carbon metabolism. J. Bacteriol. 189:4108-4119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jeans, C., S. W. Singer, C. S. Chan, N. C. Verberkmoes, M. Shah, R. L. Hettich, J. F. Banfield, and M. P. Thelen. 2008. Cytochrome 572 is a conspicuous membrane protein with iron oxidation activity purified directly from a natural acidophilic microbial community. ISME J. 2:542-550. [DOI] [PubMed] [Google Scholar]
- 37.Kim, W., T. A. Major, and W. B. Whitman. 2005. Role of the precorrin 6-X reductase gene in cobamide biosynthesis in Methanococcus maripaludis. Archaea 1:375-384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Konstantinidis, K. T., and J. M. Tiedje. 2005. Genomic insights that advance the species definition for prokaryotes. Proc. Natl. Acad. Sci. USA 102:2567-2572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kuznetsova, E., M. Proudfoot, S. A. Sanders, J. Reinking, A. Savchenko, C. H. Arrowsmith, A. M. Edwards, and A. F. Yakunin. 2005. Enzyme genomics: application of general enzymatic screens to discover new enzymes. FEMS Microbiol. Rev. 29:263-279. [DOI] [PubMed] [Google Scholar]
- 40.Lengeler, J. W., G. Drews, and H. G. Schlegel. 1999. Biology of the prokaryotes. Blackwell Science, Malden, MA.
- 41.Levican, G., J. A. Ugalde, N. Ehrenfeld, A. Maass, and P. Parada. 2008. Comparative genomic analysis of carbon and nitrogen assimilation mechanisms in three indigenous bioleaching bacteria: predictions and validations. BMC Genomics 9:581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lipton, M. S., L. Pasa-Tolic, G. A. Anderson, D. J. Anderson, D. L. Auberry, J. R. Battista, M. J. Daly, J. Fredrickson, K. K. Hixson, H. Kostandarithes, C. Masselon, L. M. Markillie, R. J. Moore, M. F. Romine, Y. Shen, E. Stritmatter, N. Tolic, H. R. Udseth, A. Venkateswaran, K.-K. Wong, R. Zhao, and R. D. Smith. 2002. From the cover: global analysis of the Deinococcus radiodurans proteome by using accurate mass tags. Proc. Natl. Acad. Sci. USA 99:11049-11054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lo, I., V. J. Denef, N. C. Verberkmoes, M. B. Shah, D. Goltsman, G. DiBartolo, G. W. Tyson, E. E. Allen, R. J. Ram, J. C. Detter, P. Richardson, M. P. Thelen, R. L. Hettich, and J. F. Banfield. 2007. Strain-resolved community proteomics reveals recombining genomes of acidophilic bacteria. Nature 446:537-541. [DOI] [PubMed] [Google Scholar]
- 44.Lowe, T. M., and S. R. Eddy. 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25:955-964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Marraffini, L. A., and E. J. Sontheimer. 2008. CRISPR interference limits horizontal gene transfer in staphylococci by targeting DNA. Science 322:1843-1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Masai, E., Y. Katayama, and M. Fukuda. 2007. Genetic and biochemical investigations on bacterial catabolic pathways for lignin-derived aromatic compounds. Biosci. Biotechnol. Biochem. 71:1-15. [DOI] [PubMed] [Google Scholar]
- 47.Matsumura, H., Y. Xie, S. Shirakata, T. Inoue, T. Yoshinaga, Y. Ueno, K. Izui, and Y. Kai. 2002. Crystal structures of C4 form maize and quaternary complex of E. coli phosphoenolpyruvate carboxylases. Structure 10:1721-1730. [DOI] [PubMed] [Google Scholar]
- 48.McDonald, W. H., R. Ohi, D. T. Miyamoto, T. J. Mitchison, and J. R. Yates. 2002. Comparison of three directly coupled HPLC MS/MS strategies for identification of proteins from complex mixtures: single-dimension LC-MS/MS, 2-phase MudPIT, and 3-phase MudPIT. Int. J. Mass Spectrom. 219:245-251. [Google Scholar]
- 49.Murphy, M. J., L. M. Siegel, S. R. Tove, and H. Kamin. 1974. Siroheme: a new prosthetic group participating in six-electron reduction reactions catalyzed by both sulfite and nitrite reductases. Proc. Natl. Acad. Sci. USA 71:612-616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.NíChadhain, S. M., J. K. Schaefer, S. Crane, G. J. Zylstra, and T. Barkay. 2006. Analysis of mercuric reductase (merA) gene diversity in an anaerobic mercury-contaminated sediment enrichment. Environ. Microbiol. 8:1746-1752. [DOI] [PubMed] [Google Scholar]
- 51.Nikodem, P., V. Hecht, M. Schlomann, and D. H. Pieper. 2003. New bacterial pathway for 4- and 5-chlorosalicylate degradation via 4-chlorocatechol and maleylacetate in Pseudomonas sp. strain MT1. J. Bacteriol. 185:6790-6800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ninfa, A. J., and P. Jiang. 2005. PII signal transduction proteins: sensors of alpha-ketoglutarate that regulate nitrogen metabolism. Curr. Opin. Microbiol. 8:168-173. [DOI] [PubMed] [Google Scholar]
- 53.Norris, P. R. 2006. Acidophile diversity in mineral sulfide oxidation, p. 199-216. In D. E. Rawlings and D. B. Johnson (ed.), Biomining. Springer, Berlin, Germany.
- 54.Parro, V., M. Moreno-Paz, and E. Gonzalez-Toril. 2007. Analysis of environmental transcriptomes by DNA microarrays. Environ. Microbiol. 9:453-464. [DOI] [PubMed] [Google Scholar]
- 55.Pukatzki, S., A. T. Ma, D. Sturtevant, B. Krastins, D. Sarracino, W. C. Nelson, J. F. Heidelberg, and J. J. Mekalanos. 2006. Identification of a conserved bacterial protein secretion system in Vibrio cholerae using the Dictyostelium host model system. Proc. Natl. Acad. Sci. USA 103:1528-1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ram, R. J., N. C. Verberkmoes, M. P. Thelen, G. W. Tyson, B. J. Baker, R. C. Blake II, M. Shah, R. L. Hettich, and J. F. Banfield. 2005. Community proteomics of a natural microbial biofilm. Science 308:1915-1920. [PubMed] [Google Scholar]
- 57.Raux, E., H. L. Schubert, and M. J. Warren. 2000. Biosynthesis of cobalamin (vitamin B12): a bacterial conundrum. Cell. Mol. Life Sci. 57:1880-1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Robart, A. R., and S. Zimmerly. 2005. Group II intron retroelements: function and diversity. Cytogenet. Genome Res. 110:589-597. [DOI] [PubMed] [Google Scholar]
- 59.Rodionov, D. A., A. A. Mironov, and M. S. Gelfand. 2002. Conservation of the biotin regulon and the BirA regulatory signal in Eubacteria and Archaea. Genome Res. 12:1507-1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rohwerder, T., T. Gehrke, K. Kinzler, and W. Sand. 2003. Bioleaching review part A: progress in bioleaching: fundamentals and mechanisms of bacterial metal sulfide oxidation. Appl. Microbiol. Biotechnol. 63:239-248. [DOI] [PubMed] [Google Scholar]
- 61.Saldanha, A. J. 2004. Java Treeview—extensible visualization of microarray data. Bioinformatics 20:3246-3248. [DOI] [PubMed] [Google Scholar]
- 62.Sand, W., K. Rohde, B. Sobotke, and C. Zenneck. 1992. Evaluation of Leptospirillum ferrooxidans for leaching. Appl. Environ. Microbiol. 58:85-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sawers, R. G. 2005. Formate and its role in hydrogen production in Escherichia coli. Biochem. Soc. Trans. 33:42-46. [DOI] [PubMed] [Google Scholar]
- 64.Segal, G., M. Feldman, and T. Zusman. 2005. The Icm/Dot type-IV secretion systems of Legionella pneumophila and Coxiella burnetii. FEMS Microbiol. Rev. 29:65-81. [DOI] [PubMed] [Google Scholar]
- 65.Simmons, S. L., G. Dibartolo, V. J. Denef, D. S. Goltsman, M. P. Thelen, and J. F. Banfield. 2008. Population genomic analysis of strain variation in Leptospirillum group II bacteria involved in acid mine drainage formation. PLoS Biol. 6:e177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Simon, J. 2002. Enzymology and bioenergetics of respiratory nitrite ammonification. FEMS Microbiol. Rev. 26:285-309. [DOI] [PubMed] [Google Scholar]
- 67.Singer, S. W., C. S. Chan, A. Zemla, N. C. Verberkmoes, M. Hwang, R. L. Hettich, J. F. Banfield, and M. P. Thelen. 2008. Characterization of cytochrome 579, an unusual cytochrome isolated from an iron-oxidizing microbial community. Appl. Environ. Microbiol. 74:4454-4462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sone, N., G. Sawa, T. Sone, and S. Noguchi. 1995. Thermophilic bacilli have split cytochrome b genes for cytochrome b6 and subunit IV. First cloning of cytochrome b from a gram-positive bacterium (Bacillus stearothermophilus). J. Biol. Chem. 270:10612-10617. [DOI] [PubMed] [Google Scholar]
- 69.Sowell, S. M., L. J. Wilhelm, A. D. Norbeck, M. S. Lipton, C. D. Nicora, D. F. Barofsky, C. A. Carlson, R. D. Smith, and S. J. Giovanonni. 2009. Transport functions dominate the SAR11 metaproteome at low-nutrient extremes in the Sargasso Sea. ISME J. 3:93-105. [DOI] [PubMed] [Google Scholar]
- 70.Stephens, B. B., S. N. Loar, and G. Alexandre. 2006. Role of CheB and CheR in the complex chemotactic and aerotactic pathway of Azospirillum brasilense. J. Bacteriol. 188:4759-4768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Strous, M., E. Pelletier, S. Mangenot, T. Rattei, A. Lehner, M. W. Taylor, M. Horn, H. Daims, D. Bartol-Mavel, P. Wincker, V. Barbe, N. Fonknechten, D. Vallenet, B. Segurens, C. Schenowitz-Truong, C. Medigue, A. Collingro, B. Snel, B. E. Dutilh, H. J. Op den Camp, C. van der Drift, I. Cirpus, K. T. van de Pas-Schoonen, H. R. Harhangi, L. van Niftrik, M. Schmid, J. Keltjens, J. van de Vossenberg, B. Kartal, H. Meier, D. Frishman, M. A. Huynen, H. W. Mewes, J. Weissenbach, M. S. Jetten, M. Wagner, and D. Le Paslier. 2006. Deciphering the evolution and metabolism of an anammox bacterium from a community genome. Nature 440:790-794. [DOI] [PubMed] [Google Scholar]
- 72.Szurmant, H., and G. W. Ordal. 2004. Diversity in chemotaxis mechanisms among the bacteria and archaea. Microbiol. Mol. Biol. Rev. 68:301-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tabb, D. L., W. H. McDonald, and J. R. Yates III. 2002. DTASelect and Contrast: tools for assembling and comparing protein identifications from shotgun proteomics. J. Proteome Res. 1:21-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Toor, N., G. Hausner, and S. Zimmerly. 2001. Coevolution of group II intron RNA structures with their intron-encoded reverse transcriptases. RNA 7:1142-1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tyson, G. W., J. Chapman, P. Hugenholtz, E. E. Allen, R. J. Ram, P. M. Richardson, V. V. Solovyev, E. M. Rubin, D. S. Rokhsar, and J. F. Banfield. 2004. Community structure and metabolism through reconstruction of microbial genomes from the environment. Nature 428:37-43. [DOI] [PubMed] [Google Scholar]
- 76.Tyson, G. W., I. Lo, B. J. Baker, E. E. Allen, P. Hugenholtz, and J. F. Banfield. 2005. Genome-directed isolation of the key nitrogen fixer Leptospirillum ferrodiazotrophum sp. nov. from an acidophilic microbial community. Appl. Environ. Microbiol. 71:6319-6324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Umeno, D., A. V. Tobias, and F. H. Arnold. 2005. Diversifying carotenoid biosynthetic pathways by directed evolution. Microbiol. Mol. Biol. Rev. 69:51-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Valdes, J., F. Veloso, E. Jedlicki, and D. Holmes. 2003. Metabolic reconstruction of sulfur assimilation in the extremophile Acidithiobacillus ferrooxidans based on genome analysis. BMC Genomics 4:51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.van der Oost, J., W. M. de Vos, and G. Antranikian. 1996. Extremophiles. Trends Biotechnol. 14:415-417. [Google Scholar]
- 80.VerBerkmoes, N. C., M. B. Shah, P. K. Lankford, D. A. Pelletier, M. B. Strader, D. L. Tabb, W. H. McDonald, J. W. Barton, G. B. Hurst, L. Hauser, B. H. Davison, J. T. Beatty, C. S. Harwood, F. R. Tabita, R. L. Hettich, and F. W. Larimer. 2006. Determination and comparison of the baseline proteomes of the versatile microbe Rhodopseudomonas palustris under its major metabolic states. J. Proteome Res. 5:287-298. [DOI] [PubMed] [Google Scholar]
- 81.Washburn, M. P., D. Wolters, and J. R. Yates III. 2001. Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nat. Biotechnol. 19:242-247. [DOI] [PubMed] [Google Scholar]
- 82.Wilmes, P., A. F. Andersson, M. G. Lefsrud, M. Wexler, M. Shah, B. Zhang, R. L. Hettich, P. L. Bond, N. C. VerBerkmoes, and J. F. Banfield. 2008. Community proteogenomics highlights microbial strain-variant protein expression within activated sludge performing enhanced biological phosphorus removal. ISME J. 2:853-864. [DOI] [PubMed] [Google Scholar]
- 83.Wilmes, P., J. P. Remis, M. Hwang, M. Auer, M. P. Thelen, and J. F. Banfield. 2009. Natural acidophilic biofilm communities reflect distinct organismal and functional organization. ISME J. 3:266-270. [DOI] [PubMed] [Google Scholar]
- 84.Wilson, C., and A. J. Dombroski. 1997. Region 1 of [sigma]70 is required for efficient isomerization and initiation of transcription by Escherichia coli RNA polymerase. J. Mol. Biol. 267:60-74. [DOI] [PubMed] [Google Scholar]
- 85.Woyke, T., H. Teeling, N. N. Ivanova, M. Huntemann, M. Richter, F. O. Gloeckner, D. Boffelli, I. J. Anderson, K. W. Barry, H. J. Shapiro, E. Szeto, N. C. Kyrpides, M. Mussmann, R. Amann, C. Bergin, C. Ruehland, E. M. Rubin, and N. Dubilier. 2006. Symbiosis insights through metagenomic analysis of a microbial consortium. Nature 443:950-955. [DOI] [PubMed] [Google Scholar]
- 86.Xie, X., S. Xiao, Z. He, J. Liu, and G. Qiu. 2007. Microbial populations in acid mineral bioleaching systems of Tong Shankou Copper Mine, China. J. Appl. Microbiol. 103:1227-1238. [DOI] [PubMed] [Google Scholar]
- 87.Zemla, A., C. E. Zhou, T. Slezak, T. Kuczmarski, D. Rama, C. Torres, D. Sawicka, and D. Barsky. 2005. AS2TS system for protein structure modeling and analysis. Nucleic Acids Res. 33:W111-W115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zhang, Y., D. M. Wolfe, E. L. Pohlmann, M. C. Conrad, and G. P. Roberts. 2006. Effect of AmtB homologues on the post-translational regulation of nitrogenase activity in response to ammonium and energy signals in Rhodospirillum rubrum. Microbiology 152:2075-2089. [DOI] [PubMed] [Google Scholar]
- 89.Zybailov, B., A. L. Mosley, M. E. Sardiu, M. K. Coleman, L. Florens, and M. P. Washburn. 2006. Statistical analysis of membrane proteome expression changes in Saccharomyces cerevisiae. J. Proteome Res. 5:2339-2347. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.