Abstract
Speciation as the result of barriers to genetic exchange is the foundation for the general biological species concept. However, the relevance of genetic exchange for defining microbial species is uncertain. In fact, the extent to which microbial populations comprise discrete clusters of evolutionarily related organisms is generally unclear. Metagenomic data from an acidophilic microbial community enabled a genomewide, comprehensive investigation of variation in individuals from two coexisting natural archaeal populations. Individuals are clustered into species-like groups in which cohesion appears to be maintained by homologous recombination. We quantified the dependence of recombination frequency on sequence similarity genomewide and found a decline in recombination with increasing evolutionary distance. Both inter- and intralineage recombination frequencies have a log-linear dependence on sequence divergence. In the declining phase of interspecies genetic exchange, recombination events cluster near the origin of replication and are localized by tRNAs and short regions of unusually high sequence similarity. The breakdown of genetic exchange with increasing sequence divergence could contribute to, or explain, the establishment and preservation of the observed population clusters in a manner consistent with the biological species concept.
THE classification of organisms into species with shared traits and niche preferences has long been fundamental to the biological sciences, yet the very existence of definable bacterial and archaeal species has been brought into question by the advent of genome sequencing. Lateral gene transfer, for example, commonly blurs the distinction between species, making evolutionary lineages less clear (Bapteste et al. 2004). More recently, environmental surveys suggest vast microbial diversity (Venter et al. 2004; Tringe and Rubin 2005; Sogin et al. 2006), possibly implying the existence of a genetic continuum surrounding the small subset of species that have been isolated and characterized physiologically. In some cases, however, sequence clustering-based measures applied to genomic data from isolated microorganisms have enabled species definitions (Palys et al. 1997) that appear to yield results consistent with ecotype models proposed for microbial species (Cohan 1994a,b). To fully evaluate the extent to which microorganisms form clusters analogous to species, it is necessary to assess the degree to which their sequences form a continuum and to identify the forces that modulate this continuum: mutation, selection, and the form and frequency of genetic exchange among closely related individuals. Here we focus on homologous recombination because it is an important form of genetic exchange between closely related microorganisms (Lawrence 2002). Its breakdown, whether due to accumulation of genetic mutations or to sudden acquisition of new genes by lateral transfer, may be a key step in speciation.
To date, most experimental studies of bacterial and archaeal recombination have focused on a few isolated bacterial pathogens, where the rate of incorporation of DNA fragments into a single genomic locus has been measured. Results suggest that recombination is quite rare (Roberts and Cohan 1993; Cohan 2001) and that the frequency for a single gene (rpoB) has a log-linear dependence on sequence divergence (Roberts and Cohan 1993; Zawadzki et al. 1995; Majewski and Cohan 1999; Majewski et al. 2000). Consequently it has been suggested that recombination occurs too infrequently to prevent purging of diversity during selection events (Cohan 2001, 2002). However, recent multistrain comparisons using genomic data have documented much higher recombination rates in Escherichia coli (Wirth et al. 2006) than previously reported. In the case of bacterial Thermotoga isolates, recombination was suggested to be sufficiently extensive to invalidate the concept of a species boundary (Nesbø et al. 2006). Multilocus sequence typing (MLST) of Neisseria isolates also revealed high rates of homologous recombination, consistent with a “fuzzy” species (Hanage et al. 2005) much like quasi-species for viruses (Eigen and Schuster 1979; Eigen 1996). For archaea, characterization of Halorubrum sp. (Papke et al. 2004), Sulfolobus islandicus (Whitaker et al. 2005), and environmental populations (Tyson et al. 2004) indicates that recombination frequencies are fast enough to unlink loci and maintain diversity during periodic selection events. However, comprehensive genomewide analyses of the decline in recombination across a species boundary are lacking. Furthermore, the dependence of recombination frequency on sequence divergence has not been quantified for archaea.
When applied to low-complexity consortia, cultivation-independent shotgun genome sequencing techniques can be used to reconstruct near-complete composite microbial genome sequences (Tyson et al. 2004; Martin et al. 2006). Furthermore, because each sequence likely derives from a different individual, variation in sequences that contribute to the composite genome can be used to evaluate population heterogeneity (Tyson et al. 2004). In this article, we use community genomic data to (1) establish the existence of discrete archaeal sequence clusters in acid mine drainage biofilms and (2) investigate the ability of homologous recombination to distribute genetic information among individuals within clusters and between them.
MATERIALS AND METHODS
Study location:
Genomic sequence data were obtained from DNA extracted from a pink biofilm floating on pH 0.7 acid mine drainage and sampled from the five-way location within the Richmond Mine, near Redding, California (supplemental Figure S1 at http://www.genetics.org/supplemental/). As described previously (Tyson et al. 2004), the biofilm community was dominated by bacteria (Leptospirillum groups II and III) and four archaeal organisms from the Thermoplasmatales lineage.
Community genomic data:
Approximately 130 Mbp of shotgun sequence data (3-kb library) from DNA extracted from the biofilm were assembled using the phredPhrap program (Ewing and Green 1998). Assemblies were comparable to those generated using the first 76 Mb of sequence data assembled with the JAZZ program, as described by Tyson et al. (2004). Sequences in the expanded data set were grouped by organism type, as described for the initial 76 Mb of sequence data (Tyson et al. 2004). As noted previously (Tyson et al. 2004), assembly of all sequencing reads from the community generates genome fragments (scaffolds) from both Ferroplasma type I [similar to the Ferroplasma acidarmanus isolate (Edwards et al. 2000)] genome and Ferroplasma type II. These assembled Ferroplasma genome fragments were manually curated to resolve errors arising from repetitive elements. Some gaps were closed by allowing for insertion/deletion of single or small groups of genes in some, but not all individuals (strain variants).
Visualization of variability, cluster analysis, and identification of recombination:
Composite Ferroplasma types I and II genome fragments were assembled from reads from coexisting individuals with similar or identical sequence types and do not capture population-level heterogeneity. To understand the degree to which sequences from these organisms form clusters, it is necessary to determine the extent to which their sequencing reads form a continuum. Genomewide comparison of sequencing reads to each other and to both composite sequences provided a means to investigate the extent of the continuum.
All reads from the community genomic data set were trimmed to remove low-quality bases (below a Phred score of 20) and then aligned to all community scaffolds using BLASTN with a cutoff e-value of 10−75. Reads better assigned to composite sequences of organisms other than Ferroplasma types I and II were excluded from subsequent analyses. For analysis of clustering, all Ferroplasma types I and II reads were aligned to the F. acidarmanus isolate genome (Edwards et al. 2000) using a very low cutoff e-value (10−25). Some reads and parts of reads were not brought into this analysis, often because they are associated with genes not present in F. acidarmanus.
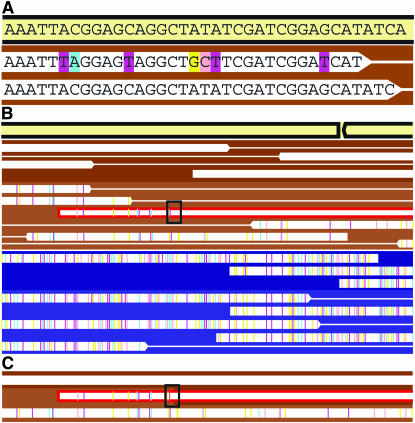
In Figure 1A, the BLASTN output has been converted into a graphical representation showing Ferroplasma type I and II sequence reads (white bars) aligned to a reference genome fragment. Colored ticks within each read represent single-nucleotide polymorphisms (SNPs) relative to the reference sequence. Cyan, pink, magenta, and yellow represent substitutions of the bases A, C, T, and G, respectively. Thinner white lines connect a read to its mate pair (the sequence from the opposite end of the same clone).
Figure 1.—
Diagram illustrating the representation of single-nucleotide polymorphisms (SNPs) that distinguish individual sequence variants within the community (white boxes signify sequencing reads, lines link mate pairs). Reads are aligned to a reference sequence that is either an isolate genome sequence or a composite sequence derived from a community genomic data set (cream-colored box). (A) Over the region shown one read has 7 SNPs relative to the reference sequence and the other is identical to it. Colored bars highlight the substituted base at each location; cyan, pink, magenta, and yellow represent substitutions of the bases A, C, T, and G, respectively. (B and C) The assembly is viewed at lower magnification without individual nucleotides labeled. The colors indicating SNPs are now small tick marks. In the region shown, sequencing reads can be easily clustered on the basis of SNP frequency. Backgrounds colored in shades of brown (similar to F. acidarmanus) and blue (similar to Ferroplasma type II scaffolds) indicate these groupings. However, within each cluster there is some fine-scale variation. In B the reads in the brown cluster are grouped into two sequence types (one with SNPs and one without SNPs). One read, outlined in red, represents a recombination of the two types. The recombinant read can be compared to the reconstructed parent sequences, as shown in C. Thus, the divergence between the parent sequences can be calculated for that recombination event.
Figure 1B illustrates a simpler rendition of data, used to examine larger genomic regions, than the one shown in Figure 1A (also see Figures 2–4). Background colors (shades of brown and violet) are used to highlight the separation of sequences into clusters based on SNP frequency (Figure 1B). For analysis of variation within these clusters, we compared reads with few SNPs (brown background in Figures 1–4 and in supplemental Figure S2 at http://www.genetics.org/supplemental/) to the very similar F. acidarmanus genome. The second cluster (violet background in Figures 1–4 and in supplemental Figure S2) has a composite sequence similar to that of the Ferroplasma type II composite genome. Consequently, for analysis of variation, these reads were aligned to the Ferroplasma type II composite genome fragments. Within clusters corresponding to the Ferroplasma type I and II populations, distinct variant types could be reconstructed on the basis of patterns of SNPs. Variants within populations are illuminated using slightly different background colors (Figure 1B).
Figure 2.—
Images showing the clear separation of the Ferroplasma type I (brown) and Ferroplasma type II (blue) genomes. (A) Reads from the community genomic data set are aligned (using BLASTN) to 2.7 kb of the isolate genome of F. acidarmanus and displayed as described in Figure 1. Reads are grouped (indicated by background shades of blue and brown) on the basis of conserved SNP patterns. Genes encoded on this fragment (bars on top from left to right) are a hypothetical protein, a putative dihydroxy-acid dehydratase, and a putative acetolactate synthase (large subunit). (B) The genomewide distribution of read sequence alignments against the F. acidarmanus genome. Every 3000 bases, all overlapping reads were assigned to one or the other Ferroplasma type and the divergence of each read from F. acidarmanus was calculated. At each point, for each type, the 5, 25, 33, 66, 75, and 95% divergence quantiles were calculated. These points were then connected to map the distributions of sequence identity genomewide. Lightly shaded regions span the 5–95% quantiles; darker regions, the 25–75% quantiles; and the darkest region, the 33–66% quantiles.
Figure 4.—
Intraspecies recombination in (A) Ferroplasma type I and (B) Ferroplasma type II. Figures are rendered as described in Figure 1. F. acidarmanus is used as a reference sequence in A and Ferroplasma type II composite sequence in B. Reads are grouped (indicated by background shades of blue and brown) into strains on the basis of conserved SNP patterns. Red outlines indicate reads containing inferred recombination points and black rectangles indicate apparent recombination end points within reads. Mate pairs assigned to different strain types are connected by a diagonal red line to highlight recombination events between strain types.
Figure 3.—
Interspecies recombination illustrated (as described in Figure 1) using the Ferroplasma type II composite genome as the reference. (A) In the region displayed, most of the reads from Ferroplasma type I (bottom cluster) share their sequence type with one of the Ferroplasma type II strains (top cluster). In A, all SNPs are displayed as gray ticks instead of using coded colors, as in Figure 1. (B) A more detailed view of the right side of A. A shaded vertical bar in B indicates a small region (∼80 bp) showing unusually high sequence similarity between the Ferroplasma types.
Some reads show discrepancies in linkage patterns. For example, the read outlined in red in Figure 1B can be split into two regions where, on the basis of SNP patterns, each region belongs to a different variant type. These mosaic patterns of SNPs, which can be viewed as a skewed distribution of polymorphic sites, are evidence of past recombination (Sawyer 1989). A small black box indicates cases where the recombination point occurs within a read (the transition between sequence types within the read is evident from the pattern of SNPs) (Smith 1999). When reads of a mate pair have different strain types, a recombination point is inferred to be present in the unsequenced part of the clone.
Sequence types related by a recombination event can be identified and reconstructed from overlapping reads (Figure 1C). For the purpose of calculating the nucleotide divergence between sequences related by a recombination event, those sequences that represent the dominant linkage pattern types were defined as “parental” (Figure 1, B and C).
Despite the general separation of sequences into the two distinct Ferroplasma types, regions exist where linkage patterns inferred from sequence similarity indicate recombination between Ferroplasma types I and II. To quantify this form of genetic exchange genomewide, we identified recombination points by investigating the BLASTN (Altschul et al. 1990) alignments of the clones that could not be clearly assigned to one or the other population.
Estimation of recombination frequency as a function of sequence divergence:
We quantitatively evaluated the dependence of recombination frequency on sequence divergence. For each clone showing intra- or interpopulation genetic exchange, we calculated, for the span of the recombinant clone, the nucleotide sequence divergence between the reconstructed parental sequences (as shown in Figure 1C). The full length of the recombinant clone was considered because, since only one recombination point is visible, there is no clear indication as to which end of the clone contains the introduced sequence. In some cases, the comparison region was reduced because one or both of the parental strain fragments did not span the full length of the recombinant clone.
Recombinant clones were counted in increments of 1% sequence divergence for intrapopulation data and 3% for interpopulation data. However, the frequency with which recombination events will be detected at any value of sequence divergence will be determined in part by how common that level of sequence divergence is across the genomes. Specifically, few recombination events are expected at high divergence due to a small fraction of the genomes showing these high divergence levels. Consequently, the numbers of recombination events as a function of sequence divergence (recombination frequencies for defined sequence divergence intervals) were normalized to account for the nonuniform distribution of sequence divergence. The distribution of sequence divergence was determined from sequence comparisons involving all reads.
Relatively few recombination events can be detected below 2% divergence because sequencing errors and recent mutations obscure already subtle differences between strain types. Due to the rarity of highly divergent variants, significant noise levels are anticipated for these cases. For these reasons, we restricted our analyses to divergences between 3 and 11%. Results were fit to an exponential model using maximum-likelihood estimation in the program R (Nelder and Mead 1965; R Development Core Team 2006). Errors in raw recombinant counts were assumed to be normal and were estimated by the maximum-likelihood optimization process.
Quantifying rates of recombination relative to mutation:
The ratio of recombination frequency to mutation rate (ρ/θ) gives an indication of how much more or less likely it is that any nucleotide will change due to recombination than due to mutation (Feil et al. 1999). Because our data differ from those published previously, we derived a new method of calculating this probability. The number of recombinant clones identified (as described above) was used as a proxy for the number of recombination events in the data set. When foreign sequence replaces existing DNA in an organism, only a fraction of positions involved will change (determined by sequence similarity, 2% for intrapopulation recombination events and 18% for interpopulation events). The observed recombination event frequency, r0, is multiplied by the estimated average recombination block size, h, and the average divergence between sequence types (as a proportion of nucleotides that are different), d, to estimate how often a single base was changed by recombination. The product r0dh is divided by the estimated mutation rate, μ0 (the average nucleotide divergence within sequence types), to give the relative frequency of recombination to mutation:
 |
(1) |
We used Watterson's segregation site-based calculation of the mutation rate to estimate θ (Watterson 1975). This method assumes no recombination, so the calculation was performed on each strain and averaged over the entire data set.
If interpopulation recombination events are rare and widely separated around the genome, the recombination block size can be determined so long as the event is evident in the majority of cells sampled and the sequence transition is captured at both ends of the block. The average block size for interpopulation recombination was estimated from these observations.
Intrapopulation, determination of the recombinant block size may be more problematic because the blocks are likely to overlap. We used several models that made use of the interpopulation observations and the frequency with which two recombination end points were detected in a single clone to estimate this value.
Given that a block of size x is detected, the probability that it lies entirely within a clone of size C is
 |
(2) |
We know the average clone size, C, to be 3.3 kb. Multiplying the above probability by the probability of having a recombination block of size x and summing that product over all possible block sizes gives the expected value of r. Working backward we can estimate the average recombination block size for a given (assumed) distribution of recombination block sizes. In model 1, we performed this calculation assuming a uniform distribution of sizes ranging from 0 to twice the average block size.
In model 2, we calculated the average block size by assuming a normal distribution of block sizes. Eight direct observations of interpopulation recombination block sizes were resampled with replacement 1 million times. Intrapopulation average block size was then estimated for all variances found in these bootstrapped data.
In model 3, we calculate the average length of sequence per observed recombination event. We compare the estimates of all three models with the block size observed for interpopulation events to estimate the likely range of average block sizes for intrapopulation recombination.
Fluorescence in situ hybridization:
Specific oligonucleotide probes were designed to target the 23S rRNA in Ferroplasma type I (fer1_23-850, CCTCACTAGACATCTCC) and Ferroplasma type II (fer2_23-242, CGATCGCCTTTAATCACGG) as previously described (Hugenholtz et al. 2002) and took into consideration accessibility of the target region (Fuchs et al. 2001). Probes were commercially synthesized and 5′ labeled with the fluorochrome fluorescein isothiocyanate (FITC), CY3, or CY5 (Operon, Alameda, CA). Paraformaldehyde-fixed samples were analyzed with both Ferroplasma probes and used in combination with general, bacterial (EUB338mix), and archaeal (ARC915) probes at optimal hybridization and wash conditions, to qualitatively determine the abundance of Ferroplasma type I and Ferroplasma type II. The optimal stringency for the probes was determined empirically using fixed AMD samples and 5% formamide increments from 10 to 50%. F. acidarmanus was used as a positive control for fer1_23-850 and a negative control for fer2_23-242. A Leica DM RX microscope was used for visualization of FISH preparations.
RESULTS
Comparative genomics of composite genome sequences:
On the basis of the number of clones analyzed and an estimate of the number of cells sampled, we obtained one clone for every 1000–10,000 Ferroplasma types I and II individuals present in the sample. A total of 24,184 clones were assigned to Ferroplasma type II (sampling depth of ∼22×) and 6820 clones were assigned to Ferroplasma type I (sampling depth of ∼6×). An additional 1027 clones could not be uniquely assigned to either population. Assembled contiguous DNA fragments (contigs) assigned to Ferroplasma type II were further assembled into a composite genome with only a small number of internal gaps. Assembly made use of mate pair information (i.e., through placement of paired sequences from the ends of clones on separate contigs; supplemental Table S1 at http://www.genetics.org/supplemental/) and accommodated the effects of heterogeneity in gene content within populations. In the refined composite Ferroplasma type II genome, the previously reported JAZZ assembly fragments have been ordered and linked (Tyson et al. 2004). Some small fragments representing strain variants are not included in the composite sequence and were not included in most analyses. The reconstructed scaffolds assigned to Ferroplasma type I contain a 16S rRNA gene identical to the gene in the F. acidarmanus isolate genome (Edwards et al. 2000). These scaffolds cover 93% of the isolate genome and share 98% nucleotide-level sequence identity. For all subsequent analyses, the F. acidarmanus isolate genome was used in place of the composite sequences as the reference for alignments.
The 16S rRNA gene sequence of Ferroplasma type II shares 99.2% identity with Ferroplasma type I (Tyson et al. 2004). The Ferroplasma type II genome encodes 1979 open reading frames (ORFs), compared to 1971 ORFs in the F. acidarmanus genome (Allen et al. 2007). The genomes share 1343 orthologs with an average ∼83% sequence identity. Gene order of the two composite genomes is largely conserved, with an average block size of ∼8 genes (see supplemental Table S1). There are 649 genes in the Ferroplasma type II genome absent in the F. acidarmanus genome compared to 610 in the F. acidarmanus (and a similar number in Ferroplasma type I) genome not present in the Ferroplasma type II genome (Edwards et al. 2000).
Clustering of sequences and evidence of recombination:
Sorting of reads based on sequence variation confirmed the division of Ferroplasma sequence into two genomic clusters that are generally represented by the original Ferroplasma type II JAZZ scaffolds and F. acidarmanus genome. Figure 2A (BLASTN e-value <10−25) clearly illustrates this separation across a 3-kb window. Comprehensive analysis showed that these clusters of reads occur genomewide (Figure 2B), with an average nucleotide identity of 82% between Ferroplasma types I and II clusters and 98% identity within each Ferroplasma cluster.
Despite the existence of clear genomic clusters, the data indicate ongoing genetic exchange between Ferroplasma types I and II (e.g., Figure 3 and supplemental Figure S2 at http://www.genetics.org/supplemental/). Of the 1027 clones that could not be uniquely assigned to one or the other type, 709 derive from three regions where the sequence type is essentially the same in all sampled individuals in both populations. In two of these regions, the single sequence type suggests recombination was followed by a sweep that selected for only those genome types with the adaptive gene variant. The other region, containing an integrase and transposases, may be the result of phage insertion into both genomes. Detailed inspection confirmed that a further 171 clones show strong evidence of recombination between the two Ferroplasma species (e.g., Figure 3 and supplemental Figure S2) at 70 locations distributed across the genomes. Twenty-one of the identified locations were associated with transposable elements. In these cases, multiple reads align to the same part of a transposable element in one genome, but their mate pairs align to unassociated genes spread randomly throughout the other genome. Clones aligning to transposable elements were removed from consideration, leaving 139 clones showing evidence of 49 recombination events. Inclusion of the three identical regions brings the total observed recombination events to 52.
Within the two Ferroplasma populations we observed finer-scale clustering to form strain variant groups. Reads within groups share >99.5% nucleotide-level sequence identity whereas strain variants typically share ∼98% sequence identity. There is evidence of extensive recombination between strain variant sequence types. Every few thousand bases across the genomes, a single clone is found to transition from one variant type to another, indicating a population characterized by many mosaic genome types (Figure 4 and Tyson et al. 2004).
By analysis of all sequencing reads assigned to Ferroplasma type I, we identified 1293 (19%) clones showing recombination between strain variants. For Ferroplasma type II we identified 1278 (24%) recombinant clones in reads assigned to seven large genome fragments derived from around the genome (a total of 564,543 bp, sampling approximately the same number of reads as for the lower coverage Ferroplasma type I data set). Because recombination events become more difficult to identify as parental sequence types become more similar, the numbers of recombination events identified in the data sets for each population are minimums.
Decline in recombination with sequence divergence:
Intrapopulation recombination frequencies for both Ferroplasma types showed a clear log-linear relationship with sequence divergence (Figure 5). The coefficient of this relationship for Ferroplasma type I is 29 and for Ferroplasma type II is 44. Between Ferroplasma types, the relationship between the rate of recombination and sequence divergence is less clear due, at least in part, to the small number of recombination events found between populations. However, the data are consistent with log-linear dependence (Figure 5C).
Figure 5.—
Observed recombination rates plotted as a function of sequence divergence. Log-linear lines were fitted to data for Ferroplasma type I (A), Ferroplasma type II (B), and interspecies (C) using maximum-likelihood estimation. Red lines were plotted from 95% confidence values of parameters. Errors, vertical blue bars, are estimated from maximum-likelihood optimization assuming the same normal error distributions for all recombinant clone counts (before normalization). Values for divergences <3% (green circles in A and B) were excluded from estimation due to difficulty in identifying recombinations. (D) All three data sets superimposed with the average value for interspecies recombination.
Recombination rates relative to mutation:
The rate of recombination relative to mutation is a useful measure of the relative strengths of two of the major forces shaping the Ferroplasma populations. To calculate this rate, an estimate of the average length of transferred sequence, h, is needed. For interpopulation recombination events, we calculated h to be 9 kb on the basis of direct observation of eight recombination blocks (e.g., Figure 3 and supplemental Figure S2 at http://www.genetics.org/supplemental/). For intrapopulation recombination a direct measurement of h was not possible. We observed that ∼1 in 16 clones contained more than one recombination end point and used this value to estimate h, assuming a uniform (model 1) and a normal (model 2) distribution of sizes. We also estimated the recombination block size using the number of recombination points found relative to the total number of sequenced bases (model 3). In conclusion, we esti mate h to be in the range of 5000–10,000 kb (supplemental Figure S2).
Using the number of recombination events (recombination points/2) per base (5.4 × 10−5 for Ferroplasma type I and 5.1 × 10−5 for Ferroplasma type II), an estimate of the mutation rate (0.006 for both populations), and the above range of average recombination lengths, we infer that the probability that a base is changed by intrapopulation recombination rather than by mutation is between 2:1 and 4:1. On the basis of the 52 observed interpopulation recombination events, we determined that the relative frequency of base change within Ferroplasma type I or II due to interpopulation recombination compared to mutation is ∼1:25.
Locations of interpopulation recombination events:
We investigated the distribution of interspecies recombination points in both Ferroplasma genomes. A total of 77% of the events clustered within 500 genes of the origin of replication, where gene order is most conserved (Figure 6). This result suggests that breakdown of synteny further from the origin contributes to the reduction of homologous recombination (Kowalczykowski 2000). Interpopulation recombination events were often associated with regions of local high sequence similarity, such as shown in Figure 3B (vertical shading; also see supplemental Figure S3 at http://www.genetics.org/supplemental/), or tRNAs (Figure 6), supporting the conclusion that short regions of sequence similarity are a critical factor in initiating homologous recombination (Majewski and Cohan 1999). Five clones show evidence for interpopulation recombination within the 23S rRNA gene, a finding that highlights the potential for recombination to skew phylogenetic inferences made from such genes.
Figure 6.—
Circular diagram (origin of replication at the top) showing a genomewide comparison of Ferroplasma type II to F. acidarmanus. The five rings of data are, progressing inward, (1) Ferroplasma type I genome showing predicted gene sequence locations color coded according to functional category (see supplemental Table S2 at http://www.genetics.org/supplemental/), (2) orthologs between Ferroplasma types I and II colored by percentage of identity (from red for 100% identity through the spectrum of colors to pale blue), (3) reconstructed Ferroplasma type II genome (colored from blue to purple as distance around the genome) highlighting rearrangements and syntenous regions, (4) tRNAs in F. acidarmanus (gray), and (5) interpopulation recombination points (red).
DISCUSSION
Community genomics and population genetics:
Our study demonstrates that unique insights into population genetics of natural microbial systems can be provided by deeply sampled metagenomic data sets. Specifically, analysis of sequence clustering and the frequency of recombination within and between clusters helps to resolve whether Ferroplasma types I and II represent different species. Additionally, the dependence of recombination on sequence identity is found to be the same as in prior experimental studies.
Are Ferroplasma types I and II different microbial species?
Genomewide analysis of sequence heterogeneity reveals two generally discrete clusters, approximately represented by the assembled composite genome sequences of Ferroplasma types I and II. These clusters have only an 87% average nucleotide identity (ANI) genomewide, significantly smaller than the 94% ANI found to correspond with the 70% DNA–DNA hybridization threshold used as part of the standard bacterial species definition (Konstantinidis and Tiedje 2005). The significant difference in metabolic potential (>600 genes are unique to each Ferroplasma type) may indicate adaptation to different niches (Coleman et al. 2006). Direct observation of phenotypic differences is not possible because Ferroplasma type II has not yet been obtained in pure culture. However, fluorescence in situ hybridization using lineage-specific probes for both Ferroplasma types suggests that the relative abundance of each can vary considerably and independently in samples from different environments (data not shown). On the basis of these findings—sequence clustering, the different gene content of these clusters, and evidence suggesting different environmental selection for the two organism types—we infer that these clusters effectively correspond to two species populations.
A general rule for recombination rates:
Our results demonstrate that the rate of genetic exchange in coexisting archaea in a natural microbial community has a log-linear dependence on sequence similarity, genomewide. This finding is consistent with experimental data for a single locus studied in isolated bacteria (Roberts and Cohan 1993; Zawadzki et al. 1995). Thus, as inferred for some isolated bacteria (Roberts and Cohan 1993), the recombination rate in archaea may also be primarily determined by the stability of the heteroduplex complex. Consequently, the log-linear relationship is far more general than previously established and may thus control on the distribution of diversity within and between population clusters.
When divergence within populations becomes sufficiently large, it can be inferred from the general log-linear dependence of recombination rates on sequence divergence that recombination rates will no longer be fast enough to spread newly acquired functions to all members prior to a selection event. In this case, it is tempting to suggest that the ancestral Ferroplasma population could have sympatrically diverged into separate species.
Fraser et al. (2007) recently used simulations to suggest that, under a model free of selection, such a scenario is possible only for organisms where the relationship between recombination and divergence is very steep (coefficient of ∼300). Although the coefficients found in this study (29 and 44) are higher than most of those previously reported (∼8–25) (Roberts and Cohan 1993; Vulic et al. 1997; Majewski et al. 2000), they are far short of this requirement. Thus, in the absence of a more sophisticated model incorporating the effects of selection, the divergence of these species may have involved some degree of physical separation.
It is perhaps not a coincidence that rates for interpopulation recombination involving the most similar sequences are lower than rates for intrapopulation events with the same sequence divergence (Figure 5D, 7–10% nucleotide divergence). These lower rates do suggest the presence of external barriers to genetic exchange. The mechanism of genetic exchange in extremely acidophilic natural archaeal populations is unknown. Uptake of free DNA from the environment is likely inhibited by its acid-promoted degradation. Genetic exchange may involve formation of cytoplasmic bridges, as described for Haloferax volcanii (Tchelet and Mevarech 1989), a conjugative mechanism documented in Sulfolobus species (Schleper et al. 1995; Grogan 1996), or vesicle-mediated genetic transfer (lipofection) (Felgner et al. 1987). For all of these mechanisms, increased physical separation due to partitioning of Ferroplasma types I and II into different microniches within the same biofilm would decrease the incidence of recombination.
The effect of short regions of similarity:
Most interspecies recombination events contained or were located near short regions of high sequence similarity that may affect the analysis of the relationship between recombination rate and sequence divergence. These short sequences are significantly shorter than the regions over which sequence similarity was measured (the length of the clone, usually ∼3 kb, see above). As a result, the measured sequence similarity will be much lower than in the short similar regions that may have contributed to the initiation of recombination. The lower slope (∼7) for the relationship between inter- vs. intrapopulation recombination and sequence divergence (Figure 5) may partly reflect this effect.
High recombination rates:
Our estimate that recombination is two to four times more likely than mutation to alter a single nucleotide is consistent with some recent observations finding higher rates of recombination than once thought, particularly among archaea (Papke et al. 2004; Whitaker et al. 2005). However, high recombination rates do not appear to be universal in archaea (Hallam et al. 2006). Thus frequent recombination within Ferroplasma populations is likely the result of many ecological and genetic factors. For example, like most archaea, both Ferroplasma types I and II lack the well-known mutS and mutL DNA mismatch repair systems and the alternative systems they employ have unknown influence on the relative rates of mutation and recombination.
Implications for species concepts:
The foundation for the biological species concept in eukaryotes is sexual reproduction. In bacteria and archaea, genetic exchange is not necessarily linked to the process of reproduction and can occur between distantly related organism types (Cohan 2002). Accordingly, the applicability of a biological species concept to bacteria and archaea is in question (Lawrence 2002; Nesbø et al. 2006). The reduced rate of genetic exchange seen between recently diverged Ferroplasma types I and II relative to the high rates within each population provides support for the concept that the breakdown of homologous recombination in these archaea serves as a species boundary. We infer a significant difference in the importance of recombination vs. mutation in sequence change within “species” populations (in the range of 2:1 to 4:1) compared to between them (∼1:25). This difference indicates that evolutionary trajectories are much more tightly linked within species groups than between them. Thus, individual populations are diverging.
Recombination between distinct organisms generates mosaic genomes with inconsistent phylogenetic affiliations at different loci around the genome, and it has been suggested that this can preclude meaningful species designations (Nesbø et al. 2006). If large fractions of genomes have inconsistent affiliations, we concur that species designations are not appropriate. This can occur if recombination is very frequent (as is observed within our Ferroplasma populations but not between them) or if recombination is rare but the block sizes are large. Consequently, it may be important to evaluate not just whether interorganism recombination occurs, but also how pervasive it is. We show that this can be achieved using comprehensive genomewide analyses made possible by deeply sampled community genomic data sets. In the case of these Ferroplasma populations (in which ∼2.6% of the genome recently participated in interpopulation recombination), a biological species definition based on the frequency of genetic exchange may be reasonable. This may not be an isolated case because, in many systems, recombination frequencies decrease in a systematic manner as sequence divergence increases. Increasing sequence divergence and sexual isolation will eventually ensure metabolic and ecological differences and distinct species identities.
Acknowledgments
We thank E. Allen, V. Denef, S. Simmons, K. Konstantinidis, P. Hugenholtz, and R. J. Whitaker for helpful comments and T. W. Arman, President, Iron Mountain Mines, R. Carver, and R. Sugarek for site access and on-site assistance. The Joint Genome Institute is thanked for providing DNA sequencing. Funding was provided by the National Science Foundation Biocomplexity Program and by grants to J.F.B. from the National Aeronautics and Space Administration and the Department of Energy Genomics:GTL project. W.M.G. was supported in part by a James S. McDonnell Foundation 21st Century Science Initiative Award.
Sequence data from this article have been deposited with the DDBJ/EMBL/GenBank Data Libraries under accession nos. AADL01000000 and AADL00000000.
References
- Allen, E. E., G. W. Tyson, R. J. Whitaker, J. C. Detter, P. M. Richardson et al., 2007. Genome dynamics in a natural archaeal population. Proc. Natl. Acad. Sci. USA 104: 1883–1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul, S. F., W. Gish, W. Miller, E. W. Meyers and D. J. Lipman, 1990. Basic local alignment search tool. J. Mol. Biol. 215: 403–410. [DOI] [PubMed] [Google Scholar]
- Bapteste, E., Y. Boucher, J. Leigh and W. F. Doolittle, 2004. Phylogenetic reconstruction and lateral gene transfer. Trends Microbiol. 12: 406–411. [DOI] [PubMed] [Google Scholar]
- Cohan, F. M., 1994. a Genetic exchange and evolutionary divergence in prokaryotes. Trends Ecol. Evol. 9: 175–180. [DOI] [PubMed] [Google Scholar]
- Cohan, F. M., 1994. b The effects of rare but promiscuous genetic exchange on evolutionary divergence in prokaryotes. Am. Nat. 143: 965–986. [DOI] [PubMed] [Google Scholar]
- Cohan, F. M., 2001. Bacterial species and speciation. Syst. Biol. 50: 513–524. [DOI] [PubMed] [Google Scholar]
- Cohan, F. M., 2002. Sexual isolation and speciation in bacteria. Genetica 116: 359–370. [PubMed] [Google Scholar]
- Coleman, M. L., M. B. Sullivan, A. C. Martiny, C. Steglich, K. Barry et al., 2006. Genomic islands and the ecology and evolution of prochlorococcus. Science 311: 1768–1770. [DOI] [PubMed] [Google Scholar]
- Edwards, K. J., P. L. Bond, T. M. Gihring and J. F. Banfield, 2000. An archaeal iron-oxidizing extreme acidophile important in acid mine drainage. Science 287: 1796–1799. [DOI] [PubMed] [Google Scholar]
- Eigen, M., 1996. On the nature of virus quasispecies. Trends Microbiol. 4: 216–218. [DOI] [PubMed] [Google Scholar]
- Eigen, M., and P. Schuster, 1979. The Hypercycle. Springer-Verlag, New York.
- Ewing, B., and P. Green, 1998. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 8: 186–194. [PubMed] [Google Scholar]
- Feil, E. J., M. C. Maiden, M. Achtman and B. G. Spratt, 1999. The relative contributions of recombination and mutation to the divergence of clones of neisseria meningitidis. Mol. Biol. Evol. 16: 1496–1502. [DOI] [PubMed] [Google Scholar]
- Felgner, P. L., T. R. Gadek, M. Holm, R. Roman, H. W. Chan et al., 1987. Lipofection: a highly efficient, lipid-mediated DNA-transfection procedure. Proc. Natl. Acad. Sci. USA 84: 7413–7417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser, C., W. P. Hanage and B. G. Spratt, 2007. Recombination and the nature of bacterial speciation. Science 315: 476–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs, B. M., K. Syutsubo, W. Ludwig and R. Amann, 2001. In situ accessibility of escherichia coli 23S rRNA to fluorescently labeled oligonucleotide probes. Appl. Environ. Microbiol. 67: 961–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grogan, D. W., 1996. Exchange of genetic markers at extremely high temperatures in the archaeon sulfolobus acidocaldarius. J. Bacteriol. 178: 3207–3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallam, S. J., K. T. Konstantinidis, N. Putnam, C. Schleper, Y. Watanabe et al., 2006. Genomic analysis of the uncultivated marine crenarchaeote cenarchaeum symbiosum. Proc. Natl. Acad. Sci. USA 103: 18296–18301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanage, W., C. Fraser and B. Spratt, 2005. Fuzzy species among recombinogenic bacteria. BMC Biol. 3: 6. [DOI] [PMC free article] [PubMed]
- Hugenholtz, P., G. W. Tyson and L. L. Blackall, 2002. Design and evaluation of 16S rRNA-targeted oligonucleotide probes for fluorescence in situ hybridization. Methods Mol. Biol. 179: 29–42. [DOI] [PubMed] [Google Scholar]
- Konstantinidis, K. T., and J. M. Tiedje, 2005. Genomic insights that advance the species definition for prokaryotes. Proc. Natl. Acad. Sci. USA 102: 2567–2572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalczykowski, S. C., 2000. Initiation of genetic recombination and recombination-dependent replication. Trends Biochem. Sci. 25: 156–165. [DOI] [PubMed] [Google Scholar]
- Lawrence, J. G., 2002. Gene transfer in bacteria: Speciation without species? Theor. Popul. Biol. 61: 449–460. [DOI] [PubMed] [Google Scholar]
- Majewski, J., and F. M. Cohan, 1999. DNA sequence similarity requirements for interspecific recombination in bacillus. Genetics 153: 1525–1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majewski, J., P. Zawadzki, P. Pickerill, F. M. Cohan and C. G. Dowson, 2000. Barriers to genetic exchange between bacterial species: Streptococcus pneumoniae transformation. J. Bacteriol. 182: 1016–1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin, H. G., N. Ivanova, V. Kunin, F. Warnecke, K. W. Barry et al., 2006. Metagenomic analysis of two enhanced biological phosphorus removal (EBPR) sludge communities. Nat. Biotechnol. 24: 1263–1269. [DOI] [PubMed] [Google Scholar]
- Nelder, J. A., and R. Mead, 1965. A simplex-method for function minimization. Comput. J. 7: 308–313. [Google Scholar]
- Nesbø, C. L., M. Dlutek and W. F. Doolittle, 2006. Recombination in thermotoga: implications for species concepts and biogeography. Genetics 172: 759–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palys, T., L. K. Nakamura and F. M. Cohan, 1997. Discovery and classification of ecological diversity in the bacterial world: the role of DNA sequence data. Int. J. Syst. Bacteriol. 47: 1145–1156. [DOI] [PubMed] [Google Scholar]
- Papke, T. R., J. E. Koenig, F. Rodriguez-Valera and F. W. Doolittle, 2004. Frequent recombination in a saltern population of halorubrum. Science 306: 1928–1929. [DOI] [PubMed] [Google Scholar]
- R Development Core Team, 2006. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna.
- Roberts, M. S., and F. M. Cohan, 1993. The effect of DNA sequence divergence on sexual isolation in bacillus. Genetics 134: 401–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawyer, S., 1989. Statistical tests for detecting gene conversion. Mol. Biol. Evol. 6: 526–538. [DOI] [PubMed] [Google Scholar]
- Schleper, C., I. Holz, D. Janekovic, J. Murphy and W. Zillig, 1995. A multicopy plasmid of the extremely thermophilic archaeon sulfolobus effects its transfer to recipients by mating. J. Bacteriol. 177: 4417–4426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, J. M., 1999. The detection and measurement of recombination from sequence data. Genetics 153: 1021–1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sogin, M. L., H. G. Morrison, J. A. Huber, D. M. Welch, S. M. Huse et al., 2006. Microbial diversity in the deep sea and the underexplored “rare biosphere”. Proc. Natl. Acad. Sci. USA 103: 12115–12120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tchelet, R., and M. Mevarech, 1989. The mechanism of DNA transfer in the mating system of an archaebacterium. Science 245: 1387–1389. [DOI] [PubMed] [Google Scholar]
- Tringe, S. G., and E. M. Rubin, 2005. Metagenomics: DNA sequencing of environmental samples. Nat. Rev. Genet. 6: 805–814. [DOI] [PubMed] [Google Scholar]
- Tyson, G. W., J. Chapman, P. Hugenholtz, E. E. Allen, R. J. Ram et al., 2004. Community structure and metabolism through reconstruction of microbial genomes from the environment. Nature 428: 37–43. [DOI] [PubMed] [Google Scholar]
- Venter, C. C., K. Remington, J. F. Heidelberg, A. L. Halpern, D. Rusch et al., 2004. Environmental genome shotgun sequencing of the sargasso sea. Science 304: 66–74. [DOI] [PubMed] [Google Scholar]
- Vulic, M., F. Dionisio, F. Taddei and M. Radman, 1997. Molecular keys to speciation: DNA polymorphism and the control of genetic exchange in enterobacteria. Proc. Natl. Acad. Sci. USA 94: 9763–9767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watterson, G. A., 1975. On the number of segregating sites in genetical models without recombination. Theor. Popul. Biol. 7: 256–276. [DOI] [PubMed] [Google Scholar]
- Whitaker, R. J., D. W. Grogan and J. W. Taylor, 2005. Recombination shapes the natural population structure of the hyperthermophilic archaeon sulfolobus islandicus. Mol. Biol. Evol. 22: 2354–2361. [DOI] [PubMed] [Google Scholar]
- Wirth, T., D. Falush, R. Lan, F. Colles, P. Mensa et al., 2006. Sex and virulence in escherichia coli: an evolutionary perspective. Mol. Microbiol. 60: 1136–1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zawadzki, P., M. S. Roberts and F. M. Cohan, 1995. The log-linear relationship between sexual isolation and sequence divergence in bacillus transformation is robust. Genetics 140: 917–932. [DOI] [PMC free article] [PubMed] [Google Scholar]