Abstract
Analysis of assembled random shotgun sequence data from a low-diversity, subsurface acid mine drainage (AMD) biofilm revealed a single nif operon. This was found on a genome fragment belonging to a member of Leptospirillum group III, a lineage in the Nitrospirae phylum with no cultivated representatives. Based on the prediction that this organism is solely responsible for nitrogen fixation in the community, we pursued a selective isolation strategy to obtain the organism in pure culture. An AMD biofilm sample naturally abundant in Leptospirillum group III cells was homogenized, filtered, and serially diluted into a nitrogen-free liquid medium. The resulting culture in the terminal dilution grew autotrophically to a maximum cell density of ∼106 cells/ml, oxidizing ferrous iron as the sole energy source. 16S rRNA-internal transcribed spacer region clone library analysis confirmed that the isolate is a member of Leptospirillum group III and that the culture is axenic. We propose the name Leptospirillum ferrodiazotrophum sp. nov. for this iron-oxidizing, free-living diazotroph. This study highlights how environmental sequence data can provide insights for culturing previously uncultured microorganisms.
Random shotgun sequencing of environmental samples has enabled recovery of genes, and in some instances near-complete genomes, from microbial communities (28, 34, 35). In addition to information about population structure and diversity, these data also reveal the metabolic potential of constituent microbial species. Many of these species have not been cultivated, and insights into their metabolism may provide clues for cultivation strategies.
In a previous study, we assembled large genomic fragments of five microbial species from a simple subsurface biofilm growing in pH 0.8, 37°C, metal rich acid mine drainage (AMD) (34). These acidophilic biofilms are essentially self-sustaining chemolithoautotrophic communities that obtain their energy by oxidizing ferrous iron and reduced sulfur compounds. They receive limited fixed carbon and nitrogen from external sources and appear to obtain most of their carbon and nitrogen requirements by fixation of atmospheric CO2 and N2.
To date, the only acidophilic microorganisms demonstrated to be capable of nitrogen fixation are Acidithiobacillus ferrooxidans and Leptospirillum ferrooxidans (21). However, these bacteria have never been found in AMD communities growing at a pH of <1 (1, 27). It was anticipated that other Leptospirillum species present in the AMD biofilm (34) would be responsible for nitrogen fixation. The genus Leptospirillum has been divided into three groups—I, II, and III—on the basis of 16S rRNA gene phylogeny (6). Representatives of groups II and III were identified in the biofilm analyzed by community genomics (34). L. ferrooxidans is a representative of group I, L. ferriphilum is a representative of group II (10). However, no cultured representatives of Leptospirillum group III have been described.
The single nif operon detected in the AMD community genomic data was assigned to a genome fragment from the Leptospirillum group III population, a relatively minor member of the biofilm (<10%). Since a near-complete genome for the Leptospirillum group II community member was obtained, we inferred that this organism lacks nitrogen-fixing genes. This suggested a clear strategy for isolating the first cultivated representative of Leptospirillum group III based on the ability to carry out nitrogen fixation. The result highlights the potential of environmental genomic data to provide insights needed to bypass the cultivation bottleneck.
MATERIALS AND METHODS
Identification of nif operon and comparative genomics.
A complete nif operon was identified in the AMD community genomic data generated in a previous study (34). Assignment of the scaffold to the Leptospirillum group III population was originally based on GC content and read depth and was confirmed by oligonucleotide frequency analysis according to the method of Teeling et al. (33). Gaps in the scaffold were filled by using primers designed using Primer 3 (http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www.cgi), and the complete scaffold was manually annotated in Artemis (26). The nif operon and genes identified in Leptospirillum ferrooxidans that were upregulated under nitrogen starvation (22) were obtained from GenBank, and orthologs in the Leptospirillum group III genome fragments were identified by using BLASTP. All annotated NifH protein sequences for all publicly available genomes were also downloaded from GenBank (http://www.ncbi.nlm.nih.gov; 262 genomes) and used in subsequent phylogenetic analyses.
Sample collection and processing.
Samples of pink biofilms growing at pH 0.7 to 1.3 and at 37 to 42°C AMD were collected from Ultra-back A and C drifts of the Richmond Mine at Iron Mountain, Calif., in November 2003. For characterization by fluorescence in situ hybridization (FISH), samples were fixed in paraformaldehyde, as previously described (6), and stored at −20°C. Samples for nucleic acid extraction were stored at −80°C.
Nucleic acid extraction.
Nucleic acids were extracted from samples with lysozyme (2 mg/ml), and cellular proteins were digested by using a STEP buffer (sodium dodecyl sulfate [0.5%], 50 mM Tris-HCl [pH 7.5], 400 mM EDTA, proteinase K [1 mg/ml], and Sarkosyl [0.5%]) at 50°C for 30 min. After two phenol-chloroform extractions, the DNA was precipitated with isopropanol and sodium acetate, and resuspended in Tris-EDTA buffer.
FISH and bright-field microscopy.
FISH was used to screen biofilm samples for abundance of Leptospirillum group III. Probes were commercially synthesized and 5′ labeled with the fluorochrome fluorescein isothiocyanate, Cy3, or Cy5 (Interactiva, Ulm, Germany). Paraformaldehyde-fixed samples were probed with Leptospirillum group III-specific probes (LF1252) used in combination with general Leptospirillum (LF655), bacterial (EUB338mix), and archaeal (ARC915) probes under standard hybridization and wash conditions (6). Gram stains were also performed on cultures and environmental AMD biofilms. A Leica DM RX microscope was used for visualization of FISH preparations. Images were captured by using the Wasabi software package (Hamamatsu).
Isolation, growth conditions, and microscopic examination.
Liquid serial dilution was used in isolation attempts due to difficulties in preparing solid media and obtaining growth on plates at a pH of ≤1.2. Serial dilution has been successfully applied in the isolation of several other organisms (24, 29). One gram of biofilm (ca. 107 to 108 cells) from Ultra-back A was homogenized by using a 16G1 needle and syringe, filtered through a 0.7-μm GD/X syringe filter (Whatman, Inc.) to remove cell aggregates, and inoculated into a modified 9K media (31). Adaptations of this medium have been used to grow other Leptospirillum species, e.g., L. ferrooxidans (2) and L. ferriphilum (10). The medium was modified to be nitrogen-free by removal of ammonium sulfate [(NH4)2SO4] and calcium nitrate [Ca(NO3)2], the only sources of fixed nitrogen in the medium. In order to mimic the pH of the natural AMD biofilm, the nitrogen-free 9K medium was prepared at pH 1.2. The sample was serially diluted to 10−10. The modified medium contains ferrous sulfate (FeSO4 · 7H2O) as the primary energy source. All serial dilutions were grown in the dark at 37°C and shaken at 150 rpm for 21 days. Cultures were periodically examined microscopically using a 2× Live/Dead BacLight staining solution (Molecular Probes, Inc.). DNA extractions and FISH were performed on terminal cultures in SD1 and SD3. After an axenic population of cells was obtained, subcultures were grown both without a nitrogen source and in the presence of fixed nitrogen [23 mM (NH4)2SO4].
16S rRNA-ITS and nifH libraries.
The universal 16S rRNA gene primer 27F (18) was used in combination with the 23S rRNA gene primer 23S-115R (18) in order to amplify the 16S internal transcribed spacer (ITS) region from DNA extracted from cultures. General nested nifH (37) primers were used to amplify nifH genes from bulk DNA extracted from cultures and environmental biofilm samples. Each 50-μl reaction mixture contained (as the final concentration) 1× reaction buffer (Invitrogen), 1.5 mM MgCl2, 200 μM concentrations of each deoxynucleoside triphosphate, 0.3 U of Platinum Taq DNA polymerase (Invitrogen), ∼50 ng of genomic DNA, and 200 ng of forward and reverse primers for each gene. Thermal cycling was carried out on a Hybaid PCR Express Thermal Cycler with an initial denaturation step of 96°C for 10 min, followed by 30 cycles at 94°C for 1 min, 50°C for 1 min, and 72°C for 2 min, followed finally by an extension step of 72°C for 5 min. The PCR products were purified with a QIAGEN QiaQuick PCR purification kit and were cloned by using the TOPO TA Cloning System (Invitrogen). Complete 16S rRNA-ITS sequences were obtained from 10 clones from each library using broadly conserved sequencing primers (27F, 530F, 1100F, 1100R, 1492R, and 115R [18]). Partial nifH sequences were obtained from 20 clones from both environmental and isolate nifH gene libraries. All sequencing was performed by the University of California Berkeley DNA Sequencing Facility on Applied Biosystems 48 capillary 3730 or Applied Biosystems 16 capillary 3100 Genetic Analyzer Systems.
Phylogenetic analysis.
The compiled sequences were aligned by using the ARB software package (19), and alignments were refined manually. Phylogenetic trees based on comparative analysis of the 16S rRNA gene and NifH protein were constructed by using neighbor joining and maximum-likelihood methods. Bootstrap values were inferred by using parsimony in PAUP* 4.0 with default settings. Bayesian inferences were generated using MRBAYES (15). A consensus tree was constructed in PAUP* 4.0. Posterior probabilities of branching points were estimated by Bayesian inference using MRBAYES v2.01 (14) with the default settings and general time-reversible model (six substitution types [n = 6]) and a gamma rate correction (rates = gamma) suggested for RNA data. A total of 100,000 generations were calculated for the data set, from which a tree was saved every 100 generations. The likelihoods of the trees had converged on a stable value after 20,000 generations, and a consensus tree was constructed from the final 80,000 generations (800 trees) in PAUP* 4.0.
RESULTS
Comparative genomics.
A complete nif operon (nifHDKENX) was identified on a 21.9-kbp scaffold in an AMD biofilm community genomic data set generated in a previous study (34). No other nif operons were detected in this data set. Based on GC content and read depth it was concluded that the nif operon belongs to Leptospirillum group III (34). The environmental nif operon and surrounding genomic context is highly syntenous with that of Leptospirillum ferrooxidans, a member of Leptospirillum group I (Fig. 1). The genome of L. ferrooxidans has not yet been sequenced; however, genes involved in nitrogen fixation in this organism were identified by using a shotgun microarray approach (22). The Leptospirillum group III nifHDKENX genes that encode the structural components of Mo-Fe nitrogenase range from 73 to 94% identity at the amino acid level to their orthologs in L. ferrooxidans. Genes flanking the nif operon (Lfe199p1, Lfe123p2, and Lfe123p3) are inferred to have some involvement in nitrogen fixation (22) and show a higher degree of sequence divergence than the nif operon genes (Fig. 1).
FIG. 1.
Comparison of the nif operon identified in the AMD biofilm community genomic data and the nif operon of L. ferrooxidans L3.2 (22). Dashed lines indicate missing sequence in L. ferrooxidans. CHP, conserved hypothetical protein.
The Leptospirillum group III genome fragments also encode a number of other genes involved in nitrogen fixation. These include the nifSU-hesB-hscBA operon necessary for Fe-S cluster assembly in the nitrogenase complex; various regulatory genes; ntrBC (two-component regulatory system), nifA (the specific activator of nif genes), nifL (oxygen/redox sensor), and glnB-like genes; and transporter genes encoding a molybdate ABC transporter (modA and modB) and a putative ammonium transporter (amtB). Orthologs of these genes were identified in L. ferrooxidans, with the exceptions of ntrB and ntrC. Interestingly, several genes involved in nitrogen assimilation via the glutamine/glutamate synthetase (GS/GOGAT) pathway, a commonly used pathway in bacteria, appear to be absent in Leptospirillum (glnE and gltBD). This suggests that this genus may assimilate ammonia via a GS pathway similar to that proposed for A. ferrooxidans (25). However, this conclusion is tentative because neither genome is complete.
Although no nif operon genes were detected in the Leptospirillum group II environmental genome, some structural and regulatory genes known to be involved in nitrogen fixation are present, including a complete nifSU-hesB-hscBA operon, glnK (PII protein), nifA, and nifL. This observation suggests that Leptospirillum group II lost the nif operon after speciation.
Phylogeny of the nif operon found in the community genomic data.
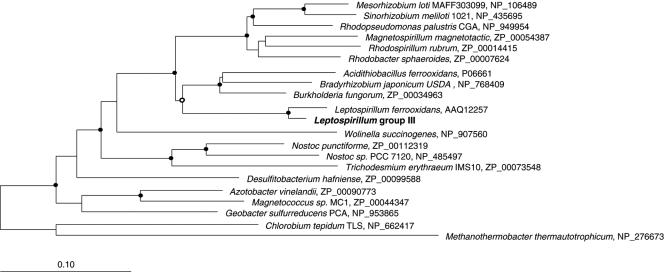
Comparative analysis of NifH and other conserved Nif proteins (not shown) indicate that the Leptospirillum species form a monophyletic group clustering within the proteobacterial radiation of the group I NifH proteins (Fig. 2). This suggests that Leptospirillum species may have obtained their nif operon via an ancient lateral gene transfer event involving a proteobacterial donor, possibly an ancestor of the acidophile Acidithiobacillus ferrooxidans. In addition, there is evidence of lateral gene transfer within proteobacterial classes (e.g., Bradyrhizobium, a member of the Alphaproteobacteria, clustering with Gammaproteobacteria [25]), suggesting that lateral transfer may have played a significant role in the evolution of nitrogen fixation (7).
FIG. 2.
Phylogenetic tree inferred from NifH protein identified in the AMD biofilm community (boldface) and reference isolate microbial genomes. Branch points supported (bootstrap values, >74%) by all inference methods used are indicated by solid circles, and those supported by most inference methods are indicated by open circles. Branch points without circles were not resolved (bootstrap values, <75%) as specific groups in different analyses. The bar represents 10% estimated sequence divergence. Outgroups are Chlorobium tepidum TLS (group II) and Methanothermobacter thermoautotrophicum (group III).
Directed isolation of a Leptospirillum group III representative.
The discovery of nitrogen fixation genes only in the Leptospirillum group III population suggested an obvious directed isolation strategy for this organism. AMD biofilm samples were screened by using FISH to identify samples with the highest relative abundance of Leptospirillum group III cells. The selected sample, Ultra-back A, was homogenized and serially diluted into nitrogen-free 9K liquid medium to obtain a pure culture. Cells actively grew in this medium and group-specific FISH confirmed that the actively growing population were members of Leptospirillum group III (Fig. 3). A final working culture of 106 Leptospirillum group III cells/ml was obtained by repeated serial dilution. Cells are gram-negative, crescent- to spirillum-shaped, approximately 0.5 μm by 2 to 3 μm, and motile by means of a single polar flagellum.
FIG. 3.
FISH images of the isolate. (A) Cells hybridized with Cy5-labeled EUB338mix; (B) FITC-labeled LF655 (Leptospirillum group I, II, and III); (C) Cy3-labeled LF1252 (only Leptospirillum group III); (D) combination of all three probes.
FIG. 4.
Maximum-likelihood dendrogram of Leptospirillum genera based on comparative analyses of 16S rRNA gene data. Group designations are bracketed on the right. Branch points supported (bootstrap values, >74%) by all inference methods used are indicated by solid circles, and those supported by most inference methods are indicated by open circles. Branch points without circles were not resolved (bootstrap values, <75%) as specific groups in different analyses. Shown in blue are the 16S rRNA gene sequence for each of the genomes recovered in the community genomics analysis (34) and in red is the 16S rRNA sequence of the Leptospirillum group III isolate. The outgroup used was Nitrospira marina. The bar represents 10% estimated sequence divergence.
Both in the presence and in the absence of fixed nitrogen, the medium changes from pale green to an orange-red color due to iron oxidation that was not observed in cell-free controls. Given that the only potential energy source in 9K medium is ferrous sulfate, active growth of the Leptospirillum group III culture confirms that this organism is capable of growth based solely on iron oxidation. The change in coloration occurred more quickly in the nitrogen-free media, indicating faster iron oxidation, possibly due to the increased need for energy for nitrogen fixation. Similar observations have been made for cultures of L. ferrooxidans (21). Since growth in all cultures occurred in the absence of a fixed carbon source, accompanied by production of ferric iron, we infer that the Leptospirillum group III culture is growing chemolithoautotrophically on ferrous iron.
Molecular characterization of isolate.
The 16S rRNA gene was directly PCR amplified from bulk DNA extracted from the terminal dilution in SD1 by using broad specificity primers and sequenced. Comparative analysis confirmed that the sequence represents a member of Leptospirillum group III (Fig. 4).
In order to check the purity of the serial dilution cultures, 16S rRNA-ITS clone libraries were prepared from the terminal dilutions in SD1 and SD3 and analyzed. Up to three nucleotide polymorphisms were detected in both the 16S rRNA and ITS regions in the SD1 library, suggesting the presence of multiple strains of a single species. By the third serial dilution (SD3), nearly all 16S-ITS sequences were identical. Only a single unduplicated polymorphism was found in the ITS region of a few clones. Since the incidence of polymorphisms is close to the expected error rate, we conclude that a pure culture has been obtained, referred to here as strain UBA1. The 16S rRNA gene sequence of the Leptospirillum group III organism identified in the AMD biofilm shotgun sequence data is very closely related but not identical to strain UBA1 (Fig. 4). We propose the name Leptospirillum ferrodiazotrophum sp. nov. (fer.ro.di.a.zo.tro′ phum. L. neut. n. ferrum iron, N.L. n. ferro- referring to ferrous ions, N.L. diazo- referring to dinitrogen, N.L. neut. adj. suffix -trophum living on, N.L. neut. adj. ferrodiazotrophum living on ferrous ions and dinitrogen.) for this isolate. This strain has been deposited in the ATCC culture collection (accession number ATCC BAA-1181).
Using broad specificity nifH primers, PCR clone libraries were constructed from both the UBA1 culture and Ultra-back A biofilm sample. The partial nifH gene (∼350 bp) amplified from the isolate was identical at the nucleotide level to the gene encoded on the Leptospirillum group III genome fragment. The Ultra-back A biofilm nifH genes were slightly divergent at the nucleotide level but identical at the amino acid level. This result suggests that Leptospirillum group III is the only detectable nitrogen fixer in this extremely acidic AMD environment. The absence of nifH in Leptospirillum group II was also confirmed by using general nifH primers on several group II enrichments (data not shown). However, it is possible that that some Leptospirillum group II members at other sites retain nitrogen fixation genes.
DISCUSSION
The ability of Leptospirillum group III to fix nitrogen provided an ideal selective advantage for the isolation of the first cultivated representative of this group, L. ferrodiazotrophum sp. nov. strain UBA1. Although acidophiles have been cultivated previously on plates using a novel overlay method at pH values of >1.6 (17), we were unable to prepare plates at the environmentally relevant conditions (pH 1.2). Consequently, our attempts to isolate Leptospirillum group III relied upon growth in liquid media. The disadvantage of liquid media is that discrete clonal colonies cannot be obtained and isolation of axenic cultures instead relies on dilution to extinction (8). Direct sequencing of 16S rRNA gene PCR products, without cloning, has been used previously to infer purity of cultures obtained via serial dilution (9, 24). However, in the present study, direct 16S rRNA gene sequencing hid the presence of multiple closely related strains after the first serial dilution. We found that multiple serial dilutions were required to ensure isolate purity.
Leptospirillum group III has only recently been recognized via culture-independent techniques and appears to be limited to acidic environments (1). Currently, there are nine full-length 16S rRNA gene sequences representing this group, with sequence divergence of up to 5%. This level of divergence suggests that Leptospirillum group III likely comprises multiple species of the same genus (32). Based on 16S rRNA gene sequence (Fig. 4), L. ferrodiazotrophum is closely related to organisms detected previously within the Richmond Mine at Iron Mountain (4, 5, 12), as well as a sequence reported from coal refuse (3).
The basic characteristics of L. ferrodiazotrophum strain UBA1 are consistent with its membership in the genus Leptospirillum (20), including morphology, motility, Gram stain, aerobic metabolism, acidophily, growth temperature, iron oxidation, and GC content. Growth of UBA1 on nitrogen-free media confirms the inference made from the environmental genomic data that members of Leptospirillum group III are nitrogen fixing chemolithoautotrophs (34). We attempted several acetylene reduction assays on UBA1 to directly demonstrate nitrogen fixation but were only able to detect picomolar amounts of ethylene production under low oxygen conditions (data not shown). This level of acetylene reduction is comparable to that reported previously for L. ferrooxidans (21). Expression of L. ferrodiazotrophum nif genes in natural AMD biofilms from the Richmond Mine has been confirmed in a recent shotgun proteome study (23a). Since nitrogenases are extremely oxygen sensitive and L. ferrodiazotrophum grows via iron oxidation coupled to reduction of molecular oxygen, the nitrogenase must be protected from oxygen. Fast respiration rates in combination with a high oxygen affinity cytochrome cbb3-type heme-copper oxidase (34) may be responsible for protecting the nitrogenase in UBA1, as has been reported for Azotobacter vinelandii (23).
Biological nitrogen fixation is an energetically expensive process, and growth in many natural environments is limited by the availability of fixed nitrogen. However, AMD microbial communities grow at the air-solution interface, in the presence of an unlimited supply of ferrous iron, their primary energy source. So long as the community contains an organism with genes necessary for nitrogen fixation, nitrogen supply may not be limiting. Leptospirillum group III is a minor component (<10%) of most AMD biofilms within the Richmond Mine. Due to this low abundance and the vital resource of fixed nitrogen that it provides, Leptospirillum group III can be considered a keystone species in this environment (34). The inferred loss of nitrogen fixation in the typically dominant Leptospirillum group II population supports this hypothesis. Removal of Leptospirillum group III would likely result in the collapse of the entire community. This is a potentially important observation, given that microbial communities in AMD systems control the rate of pyrite oxidation and thus AMD formation (30).
It is interesting to speculate as to why nitrogen fixation is strongly partitioned within the AMD biofilm communities. It is possible that the fitness of the community is enhanced by synergistic interactions among the dominant populations. For example, by analogy with rhizosphere communities, fixed nitrogen may be exported in return for certain metabolites (e.g., vitamins) or molecules involved in energy production or in return for aiding in the control of oxygen levels (11, 36). Synergistic interactions may be tested using future cultivation-b ased community assembly studies.
The isolation of members of lineages without cultivated representatives is a major challenge in microbiology (38). Directed isolation approaches have only used 16S rRNA-directed probes to track the progress of isolation attempts (14, 24) without prior knowledge of the targeted organisms physiology. Environmental genome data can provide detailed insight into the metabolic potential of uncultivated organisms. To date, genome sequences have only been used to optimize growth conditions of cultivated organisms such as Escherichia coli (13, 16). Although nitrogen fixation was an obvious basis for selective isolation, the cultivation of L. ferrodiazotrophum amply demonstrates the utility of environmental genome data for formulating isolation strategies for uncultivated organisms.
Acknowledgments
We thank Luis Rubio for help with the acetylene reduction assay, Jason Raymond for useful discussions about the phylogeny of the nif genes, H. G. Trüper for advice on etymology, and Mary Power for ecological insights. We thank Ted Arman and Richard Sugark for access to the Richmond Mine and Rudy Carver and Don Dodds for onsite assistance.
This research was funded by the U.S. Department of Energy Microbial Genome Program and the National Science Foundation Biocomplexity Program.
REFERENCES
- 1.Baker, B. J., and J. F. Banfield. 2003. Microbial communities in acid mine drainage. FEMS Microbiol. Ecol. 44:139-152. [DOI] [PubMed] [Google Scholar]
- 2.Battaglia, F., D. Morin, J. L. Garcia, and P. Ollivier. 1994. Isolation and study of two strains of Leptospirillum-like bacteria from a natural mixed population cultured on a cobaltiferous pyrite substrate. Antonie Leeuwenhoek 66:295-302. [DOI] [PubMed] [Google Scholar]
- 3.Brofft, J. E., J. V. McArthur, and L. J. Shimkets. 2002. Recovery of novel bacterial diversity from a forested wetland impacted by reject coal. Environ. Microbiol. 4:764-769. [DOI] [PubMed] [Google Scholar]
- 4.Bond, P. L., S. P. Smigra, and J. F. Banfield. 2000. Phylogeny of microorganisms populating a hick, subaerial, predominantly lithotrophic biofilm at an extreme acid mine drainage site. Appl. Environ. Microbiol. 66:3842-3849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bond, P. L., G. K. Druschel, and J. F. Banfield. 2000. Comparison of acid mine drainage microbial communities in physically and geochemically distinct ecosystems. Appl. Environ. Microbiol. 66:4962-4971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bond, P. L., and J. F. Banfield. 2001. Design and performance of rRNA targeted oligonucleotide probes for in situ detection and phylogenetic identification of microorganisms inhabiting acid mine drainage environments. Microb. Ecol. 41:149-161. [DOI] [PubMed] [Google Scholar]
- 7.Boucher, Y., C. J. Douady, R. T. Papke, D. A. Walsh, M. E. R. Boudreau, C. L. Nesbø, R. J. Case, and W. F. Doolittle. 2003. Lateral gene transfer and the origins of prokaryotic groups. Annu. Rev. Genet. 37:283-328. [DOI] [PubMed] [Google Scholar]
- 8.Button, D. K., F. Schut, P. Quang, R. Martin, and B. Robertson. 1993. Viability and isolation of marine bacteria by dilution culture: theory, procedures, and initial results. Appl. Environ. Microbiol. 59:881-891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chin, K. J., D. Hahn, U. Hengstmann, W. Liesack, and P. H. Janssen. 1999. Characterization and identification of numerically abundant culturable bacteria from the anoxic bulk soil of rice paddy microcosms. Appl. Environ. Microbiol. 65:5042-5049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Coram, N. J., and D. E. Rawlings. 2002. Molecular relationship between two groups of the genus Leptospirillum and the finding that Leptospirillum ferriphilum sp. nov. dominates South African commercial biooxidation tanks that operate at 40°C. Appl. Environ. Microbiol. 68:838-845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Denison, R. F., and E. T. Kiers. 2004. Why are most rhizobia beneficial to their plant hosts, rather than parasitic? Microbes Infect. 6:1235-1239. [DOI] [PubMed] [Google Scholar]
- 12.Druschel, G. K., B. J. Baker, T. H. Gihring, and J. F. Banfield. 2004. Acid mine drainage biogeochemistry at Iron Mountain, California. Geochem. Trans. 5:13-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Edwards, J. S., R. U. Ibarra, and B. O. Palsson. 2001. In silico predictions of Escherichia coli metabolic capabilities are consistent with experimental data. Nat. Biotechnol. 19:125-130. [DOI] [PubMed] [Google Scholar]
- 14.Huber, R., S. Burggraf, T. Mayer, S. M. Barns, P. Rossnagel, and K. O. Stetter. 1995. Isolation of a hyperthermophilic archaeum predicted by in situ RNA analysis. Nature 376:57-58. [DOI] [PubMed] [Google Scholar]
- 15.Huelsenbeck, J. P., and F. Ronquist. 2001. MRBAYES: Bayesian inference of phylogeny. Bioinformatics 17:754-755. [DOI] [PubMed] [Google Scholar]
- 16.Ibarra, R. U., J. S. Edwards, and B. O. Palsson. 2002. Escherichia coli K-12 undergoes adaptive evolution to achieve in silico predicted optimal growth. Nature 420:186-189. [DOI] [PubMed] [Google Scholar]
- 17.Johnson, D. B. 1995. Selective solid media for isolating and enumerating acidophilic bacteria. J. Microbiol. Methods 23:205-218. [Google Scholar]
- 18.Lane, D. J. 1991. 16S/23S rRNA sequencing, p. 115-148. In E. Stackebrandt and M. Goodfellow (ed.), Nucleic acid techniques in bacterial systematics. Wiley, Chichester, United Kingdom.
- 19.Ludwig, W., O. Strunk, R. Westram, L. Richter, H. Meier, A. Yadhukumar, A. Buchner, T. Lai, S. Steppi, G. Jobb, W. Forster, I. Brettske, S. Gerber, A. W. Ginhart, O. Gross, S. Grumann, S. Hermann, R. Jost, A. Konig, T. Liss, M. R. Lussmann, May, B. Nonhoff, B. Reichel, R. Strehlow, A. Stamatakis, N. Stuckmann, A. Vilbig, M. Lenke, T. Ludwig, A. Bode, and K. H. Schleifer. 2004. ARB: a software environment for sequence data. Nucleic Acids Res. 32:1363-1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Markosyan, G. E. 1972. A new iron-oxidizing bacterium: Leptospirillum ferrooxidans nov. gen. nov. sp. Biol. J. Armenia 25:26-29. (In Russian.) [Google Scholar]
- 21.Norris, P. R., J. C. Murrell, and D. Hinson. 1995. The potential for diazotrophy in iron-and sulfur-oxidizing acidophilic bacteria. Arch. Microbiol. 164:294-300. [Google Scholar]
- 22.Parro, V., and M. Moreno-Paz. 2003. Gene function analysis in environmental isolates: the nif regulon of the strict iron oxidizing bacterium Leptospirillum ferrooxidans. Proc. Natl. Acad. Sci. USA 100:7883-7888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Poole, R. K., and S. Hill. 1997. Respiratory protection of nitrogenase activity in Azotobacter vinelandii: roles of the terminal oxidases. Biosci. Rep. 17:303-317 [DOI] [PubMed] [Google Scholar]
- 23a.Ram, R. J., N. C. VerBerkmoes, M. P. Thelen, G. W. Tyson, B. J. Baker, R. C. Blake II, M. Shah, R. L. Hettich, and J. F. Banfield. 2005. Community proteomics reveals key roles for “hypothetical” proteins in a natural microbial biofilm. Science 308:1915-1920. [PubMed] [Google Scholar]
- 24.Rappe, M. S., S. A. Connon, K. L. Vergin, and S. J. Giovannoni. 2002. Cultivation of the ubiquitous SAR11 marine bacterioplankton clade. Nature 418:630-633. [DOI] [PubMed] [Google Scholar]
- 25.Rawlings, D. E., and T. Kasano. 1994. Molecular genetics of Thiobacillus ferrooxidans. Microbiol. Rev. 58:39-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rutherford, K., J. Parkhill, J. Crook, T. Horsnell, P. Rice, M.-A. Rajandream, and B. Barrell. 2000. Artemis: sequence visualization and annotation. Bioinformatics 16:944-945. [DOI] [PubMed] [Google Scholar]
- 27.Schrenk, M. O., K. J. Edwards, R. M. Goodman, R. J. Hamers, and J. F. Banfield. 1998. Distribution of Thiobacillus ferrooxidans and Leptospirillum ferrooxidans: implications for generation of acid mine drainage. Science 279:1519-1522. [DOI] [PubMed] [Google Scholar]
- 28.Schmeisser, C., C. Stockigt, C. Raasch, J. Wingender, K. N. Timmis, D. F. Wenderoth, H. C. Flemming, H. Liesegang, R. A. Schmitz, K. E. Jaeger, W. R. Streit. 2003. Metagenome survey of biofilms in drinking-water networks. Appl. Environ. Microbiol. 69:7298-7309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schut, F., E. J. de Vries, J. C. Gottschal, B. R. Robertson, W. Harder, R. A. Prins, and D. K. Button. Isolation of typical marine bacteria by dilution culture: growth, maintenance, and characteristics of isolates under laboratory conditions. Appl. Environ. Microbiol. 59:2150-2160. [DOI] [PMC free article] [PubMed]
- 30.Singer, P. C., and W. Stumm. 1970. Acidic mine drainage rate-determining step. Science 167:1121. [DOI] [PubMed] [Google Scholar]
- 31.Silverman, M. P., and D. G. Lundgren. 1959. Studies on the chemoautotrophic iron bacterium Ferrobacillus ferrooxidans. I. An improved medium and a harvesting procedure for securing high cell yields. J. Bacteriol. 77:642-647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stackebrandt, E., and B. M. Goebel. 1994. A place for DNA-DNA reassociation and 16S ribosomal-RNA sequence analysis in the present species definition in bacteriology. Int. J. Syst. Evol. Microbiol. 44:846-849. [Google Scholar]
- 33.Teeling, H., A. Meyerdierks, M. Bauer, R. Amann, F. O. Glockner. 2004. Application of tetranucleotide frequencies for the assignment of genomic fragments. Environ. Microbiol. 6:938-947. [DOI] [PubMed] [Google Scholar]
- 34.Tyson, G. W., J. Chapman, P. Hugenholtz, E. E. Allen, R. J. Ram, P. M. Richardson, V. V. Solovyev, E. M. Rubin, D. S. Rokhsar, and J. F. Banfield. 2004. Community structure and metabolism through reconstruction of microbial genomes from the environment. Nature 428:37-43. [DOI] [PubMed] [Google Scholar]
- 35.Venter, J. C., K. Remington, J. F. Heidelberg, A. L. Halpern, D. Rusch, J. A. Eisen, D. Wu, I. Paulsen, K. E. Nelson, W. Nelson, D. E. Fouts, S. Levy, A. H. Knap, M. W. Lomas, K. Nealson, O. White, J. Peterson, J. Hoffman, R. Parsons, H. Baden-Tillson, C. Pfannkoch, Y. H. Rogers, and H. O. Smith. 2004. Environmental genome shotgun sequencing of the Sargasso Sea. Science 304:66-74. [DOI] [PubMed] [Google Scholar]
- 36.West, S. A., Kiers. E. T., E. L. Simms, R. F. Denison. 2002. Sanctions and mutualism stability: why do rhizobia fix nitrogen? Proc. R. Soc. Lond. B Biol. Sci. 269:685-694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zehr, J. P., and L. A. McReynolds. 1989. Use of degenerate oligonucleotides for amplification of the nifH gene from the marine cyanobacterium Trichodesmium thiebautii. Appl. Environ. Microbiol. 55:2522-2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zengler, K., G. Toledo, M. Rappe, J. Elkins, E. J. Mathur, J. M. Short, and M. Keller. 2002. Cultivating the uncultured. Proc. Natl. Acad. Sci. USA 99:15681-15686. [DOI] [PMC free article] [PubMed] [Google Scholar]